Abstract
DNA double-strand breaks (DSBs) are among the most lethal lesions associated with genome stability, which, when destabilized, predisposes organs to cancers. DSBs are primarily fixed either with little fidelity by non-homologous end joining (NHEJ) repair or with high fidelity by homology-directed repair (HDR). The phosphorylated form of H2AX on serine 139 (γ-H2AX) is a marker of DSBs. In this study, we explored if the protein phosphatase PP6 is involved in DSB repair by depletion of its expression in human cancer cell lines, and determined PP6 expression in human breast cancer tissues by immunohistochemistry staining. We found that bacterially produced PP6c (the catalytic subunit of PP6)-containing heterotrimeric combinations exhibit phosphatase activity against γ-H2AX in the in vitro phosphatase assays. Depletion of PP6c or PP6R2 led to persistent high levels of γ-H2AX after DNA damage and a defective HDR. Chromatin immunoprecipitation assays demonstrated that PP6c was recruited to the region adjacent to the DSB sites. Expression of PP6c, PP6R2 and PP6R3 in human breast tumors was significantly lower than those in benign breast diseases. Taken together, our results suggest that γ-H2AX is a physiological substrate of PP6 and PP6 is required for HDR and its expression may harbor a protective role during the development of breast cancer.
Key words: protein phosphatase, PP6, γ-H2AX, DNA double-strand break, homology-directed repair
Introduction
Reversible phosphorylation is the most significant posttranslational modification found within proteins and plays an essential role in most, if not all, cellular events, including the cellular responses to DNA double-strand breaks (DSBs). DSBs represent the most lethal lesions associated with genome stability, which, when destabilized, predisposes organs to cancers.1 DSBs are primarily repaired by non-homologous end joining (NHEJ), in which the ends are ligated without the use of extensive homology. This type of repair takes place with little fidelity, leading to mutations, deletions and/or translocations.2 DSBs are also repaired by homology-directed repair (HDR), which utilizes a homologous template for gene conversion (GC) through strand-invasion and nascent DNA synthesis.2 HDR is the most precise when an identical sister chromatid is used as the template for repair. Accordingly, factors that are essential for HDR (e.g., BRCA1) are critical for maintenance of genome stability.
Phosphorylation is executed by protein kinases. The human genome encodes about 520 protein kinases (also known as the kinome), more than 80% of which phosphorylate serine/threonine residues.3 It is estimated that 20–30% of human proteins are phosphorylated in an unperturbed cell and more than 98% of phosphorylation occurs on serine or threonine residues. Dephosphorylation is executed by protein phosphatases. The human genome encodes only about 147 protein phosphatases, 40 of which are serine/threonine phosphatases.3 The PP1 and PP2A family members (13 in total in the human genome) are the most abundant and well studied protein phosphatases.3,4
H2AX phosphorylated on Ser-139 (γ-H2AX) is a marker for DSBs. It is quickly phosphorylated predominately by ATM/ATR/DNA-PKcs (ataxia telangiectasia mutated/ataxia telangiectasia and Rad3 related/DNA-dependant protein kinase catalytic subunit) in response to DSBs.5,6 The closely-related type 2A protein phosphatases PP2, PP4 and PP6c and the Mn2+/Mg2+-dependent phosphatase PPM1D have been reported to dephosphorylate γ-H2AX both in vitro and in vivo.7–15 It is of great interest to understand why several protein phosphatases would target the same substrate. We speculate that temporal and spatial regulation of these phosphatases by their regulatory/scaffolding subunits confers specific recognition of γ-H2AX generated under different stresses. Indeed, we found that the PP4c-PP4R2-PP4R3β complex specifically dephosphorylates γ-H2AX generated during DNA replication and is required for DNA DSB repair.8
PP6 appears to be ubiquitously expressed in all tissues tested with highest expression levels in testis, heart, kidney, brain, stomach, liver and skeletal muscle and lowest in placenta, lung, colon and spleen.16,17 The PP6 holoenzyme has been proposed to be a heterotrimeric complex formed by the catalytic subunit, a SAPS (SIT4 phosphatase-associated proteins) domain-containing subunit (PP6R) and an ankyrin repeat-domain containing regulatory subunit (ARS).17,18 PP6 is a component of a signaling pathway regulating cell cycle progression in response to IL-2 receptor stimulation.19 Its N-terminal domain restricts G1 to S phase progression in cancer cells, in part through control of cyclin D1.20 A recent report demonstrated that PP6 is required for efficient activation of DNA-PK,21 which is essential for NHEJ-mediated repair of DSBs.22,23
We recently demonstrated that PP6, like PP2 and PP4, may dephosphorylate γ-H2AX induced by IR and that PP6 is recruited to DSB sites by DNA-PK.11 In this report, we provide biochemical evidence that certain PP6c-containing heterotrimeric combinations dephosphorylate γ-H2AX directly, dynamic association of PP6c and its targeting/scaffold subunits with DSBs is important for HDR and expression of PP6 may harbor a protective role during the development of breast cancer.
Results
Depletion of PP6 leads to sustained high levels of γ-H2AX after CPT treatment.
We recently demonstrated that PP6c and PP6R1 regulate dephosphorylation of IR-induced γ-H2AX.11 γ-H2AX is frequently regarded as a marker of unrepaired DSBs.24 We wanted to determine if other PP6 subunits are required for this process. We used camptothecin (CPT) to induce DNA damage. CPT is different from IR in that CPT-induced DSBs are replication-dependent because it is a DNA topoisomerase I inhibitor.25,26 MCF-7 cells transfected with control or siRNAs against PP6c or its subunits were treated with CPT for 1 h and then washed free of drug, γ-H2AX levels were analyzed thereafter by immunoblotting (Fig. 1). In the presence of control siRNA or siRNAs against PP6R1 or PP6R3, γ-H2AX was not detected in cells untreated with CPT. γ-H2AX began to be detected immediately after adding CPT, peaked at the time of CPT removal and returned to background levels by 4 h. However, in the presence of siRNAs against PP6c or PP6R2, γ-H2AX levels were sustained at significantly higher levels and were only slightly decreased 8 h after CPT removal. Persistence of high γ-H2AX levels in PP6 knockdown cells reveals the existence of unrepaired DSBs, indicating that PP6C and PP6R2 are required for efficient repair of CPT-induced DSBs.
Figure 1.
Inhibition of PP6c or PP6R2 expression induced sustained levels of γ-H2AX in CPT-treated cells. MCF-7 cells were transfected with si-CONTROL or siRNA oligos against PP6 subunits. Transfectants were treated with CPT (10 µM) for 1 h, washed free of drug (0 time point) and harvested at various times thereafter. Total cell lysate was immunoblotted for γ-H2AX, which is a marker for damaged DNA not yet repaired and other proteins as indicated.
To further support this conclusion, we perform neutral comet assays to directly measure the extent of the CPT-induced DNA damage in U2OS cells and MCF-7 cells depleted of PP6c or its subunits by siRNA. As expected, 8 h after removal of CPT, depletion of PP6c or PP6R2 resulted in significant fractions of CPT-induced DSBs unrepaired in U2OS cells, whereas most CPT-induced DSBs were repaired in PP6R1-, PP6R3- or mock-depleted U2OS cells (Fig. 2). Similar results were obtained in MCF-7 cells (data not shown).
Figure 2.
PP6 is required for repair of CPT-induced DSBs. (A) U2OS cells were transfected with si-CONTROL or siRNA oligos against PP6 subunits. Transfectants were untreated, treated with CPT (10 mM) for 1 h or washed free of drug after 1 h-CPT treatment and then incubated for 8 h. Cells were collected for neutral comet assays. A representative image of cells under each condition is presented. (B) Quantification of the tail lengths from the experiment for which results are shown in (A). The tail length for each condition was calculated from a minimum of 100 cells for each data point.
We would thus expect that depletion of PP6c or PP6R2 would sensitize cells to CPT treatment. Indeed, in the MTT-based cell proliferation assays, PP6c- or PP6R2-depleted MCF-7 cells, when treated with CPT, exhibited less cell proliferation in comparison to mock-depleted MCF-7 cells (Sup. Fig. 1). However, it was unexpected that depletion of PP6R1 or PP6R3 also resulted in decreased cell proliferation in comparison to control after CPT treatment. This raises a possibility that PP6R1 and PP6R3 may play a role in response to CPT-induced transcription-associated lesions other than in response to CPT-induced replication-dependent DSBs.
PP6c-containing heterotrimeric complexes dephosphorylate γ-H2AX.
It has been demonstrated that both PP2c and PP4c require additional regulatory subunits and/or targeting subunits for their catalytic activity, sub-cellular localization and substrate recognition.4,8 It was recently proposed that the PP6 holoenzyme is a heterotrimeric complex, in which SAPS domain-containing proteins act as scaffold factors, whereas ankyrin repeat-containing proteins are regulatory or targeting subunits.18 We demonstrated that depletion of PP6c leads to sustained high levels of γ-H2AX after IR11 or CPT treatment (Fig. 1), suggesting that γ-H2AX is likely one of PP6 substrates. Indeed, we found that wild-type PP6c, but not catalytic inactive PP6c (D84N) produced in the transcription/translation reticulocyte system was able to dephosphorylate γ-H2AX in vitro (Fig. 3A). Under this situation, regulatory/targeting subunits required for the PP6c activity were likely provided in the reticulocyte lysates.
Figure 3.

PP6c-containing heterotrimeric combinations dephosphorylate γ-H2AX in vitro. (A) In vitro transcribed/translated PP6c dephosphorylates γ-H2AX. HA-tagged phosphatase-dead PP6 (lanes 2 and 3), wt-PP6 (lanes 4 and 5) or vector alone (lane 1) was produced by in vitro transcription/translation in the rabbit reticulocyte system. 1x (lanes 2 and 4) or 4x (lanes 1, 3 and 5) products were immunoprecipitated with an anti-HA antibody and incubated with phosphorylated γ-H2AX as described in Materials and Methods. Immunoblots were probed for either HA (PP6), γ-H2AX or total H2AX as indicated. Signals were quantitated. Result is representative of three separate experiments. (B) In vitro phosphatase assays using PP6c-containing heterotrimeric complexes. Bacterially produced His-HA-PP6c mixed with two of the bacterially-produced GST-tagged PP6-interacting proteins was incubated with acidic histone extracts derived from HeLa cells treated with nocodazole overnight at 30°C for 30 minutes. The mixtures were resolved on 4–15% SDS-PAGE and immunoblotted with antibodies as indicated. Lane 1: PP6c alone; lane 2: PP6c + PP6R1 + PP6R2; lane 3: PP6c + PP6R1 + PP6R3; lane 4: PP6c + PP6R1 + ARS − A; lane 5: PP6c + PP6R2 + PP6R3; lane 6: PP6c + PP6R2 + ARS-A; lane 7: PP6c + P6R3 + ARS − A. Images for individual antibody detection were cropped from one single exposure on the same blot.
Furthermore, we produced and purified soluble His-HA double tagged PP6c and GST fusions of its subunits in E. coli. In vitro phosphatase assays were performed using histone extracts from HeLa cells treated with nocodazole overnight as the substrate and PP6c in combination with two of regulatory subunits as catalytic complexes (Fig. 3B). We found that (1) PP6R2-containing heterotrimeric combinations (with the exception of PP6c-PP6R1-PP6R2, Fig. 3B, lane 2) showed enzymatic activity against γ-H2AX; (2) PP6R1-containing heterotrimeric combinations (with the exception of PP6c-PP6R1-ARS-A, which had weak activity, Fig. 3B, lane 4) showed no enzymatic activity against γ-H2AX (Fig. 3B, lanes 2 and 3). The enzymatic activities associated with these heterotrimeric combinations are unlikely due to a bacterial phosphatase inadvertently co-purified because every subunit tested and certain heterotrimeric combinations showed no activity against γ-H2AX (Fig. 3B and data not shown). It is noted that protein folding, conformation and post-translational modifications in bacterially produced proteins and optimal stoichiometry of subunits may compromise the PP6 catalytic activity in vitro. Nonetheless, these in vitro data provide biochemical evidence that certain PP6c-containing heterotrimeric combinations exhibit phosphatase activity. Taken together (Figs. 1–3), these data demonstrate that γ-H2AX is likely one of the PP6 physiological substrates.
Depletion of PP6c or PP6R2 compromises HDR of DSBs.
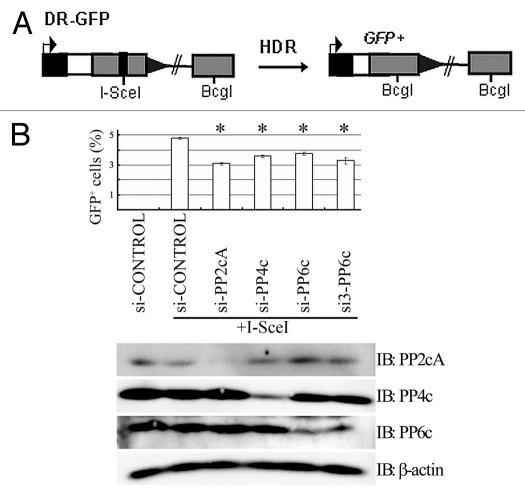
We previously showed that depletion of PP6c confers radiation sensitivity but no obvious effect on repair of IR-induced DSBs using comet assay.11 However, PP6 is required for dephosphorylation of H2AX after both IR11 and CPT (Fig. 1). In human cells, DSBs resulting from replication blockage, for example after CPT treatment, are fixed primarily by HDR.27 We therefore examined more carefully if PP6 plays a role in HDR using a well established GFP reporter system.28 HDR utilizes a homologous template for GC through strand invasion and nascent DNA synthesis.2 It ensures high fidelity of DNA repair. Thus, factors that are essential for HDR are expected to maintain genome stability. DRGFP U2OS cells have one single copy of the DRGFP gene stably integrated into their genome.28,29 The DRGFP construct carries a tandem repeat of the GFP gene, in which one copy is inactivated by the I-SceI sequence and the other by truncations at the N- and C-termini. These cells transiently transfected with the I-SceI expression construct were used for this assay. A functional GFP gene can be reconstituted if the DSB is repaired by HDR using another partial GFP gene as a template and detected by FACS (Fig. 4A). Thus, the number of GFP positive cells is a measure of HDR of I-SceI-induced DSBs. Depletion of PP6c or its regulatory subunits in U2OS cells did not obviously alter the cell cycle profile, particularly the S/G2/M profile (data not shown). When DRGFP U2OS cells were treated with KU55933 at a final concentration of 10 µM, which is sufficient to inhibit activity of both ATM and DNA-PK, but not ATR, HDR of I-SceI induced DSBs was compromised (Sup. Fig. 2). DRGFP U2OS cells were transfected with a si-CONTROL or siRNA oligos against PP2cA, PP4c or two independent sets of siRNA oligos against PP6c (si-PP6c and si3-PP6c) along with the I-SceI expression construct. It has been established that PP2cA and PP4c are required for HDR.7,8 As expected, depletion of PP2cA or PP4c led to a defective HDR (Fig. 4B). Furthermore, there was a significant reduction in HDR efficiency in cells with PP6c siRNA compared to the control (Fig. 4B).
Figure 4.
PP6 is required for HDR of I-SceI-induced DSBs. (A) Simplified diagram of the HDR reporter system. (B) Depletion of PP6c results in a defective HDR of I-SceI-induced DSBs. PP2cA-, PP4C- or mock-depleted DRGFP U2OS cells were transfected with I-SceI. PP2cA- or PP4C-depleted cells were positive controls. Two days later, cells were collected and analyzed for GFP expression by FACS. All data are the average of three independent experiments, bars are SD. *p < 0.05.
The PP6 holoenzyme is likely a heterotrimeric complex. Different combinations of scaffolding subunits and regulatory (targeting) subunits may yield many potential heterotrimeric complexes for a variety of substrates. We sought to determine which heterotrimeric combination(s) is essential for HDR of DSBs. To this end, we depleted PP6c or its subunits one at a time and determined HDR of the I-SceI-induced DSB in DRGFP U2OS cells. Depletion of PP6c resulted in a modest decrease of PP6R1, PP6R2 and PP6R3 (Fig. 5A). This is consistent with the earlier report that inhibition of PP6c expression led to a decrease of protein levels of PP6R1 and, to a lesser degree, PP6R3.17 This indicates that PP6R1, PP6R2 and PP6R3 form a stable complex with PP6c. We found that depletion of PP6c or PP6R2 resulted in defective HDR of I-SceI-induced DSBs, whereas depletion of PP6R1 or PP6R3 did not compromise the HDR efficiency (Fig. 5B). This indicates that PP6c and PP6R2 play a role in HDR.
Figure 5.
PP6c and PP6R2 are required for the HDR of I-SceI-induced DSBs. (A) Depletion of PP6 subunits in DRGFP U2OS cells. DRGFP U2OS cells were transfected with si-CONTROL or siRNA oligos against PP6c subunits. Total cell lysates were harvested 48 h after transfection and immunoblotted with antibodies as indicated. (B) DRGFP U2OS cells were transfected with si-CONTROL or siRNA oligos against PP6c or its subunits. One day later, target- or mock-depleted cells were transfected with the I-SceI expression construct. Cells were collected and analyzed for GFP expression by FACS 2 days after I-SceI transfection. All data are the average of three independent experiments, bars are SD. *p < 0.05.
PP6 is recruited to the DSB site.
We then asked whether PP6c or any of its subunits is recruited to the DSB site. ChIP assays were used to evaluate if a subunit is recruited to the adjacent region to the I-SceI-induced DSB in DRGFP U2OS cells. We found that PP6c (Fig. 6, lanes 11 vs. 12; Sup. Fig. 3, lanes 5 vs. 6) was enriched at the DSB site upon expression of I-SceI, whereas PP6R2 (Fig. 6, lanes 7 vs. 8) was dissociated from that region. Signal itself of PP6R1 enrichment at the DSB site was too weak to make any conclusive remark, however, it served as an excellent negative control. This suggests that dynamic recruitment/dissociation of protein phosphatase complexes occurs at the site of damaged DNA. The significance of PP6R2 dissociation from the DSBs warrants further investigation.
Figure 6.
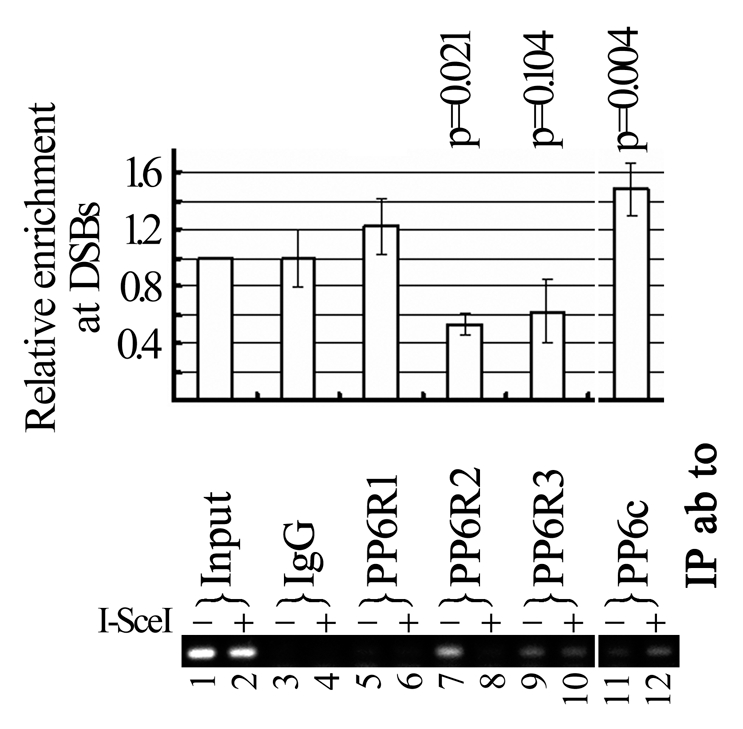
PP6c is recruited to the DSB site. DR GFP U2OS cells were transfected with the I-SceI expression construct or a control vector. One day later, transfectants were harvested for ChIP analysis. ChIP enriched DNA was used as template for PCR (top part) or real-time PCR (bottom part) using a pair of DNA primers adjacent to the DSB site induced by the I-SceI expression. Three independent real-time PCR were performed, relative enrichment of a target on the DSB site was the ratio of with and without I-SceI expression, bars are SD.
Expression of PP6, PP6R2 and PP6R3 in breast cancer and breast benign disease.
DSB repair capacity in cancer cells may associate with efficacy of chemotherapy and/or radiotherapy. Thus, we determined expression status of PP6c and its subunits in a cohort of 157 patients with operable primary breast cancer with the median follow-up of 72 months (range from 3 months to 98 months) and a cohort of 52 patients with benign breast disease. We found that antibodies against PP6c, PP6R2 and PP6R3 were suitable for immunohistochemistry (IHC) staining, whereas PP6R1 antibody was not suitable for IHC staining, on formalin-fixed and paraffin-embedded tissues (data not shown). Immunoreactive signals of PP6c, PP6R2 and PP6R3 were evenly distributed mainly in the cytoplasm and weakly detected in the nucleus (Fig. 7). We started with a small cohort of 86 breast cancer patients (69 cases of invasive ductual carcinoma (IDC), 11 cases of invasive lobular carcinoma (ILC), four cases of medullary carcinoma (MC) and one case of mucinous carcinoma) for PP6c, PP6R2 and PP6R3 and expanded to a cohort of 157 breast cancer patients (126 cases of IDC, 19 cases of ILC, eight cases of MC and three cases of mucinous carcinoma) for PP6R2 and PP6R3. The expression levels of PP6c, PP6R2 and PP6R3 were not significantly associated with tumor size, lymph nodes involvement, estrogen receptor, progesterone receptor and Her-2 status in this cohort of patients (Sup. Tables 1–3); furthermore, no statistically significant associations between the expressions of these proteins and clinical outcomes were observed in this cohort (Sup. Figs. 4–6), possibly due to a relative small sample size, further studies with a larger sample size are warranted. Nevertheless, the expression levels of PP6c, PP6R2 and PP6R3 were significantly higher in patients with benign disease in their breast specimens compared with patients with breast cancer in their breast tumors (Table 1). These findings suggested that PP6 might harbor a protective role during the development of breast cancer.
Figure 7.
Expression of PP6c, PP6R2 and PP6R3 in human breast tumors and benign breast disease. Formalin-fixed, paraffin-embedded human breast sections were immunostained with rabbit anti-human polyclonal PP6c, PP6R2 or PP6R3 antibodies (1 µg/µl) at a 1:200 dilution. The evaluation of staining intensity was performed by two independent pathologists. Representative immunohistochemical stainings in IDC and benign disease are shown. Bar: 25 µm.
Table 1.
Expression of PP6c, PP6R2 and PP6R3 in human benign breast disease and human breast cancer
| n | Protein expression | p* | ||
| Negative | Positive | |||
| n (%) | N (%) | |||
| PP6R2 | 209 | |||
| Breast cancer | 157 | 98 (62.4) | 59 (37.6) | <0.001 |
| Benign | 52 | 4 (7.7) | 48 (92.3) | |
| PP6R3 | 205 | |||
| Breast cancer | 153 | 37 (24.2) | 116 (75.8) | 0.01 |
| Benign | 52 | 4 (7.7) | 48 (92.3) | |
| PP6c | 138 | |||
| Breast cancer | 86 | 32 (37.2) | 54 (62.8) | <0.001 |
| Benign | 52 | 5 (9.6) | 47 (90.4) | |
Discussion
This is the first biochemical demonstration that heterotrimeric combinations of the catalytic subunit PP6c with a SAPS scaffold subunit and an ankyrin repeat-containing regulatory subunit exhibit phosphatase activity, supporting the hypotheses that the PP6 holoenzyme is a heterotrimeric complex.18
A recent report found that activation of DNA-PK by IR is mediated by PP6c-PP6R1 (other PP6 subunits were not examined). When PP6c and PP6R1 were individually depleted, glioblastoma cells were sensitized to IR.21 This is a reminder of an early report that PP5 is required for ATM/ATR activation after DNA damage.30,31 However, our recent results suggest that DNA-PKcs could be a targeting subunit to recruit PP6 to the DSB sites, rather than a substrate for PP6.11 Our previous results also show that depletion of PP6c or PP6R1 does not obviously compromise repair of IR-induced DSBs in the neutral comet assays.11 However, in the current report, we did find that both PP6c and PP6R2, but not PP6R1 or PP6R3, are required for HDR of CPT-induced DSBs or I-SceI-induced DSBs. This is a clear demonstration that different scaffolding/regulatory/targeting subunits may target PP6c to and dephosphorylate the same substrate generated under different contexts.
It is not a surprise to find that PP6R1 was not enriched at the I-SceI induced DSBs since it is not essential for repair of DSB induced by I-SceI (Fig. 5) or ionizing radiation.11 It is interesting to find that PP6c was enriched at the DSB site and surprisingly, at the same time PP6R2 dissociated from the DSB site (Fig. 6). Though PP6c-PP6R2 combinations exhibited enzymatic activity against γ-H2AX in vitro (Fig. 3), these combinations in vivo are apparently not the PP6 holoenzymes responsible for dephosphorylation of γ-H2AX near DSBs. We speculate that the dissociation of PP6R2 from the DSBs may be required for the recruitment/enrichment of PP6c on the DSB sites. Alternatively, PP6R2 dissociation from chromatin may facilitate activation of PP6c present in the nucleoplasm. Nonetheless, we propose that dynamic association of PP6c and PP6R2 with DSBs may play an important role in repair of DSBs.
PP2, PP4, PP6 and PPM1D are all capable of dephosphorylating γ-H2AX. PP2cA was reported to dephosphorylate γ-H2AX mainly generated by IR,7 while PP4c dephosphorylated γ-H2AX mainly generated during replication stress;8 PP6c-PP6R1 is responsible for elimination of γ-H2AX after IR treatment, PP6c and PP6R2 are required for dephosphorylation of γ-H2AX after CPT treatment or after formation of I-SceI-induced DSBs. This suggests that DSBs generated by different stresses (IR, replication, CPT, enzymatic cut by I-SceI) may have distinct DNA ends, thus recruiting different phosphatases or different phosphatase complexes. It is also highly possible that tissue or cell type-specific expression of PP6c and its regulatory subunits will allow a variety of combinations of phosphatase holoenzymatic complexes, thus conferring dephosphorylation of the same substrate by different phosphatase complexes in a tissue or cell type-specific manner.
The type 2A phosphatases are required for HDR of DSBs. Depletion of expression of any of these phosphatases may tip the balance of phosphorylation vs. dephosphorylation or change the phosphorylation status of essential factors for HDR, thus leading to a defective HDR. Though these phosphatases have a common physiological substrate, γ-H2AX, this substrate is generated under different conditions and recognized by different phosphatase complexes. Furthermore, many more physiological substrates of these phosphatases essential for HDR are awaiting further exploration. Therefore, we have many reasons to believe that PP2, PP4 and PP6 have very unique functions in HDR of DSBs, as well as in other signaling pathways.
An intact HDR is essential for high fidelity of cell divisions and maintenance of genome stability, which, when destabilized, can favor tumorigenesis. Therefore, most HDR factors are expected to be involved in tumorigenesis of human cells. In this study, though expression of PP6c and PP6R2 is not a biomarker for breast tumor prognosis in a relative small cohort of breast cancer patients, their expression overall decreases in breast tumors in comparison to that in benign breast diseases. This indicates that PP6 may have a protective role in breast cancer development by modulating HDR. It will be of great interest to determine if and how PP6 is involved in tumorigenesis in animal models.
In summary, we have demonstrated that γ-H2AX is a physiological substrate of PP6, dynamic association of PP6c and PP6R2 with chromatin surrounding DSBs is important for HDR of DSBs and PP6 may play a protective role in the development of breast cancer.
Materials and Methods
Cell lines, plasmids, siRNA oligos and antibodies.
Human cervical cancer cell line HeLa, human osteosarcoma cell line U2OS and human breast cancer cell line MCF-7 were obtained from the American Type Culture Collection (Rockville, MD). DRGFP U2OS cells with one copy of the DRGFP gene stably integrated into its genome and the I-SceI expression construct were gifts from Maria Jasin (Memorial Sloan Kettering Cancer Center).28 These cell lines were cultured in DMEM medium (Hyclone, Logan, UT) with 10% fetal bovine serum (FBS, Hyclone, Logan, UT). All cell lines were grown at 37°C in the presence of 5% CO2.
Cloning details of human PP6c, PP6R1, PP6R2 and PP6R3 for expression in E. coli or human cancer cell lines are available upon request. The phosphatase-inactive construct pcDNA-HA-PP6cres (D84N) were generated using the QuickChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA).
All siRNA oligo duplexes (OnTarget plus option) were purchased from Dharmacon (Lafayette, CO). siRNAs against human PP6c, PP6R1, PP6R2 and PP6R3 were a mixture of four predesigned OnTarget plus siRNA oligonucleotide duplexes. Sequences are available upon request. The control siRNA oligo (si-CONTROL) was CGU ACG CGG AAU ACU UCG ADT DT, and the sequence of si3-PP6c was ACA CUG GAU CAA AUU CGA ADT DT. Si-PP6c and si3-PP6c are independent of each other in terms of their targeting sequence on PP6c.
Antibodies against each PP6 subunit were affinity-purified from the sera of rabbits immunized with one of 3–5 peptide sequences chosen for each target (Bethyl Laboratories Inc., Montgomery, TX). Each antibody was characterized by immunoblotting, immunoprecipitation and indirect immunofluorescence, and one antibody was selected for each target based on highest specificity and a high titer (PP6R1, PP6R2, PP6R3 and PP6C). Rabbit polyclonal antibodies against PP2cA, H2AX, γ-H2AX, GST and HA were obtained from Bethyl. Mouse monoclonal antibody against β-actin (clone AC15) and γ-H2AX were purchased from Sigma (St. Louis, MO) and Millipore (Billerica, MA), respectively. Peroxidase-conjugated secondary antibodies were from JacksonImmuno Research (West Grove, PA).
In vitro phosphatase assay.
Acidic histone extract was prepared essentially as described (www.stanford.edu/group/gozani/protocols.html). In vitro phosphatase assay using in vitro transcribed/translated PP6c in reticulocyte lysates was performed as described before.8 All the GST-fusion proteins and His-HA-PP6c were extracted from E. coli using standard procedures. In vitro phosphatase assay was performed by mixing acidic histone extract, HIS-HA-PP6c and GST fusions in dephosphorylation buffer (50 mM Tris-HCl, pH 7.4, 5 mM DTT, 0.1 mM EDTA, 0.01% Brij35) and incubating at 30°C for 30 min. Reaction mixtures were resolved in 4–18% SDS-PAGE, and relative phosphatase activity was determined by loss of γ-H2AX immunoreactivity.
Western blot analysis and immunoprecipitation.
It was performed as described previously in reference 32.
Assays for HDR-mediated repair of DSBs.
Assay for HDR of DSBs and the chromatin immunoprecipitation (ChIP) assay were performed essentially as described before in reference 32. ChIP PCR primers are HRChIP.S: TCT TCT TCA AGG ACG ACG GCA ACT and HRChIP.R: TTG TAG TTG TAC TCC AGC TTG TGC, annealing temperature was 55°C, the size of PCR products was 145 bps.
Neutral comet assay.
U2OS cells were untreated, treated with CPT (10 µM) for 1 h or washed free of CPT after 1 h-CPT treatment and then incubated for 8 h. Cells at a concentration of 1 × 105/mL were mixed gently with pre-melted low-temperature-melting agarose at a volume ratio of 1 to 10 (v/v) and spread on glass slides. The slides were then submerged in precooled neutral lysis buffer at 4°C for 30 min. After rinsing, the slides were equilibrated in Tris-borate EDTA solution, electrophoresed at 1.0 V/ cm for 20 min, and then stained with PI. Fluorescence images for at least 100 nuclei were captured using an Olympus IX-81 microscope and analyzed by CASP-1.2.2 software (University of Wroclaw) for tail moment.
Immunohistochemistry (IHC) staining.
The cohort of 157 patients with operable primary breast cancer was treated at Peking University School of Oncology from March 2001 to December 2003, the median follow-up was 72 months (range from 3 months to 98 months). Fifty-two patients with breast benign diseases were treated at Quanzhou People's Hospital. This study was approved by the Research and Ethical Committee of Peking University School of Oncology and Quanzhou People's Hospital, respectively.
The expression of PP6c, PP6R2 and PP6R3 in breast cancers from 157 primary breast cancer patients and breast samples from 52 benign disease was determined by IHC as described elsewhere in reference 32. Those proteins were predominantly expressed in the cytoplasm, therefore, the staining intensity in the cytoplasm was evaluated. Scoring for staining was graded as follows: no staining or staining observed in <10% of tumor cells was given a score 0; faint/barely perceptible staining detected in ≥10% of tumor cells was scored as 1+; a moderate or strong complete staining observed in ≥10% of tumor cells was scored as 2+ or 3+, respectively. A score of 0 and 1+ was considered negative whereas 2+ and 3+ were considered positive.
Statistical analysis.
Student's t-test was performed to assess if the means of two groups of experiments (not less than three independent experiments in each group) were statistically different from each other. The significance level was defined as p < 0.05. The associations between PP6, PP6R2 or PP6R3 expression and clinicopathologic characteristics in this cohort of 157 patients were determined using Pearson's X2 test. Disease-free survival (DFS) was defined as the time from date of diagnosis to first recurrence (local or distant) or death from breast cancer without a recorded relapse. Overall survival (OS) was defined as the time from date of diagnosis to death for any causes. Survival curves were derived from Kaplan-Meier estimates, and the curves were compared by log-rank tests. All statistical tests were two-sided and p values less than 0.05 were considered as statistically significant. The statistical analyses were performed using the SPSS 13.0 software.
Acknowledgments
We thank Eric W. McIntush from the Bethyl Laboratories for antibodies against PP6c and its interacting proteins, and Gerd p Pfeifer from the City of Hope National Medical Center and Susan Lees-Miller from University of Calgary for comments on this manuscript. We thank other members of the Xu laboratory for help. This work was supported by the startup fund from Capital Normal University, National Natural Science Foundation of China (30570371, 90608014, 30711120570 and 31071190), the Program for New Century Excellent Talents in University (NCET-06-0187), Beijing Natural Science Foundation Program and Scientific Research Key Program of Beijing Municipal Commission of Education (KZ200810028014), the 973 project 2010CB911904, and Funding Project for Academic Human Resources Development in Institutions of Higher Learning Under the Jurisdiction of Beijing Municipality (PHR20110508) to X.X. J. Li was supported by National Natural Science Foundation of China (30700420) & Beijing Nova Program (2007B062). J. Liao was supported by Beijing Natural Science Foundation Program and Scientific Research Program of Beijing Municipal Commission of Education (KM200910028012). Work in Dr. Stark's lab was supported by a National Institutes of Health grant RO1CA120954.
Authors' Contributions
Conceived and designed the experiments: J. Li, J Liao, Y.X., X.X. Performed the experiments: J.Z., J.Liao, X.L., J. Liu, W.H., L.Y., P.W., B.Z. Analyzed the data: J.Z., Y.X., X.X. Contributed reagents/materials/analysis tools: J.W., J.M.S. Wrote the paper: J.Z., Y.X., X.X. All authors read and approved the final manuscript.
Supplementary Material
References
- 1.Harper JW, Elledge SJ. The DNA damage response: Ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Ohnishi T, Mori E, Takahashi A. DNA double-strand breaks: Their production, recognition and repair in eukaryotes. Mutat Res. 2009;669:8–12. doi: 10.1016/j.mrfmmm.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 3.Moorhead GB, Trinkle-Mulcahy L, Ulke-Lemee A. Emerging roles of nuclear protein phosphatases. Nat Rev Mol Cell Biol. 2007;8:234–244. doi: 10.1038/nrm2126. [DOI] [PubMed] [Google Scholar]
- 4.Eichhorn PJ, Creyghton MP, Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta. 2009;1795:1–15. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 7.Chowdhury D, Keogh MC, Ishii H, Peterson CL, Buratowski S, Lieberman J. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury D, Xu X, Zhong X, Ahmed F, Zhong J, Liao J, et al. A PP4-phosphatase complex dephosphorylates gamma-H2AX generated during DNA replication. Mol Cell. 2008;31:33–46. doi: 10.1016/j.molcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 10.Nakada S, Chen GI, Gingras AC, Durocher D. PP4 is a gamma H2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep. 2008;9:1019–1026. doi: 10.1038/embor.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douglas P, Zhong J, Ye R, Moorhead GB, Xu X, Lees-Miller SP. Protein phosphatase 6 interacts with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and dephosphorylates {gamma}-H2AX. Mol Cell Biol. 2010;30:1368–1381. doi: 10.1128/MCB.00741-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cha H, Lowe JM, Li H, Lee JS, Belova GI, Bulavin DV, et al. Wip1 directly dephosphorylates gammaH2AX and attenuates the DNA damage response. Cancer Res. 70:4112–4122. doi: 10.1158/0008-5472.CAN-09-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macurek L, Lindqvist A, Voets O, Kool J, Vos HR, Medema RH. Wip1 phosphatase is associated with chromatin and dephosphorylates gammaH2AX to promote checkpoint inhibition. Oncogene. 29:2281–2291. doi: 10.1038/onc.2009.501. [DOI] [PubMed] [Google Scholar]
- 14.Moon SH, Lin L, Zhang X, Nguyen TA, Darlington Y, Waldman AS, et al. Wild-type p53-induced phosphatase 1 dephosphorylates histone variant gammaH2AX and suppresses DNA double strand break repair. J Biol Che. 285:12935–12947. doi: 10.1074/jbc.M109.071696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moon SH, Nguyen TA, Darlington Y, Lu X, Donehower LA. Dephosphorylation of gammaH2AX by WIP1: An important homeostatic regulatory event in DNA repair and cell cycle control. Cell Cycle. 2010;9:2092–2096. doi: 10.4161/cc.9.11.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bastians H, Ponstingl H. The novel human protein serine/threonine phosphatase 6 is a functional homologue of budding yeast Sit4p and fission yeast ppe1, which are involved in cell cycle regulation. J Cell Sci. 1996;109:2865–2874. doi: 10.1242/jcs.109.12.2865. [DOI] [PubMed] [Google Scholar]
- 17.Stefansson B, Brautigan DL. Protein phosphatase 6 subunit with conserved Sit4-associated protein domain targets IkappaBepsilon. J Biol Chem. 2006;281:22624–22634. doi: 10.1074/jbc.M601772200. [DOI] [PubMed] [Google Scholar]
- 18.Stefansson B, Ohama T, Daugherty AE, Brautigan DL. Protein phosphatase 6 regulatory subunits composed of ankyrin repeat domains. Biochemistry. 2008;47:1442–1451. doi: 10.1021/bi7022877. [DOI] [PubMed] [Google Scholar]
- 19.Filali M, Li S, Kim HW, Wadzinski B, Kamoun M. Identification of a type 6 protein ser/thr phosphatase regulated by interleukin-2 stimulation. J Cell Biochem. 1999;73:153–163. [PubMed] [Google Scholar]
- 20.Stefansson B, Brautigan DL. Protein phosphatase PP6 N terminal domain restricts G1 to S phase progression in human cancer cells. Cell Cycle. 2007;6:1386–1392. doi: 10.4161/cc.6.11.4276. [DOI] [PubMed] [Google Scholar]
- 21.Mi J, Dziegielewski J, Bolesta E, Brautigan DL, Larner JM. Activation of DNA-PK by ionizing radiation is mediated by protein phosphatase 6. PLoS One. 2009;4:4395. doi: 10.1371/journal.pone.0004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, et al. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurimasa A, Kumano S, Boubnov NV, Story MD, Tung CS, Peterson SR, et al. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol Cell Biol. 1999;19:3877–3884. doi: 10.1128/mcb.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeggo P, Lavin MF. Cellular radiosensitivity: How much better do we understand it? Int J Radiat Biol. 2009;85:1061–1081. doi: 10.3109/09553000903261263. [DOI] [PubMed] [Google Scholar]
- 25.Ben-Yehoyada M, Gautier J, Dupre A. The DNA damage response during an unperturbed S-phase. DNA Repair (Amst) 2007;6:914–922. doi: 10.1016/j.dnarep.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Pommier Y. Camptothecins and topoisomerase I: A foot in the door. Targeting the genome beyond topoisomerase I with camptothecins and novel anticancer drugs: Importance of DNA replication, repair and cell cycle checkpoints. Curr Med Chem Anticancer Agents. 2004;4:429–434. doi: 10.2174/1568011043352777. [DOI] [PubMed] [Google Scholar]
- 27.Hochegger H, Sonoda E, Takeda S. Post-replication repair in DT40 cells: Translesion polymerases versus recombinases. Bioessays. 2004;26:151–158. doi: 10.1002/bies.10403. [DOI] [PubMed] [Google Scholar]
- 28.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–540. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ali A, Zhang J, Bao S, Liu I, Otterness D, Dean NM, et al. Requirement of protein phosphatase 5 in DNA-damage-induced ATM activation. Genes Dev. 2004;18:249–254. doi: 10.1101/gad.1176004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Bao S, Furumai R, Kucera KS, Ali A, Dean NM, et al. Protein phosphatase 5 is required for ATR-mediated checkpoint activation. Mol Cell Biol. 2005;25:9910–9919. doi: 10.1128/MCB.25.22.9910-9919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, Zhao A, Chen L, Zhong X, Liao J, Gao M, et al. Human RIF1 encodes an anti-apoptotic factor required for DNA repair. Carcinogenesis. 2009;30:1314–1319. doi: 10.1093/carcin/bgp136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.