Abstract
Previous studies of the association of the C17T polymorphism of the mu opiate receptor gene with substance dependence compared cases with substance dependence to controls and usually found no significant association. However, the studies were limited by small sample size - no study had more than 12 subjects with the TT genotype, a genotype that is rare in white and Asian subjects. Moreover, drug use is not dichotomous but follows a spectrum from non-use to modest, intermittent use, to use several times daily. We asked whether the Kreek-McHugh-Schluger-Kellogg (KMSK) scales for alcohol, cocaine, opiates, and tobacco that quantify substance use during the time of a subject's maximal use might be more sensitive measures than dichotomous outcomes. We administered the KMSK scales and completed C17T genotyping on 1009 HIV-infected and 469 HIV-uninfected women in The Women's Interagency HIV Study (WIHS), an ongoing study of HIV in women. Forty-two of 697 African-American, 1 of 182 Hispanic, and none of 161 white women had the TT genotype. KMSK cocaine, alcohol, and tobacco scores were significantly higher in African-American women with the TT genotype (p =0.008, 0.0001, and 0.006 respectively) but opiate scores were not. Ordinal regression models controlling for HIV-serostatus, age, education, and income had odds ratios for the TT genotype for predicting alcohol, tobacco, cocaine, and opiates scores of 2.1 (p = 0.02), 2.4 (p = 0.0004), 2.0 (p = 0.03), and 1.9 (p = 0.07). We conclude that the TT genotype of OPRM1 may increase the risk of substance use and abuse.
Keywords: C17T polymorphism, HIV, mu opioid receptor gene, quantitative measures, substance abuse, substance dependence
Many studies of the genetics of substance abuse have focused on the mu opiate receptor. The two polymorphisms in exon 1 that have received the most attention are C17T that results in an alanine to valine substitution and A118G that results in an asparagine to aspartic acid substitution (Bergen et al. 1997; Berrettini 1997; Bond et al. 1998; Gelernter et al. 1999; Hoehe et al. 2000; Kapur et al 2007; reviewed in Kreek et al. 2005). The frequency of the minor T allele at position 17 varies from about 1% in white (Rommelspacher et al. 2001) and Eastern Asian cohorts (Tan et al. 2003) to 15- 20% in cohorts of African-Americans (Crowley et al. 2003) or of Northern Indians (Kapur et al. 2007). Although several studies have looked at the association of C17T and substance dependence, only three have investigated African-American or Northern Indian cohorts that contain over 50 cases and 50 controls (Hoehe et al, 2000; Crowley et al. 2003; Kapur et al. 2007). One study with African-Americans found the TT genotype in 6 cases with cocaine or opiate dependence and in no controls (Hoehe et al, 2000), the other found the TT genotype in 7 controls and 3 cases with opiate dependence (Crowley et al, 2003). Kapur et al. found the TT genotype in 12 cases with opiate dependence and in no controls in a cohort of men from Northern India (Kapur et al. 2007).
All of the previous association studies of the C17T polymorphism have compared cases with substance dependence to a “normal” control group. However, in many cohorts and for a variety of substances, drug use is not dichotomous but follows a spectrum ranging from non-users through those with modest, intermittent use, to those who use a great deal of drugs almost all of the time. Accordingly, quantitative measures of drug use may be more informative than dichotomous outcomes. The lifetime Kreek-McHugh-Schluger-Kellogg (KMSK) scales (Kellogg et al. 2003) quantify use of alcohol, tobacco, opiates, and cocaine during the time of an individual's maximal use. We hypothesized that there would be differences in KMSK score associated with C17T polymorphisms.
To test this hypothesis required a cohort with two characteristics: a broad spectrum of substance use and a relatively high frequency of the T allele. The Women's Interagency HIV Study (Bacon et al. 2005; Barkan et al. 1998) provided such a cohort. Over 50% of the participants are of African descent and drug use was prevalent.
Methods
Study Design
The data presented in this paper were collected from a cross-sectional study of genetics predictors of substance abuse that was nested within the overall WIHS study. We administered the KMSK along with instruments that assessed cognition (Stroop 1935), depression (Spitzer et al. 1999; Gilbody et al. 2007), and family history of origin as well as other measures in a single 45 minute interview that was either completed at the end of the routine study follow-up or in a separate visit. The study was approved by the Institutional Review Boards at the six clinical sites, and informed consent was obtained. Although the WIHS is a longitudinal study, the KMSK was only administered once. Interviews to administer the KMSK started with the beginning of visit 24 in April 2006 and were completed in September 2009 during visit 30.
Cohort
The Women's Interagency HIV Study (Bacon et al. 2005) is an ongoing prospective study of HIV in women. The WIHS began in 1994 and has enrolled a total of 3766 women across six sites in SF, LA, Chicago, Washington, DC, Brooklyn and the Bronx (New York). WIHS initially recruited 2054 HIV-infected and 569 HIV-uninfected women in 1994-95, and an additional 737 HIV-infected and 406-HIV uninfected women in 2001-2002. Participants are evaluated every six months with an extensive interview that includes history of interval illnesses and interval substance abuse, current medications and medication adherence, physical exam, and blood and gynecological specimen collection. At each visit, women are asked whether they have used heroin or other opiates, cocaine, tobacco, and alcohol in the previous six months and the amount of alcohol use is quantified.
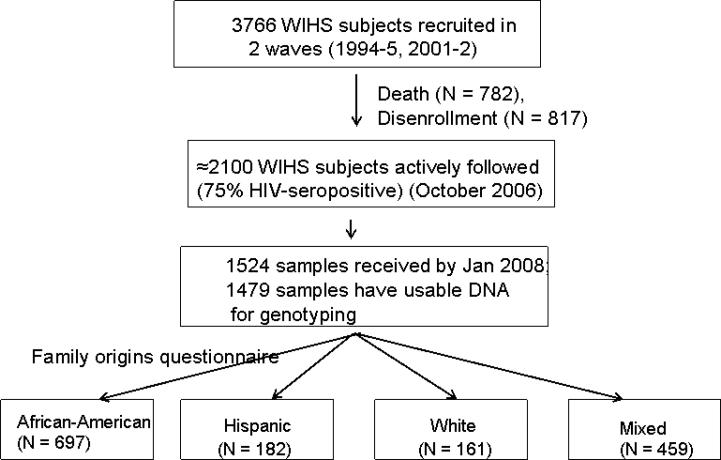
When enrollment for this sub-study began in October 2006, about 2100 of the original 3766 women were still actively followed in WIHS. Attrition from the original sample resulted from death (N =782), or disenrollment and loss-to-follow-up (N=817). Over the next 3.5 years, recruitment for this sub-study, psychological testing, and genotyping were ongoing. Throughout the study, at least 96% of available subjects agreed to participate in the study. We received 1524 specimens from WIHS by Jan 31, 2008. Usable DNA was recovered from 1478: 1009 HIV-infected and 469 HIV-uninfected participants. Figure 1 illustrates recruitment and stratification of participants.
Figure 1.
Participant recruitment
Measures
Outcome variables - The KMSK scales summate ordinal measures of the amount, frequency, and duration (ranging from six months to 18 months) of alcohol, tobacco, opiate, and cocaine use at the time of an individual's maximal drug use. It was developed to substantiate the concept that substance dependence and the degree of self-exposure a subject has to a drug are correlated. Scores range from 0 to 12 for tobacco, 0 to 13 for alcohol and for opiates (non-prescription), and from 0 to 16 for cocaine. Those who never used drug receive a score of 0. The scale takes less than five minutes to complete and has been validated in a study in a cohort of 100 subjects using the Structured Clinical Interview for DSM-IV as the gold standard (Kellogg et al. 2003).
Use of KMSK to categorize participants into groups
We used previously published cut-scores to identify participants likely to have met DSM-IV criteria for substance dependence at the time of their maximal drug use (Kellogg et al, 2003). We labeled this group as “possible opiate dependence,” “possible cocaine dependence,” etc. Previous work had shown sensitivity and specificity of 90% and 90% for alcohol, 97% and 94% for cocaine, and 100% and 99% for opiates against DSM-IV diagnoses (Kellogg et al, 2003). Because there is no DSM-IV diagnosis of tobacco abuse, we did not dichotomize the KMSK tobacco scale.
A sample of 157 women from WIHS who had been administered the KMSK scales also completed the computerized version of the DSM-IV Composite International Diagnostic Interview (CIDI). We used KMSK cut-score grouping to determine the relative risk for CIDI diagnoses of alcohol abuse or dependence, cocaine abuse or dependence, and heroin abuse or dependence. With the exception of opiate dependence, all of the relative risks were significant at p <0.001 with values ranging from 2.2 to 4.5. The relative risk for opiate dependence was 1.8 and not significant (J Cook and H Crystal, unpublished data, 2009).
For the control group to complement the “possible drug dependence” group we selected subjects free of any substance abuse. We required no cocaine use (KMSK cocaine = 0), and no opiate use (KMSK opiate = 0), and less than 4 drinks per day, 7 days a week as operationalized by KMSK alcohol < 11. Tobacco use did not exclude subjects from this control group. A score of 10 on the KMSK alcohol scale would result from daily drinking for 18 months of 2 to 3 drinks a day (Kellogg et al, 2003). The KMSK alcohol scale does not distinguish between 2 or 3 drinks per day in calculating total score.
Percentage of visits reporting drug use
We used data from all of a participant's visits to calculate the percentage of visits where a woman reported using each substance in the previous 6 months. Women recruited in the first wave in 1994 could have come to up to 29 consecutive visits, those recruited in the second wave in 2001 could have come to 15 consecutive visits; but some participants missed an occasional follow-up. For example, if a woman came to 24 out of 29 possible visits, and reported having used cocaine in the previous 6 months for 6 of these 24 visits, her percentage of reporting cocaine use would be 25%. Any use of cocaine or heroin in the previous 6 months was categorized as drug use. For alcohol, we identified a group of ‘heavy alcohol users’ who reported drinking more than 21 drinks a week at any time during the previous 6 months. We then used this figure to define a new dichotomous outcome variable – ‘frequent drug abuse.’ For each drug, we determined the cumulative frequency of the percentage visits reporting substance use for cocaine, tobacco, and heroin, and for ‘heavy alcohol use.’ We then selected the top 10% for each drug as ‘frequent drug users.’ The complementary group – ‘not drug users’ was defined as participants with no cocaine and no heroin and no ‘heavy alcohol use’ at any of up to 29 visits. Tobacco use did not exclude a subject from the control groups for heroin, cocaine, or alcohol.
Covariates
Income was rated on an 8 point scale: 1= $6000 or less, 2= $6001-$12000, 3= $12001-$18000, 4= $18001-$24000, 5= $24001-$30000, 6= $30001-$36000, 7= $36001-$75000, 8= > $75000. Education was rated on an 8 point scale ranging from 1 – no education to 8 – completed doctoral degree.
To assess whether a participant's knowledge that she was HIV-seropositive might have influenced when she used drugs the most, we calculated participants’ age when they estimated they used each drug the most and then compared that to their estimated age at HIV-diagnosis. The estimated age at HIV-diagnosis was calculated based on self-report of the year of first positive HIV test or on documented seroconversion during the period of WIHS follow-up. To be conservative, if participants couldn't recall the date they learned they were HIV-seropositive, the date of study entry was used as date of HIV-diagnosis.
Race/ethnicity
In order to control for heterogeneity in allele frequencies among different racial/ethnic groups, subjects were stratified into groups based on what they had reported as the race/ethnicity of their grandparents. Subjects classified as African-American had four grandparents of African-American or African descent (N = 697). Hispanic (N = 182) and white participants (N = 161) were also classified in this way. In these analyses, all participants with mixed inheritance (at least one grandparent from a different racial/ethnic group) were excluded. Our previous work has demonstrated that this method of classification agrees with classification based on genetic ancestral markers in 99% of cases in a sample that included African-Americans and whites, but not Hispanics (Levran et al, 2009).
Genetics
DNA was isolated from cryopreserved peripheral blood mononuclear cell (PBMC) pellets, containing one million cells using PureGene® Genomic DNA purification kit (Gentra Systems, Minneapolis, MN), which includes RNase A treatment. The DNA concentrations were measured using PicoGreen® dsDNA quantitation assay ((Invitrogen, Carlsbad, CA). The average yield of DNA was 3.8 μg per million cells. A random subset of the DNA samples were run on 1% Agarose E-Gels® 96 (Invitrogen) to assess their quality. Prior to storage at -80°C, the DNA was transferred to 96-well plates and the concentrations adjusted to a uniform 50 ng/μl.
TaqMan primers (GAAAAGTCTCGGTGCTCCTGG - forward and CAAGGCATCAGTGCAATTGC - reverse) and TaqMan probes (AGCGCTGCCCCCA, FAM-labeled and AGCGCTGTCCCCA, VIC-labeled) for the genotyping of the OPRM1 C17T (rs1799972) polymorphism were designed, tested, and evaluated in our laboratory (MJK). The primer sets were selected such that they would result in the shortest possible amplicon (110 bp) suiTable for successful TaqMan assay and yet be able to be used as primers for verification of the assays using Sanger sequencing.
TaqMan amplification was done in 5 μl volumes using 384-well plates. Each reaction mix contained 3 ng of the genomic DNA, 1x TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA), 4.5 pmole of each primer and 1 pmole of each TaqMan probe. The cycling was performed in a GeneAmp® PCR system 9700 with a dual 384-well sample block (Applied Biosystems), using the following conditions: two initial holds first at 50°C for 2 minutes then at 95°C for 10 minutes, followed by 50 cycles of 95°C for 15 seconds and 60°C for 1 minute. Upon completion of cycling, analysis was performed on the ABI Prism® 7900 Sequence Detection System using SDS v2.2 software (Applied Biosystems).
A total of 205 unique DNA specimens were Sanger sequenced, where the resulting sequence data covered this variation in both forward and reverse directions. Of the 205 DNA specimens there was only a single DNA specimen where the sequencing genotype result conflicted with the TaqMan result (1 out of 205, 0.49%).
Prior to electrophoresis on an ABI 3730xl DNA Analyzer, the sequencing reactions were purified using BigDye® XTerminator™ purification kit (the sequencing and purification kits, as well as the 3730xl instrument are from Applied Biosystems). Six SNPs for OPRM1 (rs17174638, rs1799972, rs1799971, rs17174801, rs1799974, and rs17174822) were studied.
All subjects were genotyped in duplicate. There were 18 samples for which the duplicate failed and they were all confirmed by sequencing. There were 14 samples (0.9%) with conflicting TaqMan data, where one of the replicate was called heterozygotes (C/T) and the other homozygotes (C/C). These samples were resolved by Sanger sequencing.
Statistical methods
Our primary aim was to evaluate the association of the TT genotype with measures of substance abuse. The main outcome variables were the KMSK scales that are created by summing 3 ordinal subscales. Values for the KMSK scales are not normally distributed and we used ordinal regression as a method of analysis. We performed ordinal regression in R (http://www.r-project.org/) from the Design library using the lrm (logistic regression model) function using the logit link where the each of the KMSK variables was the ordered response variable and TT versus CT/CC genotype was the explanatory variable. To test the validity of the proportional odds assumption we performed a score test. We failed to reject the proportional odds assumption (p-value >0.20).
For the ordinal regression analyses, we report the odds ratio someone with the TT genotype (as compared with someone with the CC or the CT genotype) would have a higher KMSK score given that all other variables in the model were held constant. The odds ratio remains the same at every value of the KMSK scale. For example, in table 3, in the 2nd row, right-most column, we report that the odds ratio in the fully adjusted model for predicting the KMSK alcohol score is 2.18. This means that someone with the TT genotype is 2.18 times as likely to have a score of 5 than a score of 4 or lower as someone with the CC or CT genotypes. Because the odds ratio remains the same at each value of the KMSK scale, someone with the TT genotype would also be 2.18 times as likely to have a score than a score of 5 or lower as someone with the CC or CT genotype.
Table 3.
Odds ratios for the TT genotype for the prediction of Kreek–McHugh–Schluger–Kellogg (KMSK) score in African American women. Values in each cell represent odds ratios, confidence intervals and significance levels derived from ordinal regression models.
| Only TT | TT, HIV serostatus | TT, HIV serostatus, age, income, education | |
|---|---|---|---|
| KMSK alcohol | 2.17 (1.16–4.04) 0.015 | 2.20 (1.18–4.10) 0.013 | 2.14 (1.15–4.00) 0.017 |
| KMSK tobacco | 2.38 (1.31–4.33) 0.0044 | 2.42 (1.33–4.40) 0.0038 | 2.39 (1.32–4.35) 0.0004 |
| KMSK cocaine | 2.17 (1.15–4.06) 0.016 | 2.20 (1.18–4.12) 0.013 | 2.03 (1.08–3.83) 0.028 |
| KMSK opiates | 1.77 (0.89–3.53) 0.10 | 1.85 (0.92–3.68) 0.08 | 1.93 (0.95–3.93) 0.061/9 |
We used logistic regression to determine the odds ratio for the TT vs. the CC/CT genotype at predicting the dichotomous outcomes of ‘likely substance abuse’ vs. controls or of ‘frequent substance abuse’ vs. controls (see above for definitions). Logistic Regression was performed using glm (generalized linear models) in R where we defined the family as binomial and logit as the link function.
For all analyses, we tested three models: a) only including TT vs. CC/CT genotypes and uncorrected for other variables; b) adjusted for HIV status, and c) corrected for HIV status, age, income, education level, and when appropriate, for race/ethnicity.
We addressed issues of racial/ethnic stratification in our analyses in several ways. First, we compared substance use across racial/ethnic groups as stratification is critical when both disease frequency and gene frequency is greater in one ethnic/racial group. Second, because the genotype that is the focus of this report occurred in 6% of African-Americans, in 0.9% of Hispanics, and in no whites, we performed all analyses in two ways - first limited to African-American participants, and then in the entire cohort of whites, Hispanics, and African-Americans considered together using race/ethnicity as a cofactor. Results were consistent when analyses were restricted to the African-American group and when the entire cohort was considered with race/ethnicity as a covariate. In the results section, for the first ordinal regression analyses we describe results both for the African-American only group and for the group overall. Thereafter, we only report results for the African-American cohort. Results for the combined cohort are available on request.
We evaluated differences in mean scores for normally distributed values (age, education, and income) between HIV-infected and HIV-uninfected participants with the Student t-test, and among the 3 ethnic/racial groups with ANOVA followed by the Dunnett T3 test for post-hoc testing. We evaluated differences in mean scores for non-normally distributed values (all of the KMSK scales) with the Mann-Whitney test or the Kruskal-Wallis test. When a significant difference was found with Kruskal-Wallis test, comparisons between two groups were made with the Mann-Whitney test. Summary statistics for KMSK scores, that are not normally distributed, are listed for the median, 25th and 75th percentiles. When the distribution of values was such that the 25th, 50th, or 75th percentile lay between 2 values, an interpolated, weighted value was created to indicate this. Significance levels for differences in frequency among categorical variables were assessed with the chi-square test. We used the likelihood ratio test to test for deviations from Hardy-Weinberg equilibria.
We estimated power using PAWE (Power for Association with Error) assuming a significance level of 0.05 and using the allelic frequencies and sample sizes from our observed data (Gordon et al., 2002).
We used a significance level of p <.05, and did not correct for multiple comparisons.
RESULTS
Demographics
KMSK scores and demographic data for study participants are shown in Table 1. Subjects were divided into six groups based on their ethnicity/race as identified by the family of origins questionnaire and by their HIV-serostatus. HIV-seropositive white women were significantly older (F = 27.9, p <0.0001) than their Hispanic (p = 0.002) or African American counterparts (p < 0.001). HIV-seropositive white women were 9 years older than seronegatives (t = 36.3, p <0.0001); HIV-seropositive Hispanic women were 8 years older than Hispanic HIV-seronegatives (t = 19.7, p <0.0001), and HIV- seropositive African-American women were 3 years older than seronegatives (t = 19.8, p <0.0001). HIV -seropositive white women had more education (F = 38.8, p < 0.0001) than Hispanics (p < 0.001) or African-Americans (p < 0.001). HIV-seronegative white women had more education (F = 14.5, p = 0.0002) than Hispanics (p < 0.001) or African-Americans (p < 0.001). There were no significant differences in income among the 3 ethnic/racial groups.
Table 1.
Demographics and Kreek–McHugh–Schluger–Kellogg (KMSK) scores in study subjects.
| HIV+ |
HIV- |
|||||||
|---|---|---|---|---|---|---|---|---|
| White (n = 115) | Hisp (n = 114) | AA (n = 484) | F or χ2, P | White (n = 46) | Hisp (n = 68) | AA (n = 213) | F or χ2, P | |
| Age | 45.8 (8.8) | 41.7 (8.8) | 40.7 (8.7) | 27.8, < 0.0001 | 36.3 (8.4) | 33.7 (9.4) | 37.6 (10.6) | 2.6, 0.11 |
| Education | 4.8 (1.1) | 3.8 (0.9) | 3.4 (1.0) | 38.8, < 0.0001 | 4.8 (1.0) | 3.8 (0.9) | 4.0 (0.9) | 14.5, 0.0002 |
| Income | 4.3 (2.6) | 3.1 (2.0) | 3.1 (2.0) | 0.45, 0.50 | 4.3 (2.3) | 4.1 (2.1) | 3.41 (2.1) | 0.24, 0.62 |
| KMSK alcohol | 9 (5,11) | 6 (3,10) | 7 (2,11) | 13.3, 0.0013 | 10 (9,12) | 6 (3,9) | 7 (2,10) | 21.3, <0.0001 |
| KMSK cocaine | 4 (0,11) | 4 (0,13) | 6 (0,15) | 3.1, 0.21 | 6.5 (0,12) | 3 (0,12) | 0 (0,11) | 2.9, 0.23 |
| KMSK opiates | 0 (0,4) | 0 (0,8) | 0 (0,3) | 6.42, 0.04 | 0 (0,5.75) | 0 (0,4.5) | 0 (0,0) | 9.7, 0.008 |
| KMSK tobacco | 9 (0,11) | 7 (0,11) | 9 (0,11) | 1.46, 0.48 | 9.5 (5,11) | 8 (3,11) | 8 (3,10) | 5.1, 0.08 |
Means (and standard deviations) are shown for age, education, income and PHQ9. Medians (and 25th and 75th percentiles) are shown for KMSK scores. ANOVA was used for comparing normally distributed scores: the Kruskal–Wallis test was used for comparing KMSK scores.
KMSK alcohol scores were significantly higher in white HIV-seropositives (χ2 = 13.3, p = 0.0013) than in Hispanics (p = 0.003) or African-Americans (p = 0.024). White HIV-seronegative women had higher KMSK alcohol scores (χ2 = 21.2, p <0.0001) than Hispanics (p < 0.001) or African-Americans (p < 0.001). In HIV-seropositive African-Americans, KMSK opiate scores were significantly lower (χ2 = 6.4, p = 0.04) in Hispanics (p = 0.01), but not whites (p = 0.26). In HIV-seronegativeAfrican-American women, KMSK opiate scores were significantly lower (χ2 = 9.7, p = 0.008) in whites (p = 0.002), but not in Hispanics. KMSK cocaine or tobacco scores did not differ significantly different among the 3 ethnic/racial groups in either HIV-seropositive or HIV-seronegative participants.
When contrasting KMSK scores between HIV-seropositive and seronegative participants within the same ethnic/racial group only 2 significant differences were found among 12 possible comparisons (4 KMSK scales × 3 ethnic/racial groups). Seropositive African-American women had higher KMSK cocaine (u = 14.3, p = 0.0008) and opiate scores (u = 6.5, p = 0.04) than seronegative women.
Gene frequency
Genotype frequencies for the C17T OPRM1 polymorphism in whites, Hispanics, and African-Americans are shown in Table 2. The CT genotype was found in 35.7% of African-Americans, 12.1% of Hispanics, and 1.2% of whites (χ2 = 84.4, p <0.0001). The TT genotype was found in 6.0% of African-Americans, 0.5% Hispanics, and in no white women. Genotype frequencies did not deviate from Hardy-Weinberg equilibrium in either the cases or the controls. Genotype frequencies for the five other SNPs we studied are shown in Table S1.
Table 2.
C17T genotype frequencies, HIV-serostatus, and ethnic/racial group.
| HIV-seropositive | HIV-seronegative | |||||
|---|---|---|---|---|---|---|
| White | Hispanic | African-American | White | Hispanic | African-American | |
| CC | 114 (100%) | 94 (84.7%) | 290 (60.3%) | 45 (97.8%) | 64 (94.1%) | 143 (67.1%) |
| CT | 0 (0%) | 16 (14.4%) | 162 (33.7%) | 1 (2.2%) | 4 (5.9%) | 57 (26.8%) |
| TT | 0 (0%) | 1 (0.9%) | 29 (6.0%) | 0 (0.0%) | 0 (0.0%) | 13 (6.1%) |
Association of KMSK scores with C17T polymorphisms and with other SNPs
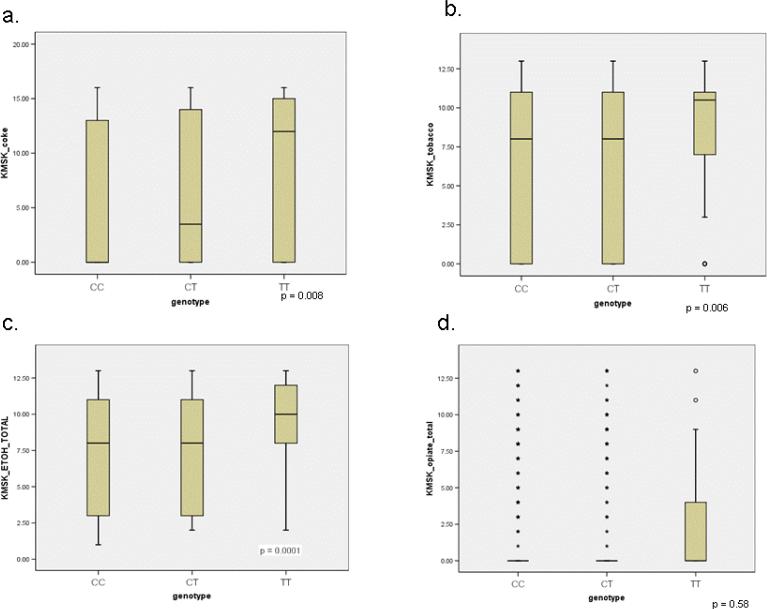
Of the 6 SNPs we genotyped, only the C17T polymorphism was significantly associated with KMSK scores. Figure 2 displays KMSK scores for cocaine, alcohol, tobacco, and opiates for the CC, CT, and TT genotypes of OPRM1 in African-American subjects. Differences in scores between the CC and CT genotypes were small, and not statistically significant. However, the distribution of KMSK cocaine, alcohol, and tobacco scores for participants with the TT genotype were greater than for either the CC or CT genotypes. The distribution of KMSK opiate scores did not differ among the 3 genotypes.
Figure 2. C17T genotype and KMSK score.
Box and whisker plots displaying distribution of KMSK scores for the CC, CT, and TT genotypes among African-Americans. Twenty-fifth percentile, 50th, and 75th percentiles are represented. Open circles represent single cases of outliers (1.5-3 interquartile ranges from the end of the box). Stars represent single cases of extreme values (> 3 interquartile ranges from end of the box). The 25th and 50th percentiles for the CC genotype for the KMSK cocaine scale overlap. Significance was determined with the Kruskal-Wallis test.
Association of the TT genotype with KMSK scores
Table 3 shows the effect of TT genotype on KMSK scores in African-American women. Odds ratios for the effect of the TT genotype on the alcohol, tobacco, and cocaine scales were approximately 2 for all models and all were significant at least at p <0.03. There was a statistically non-significant higher KMSK opiate score (OR 1.93, p=0.069) among African-American women. Analyses in the combined group of African American, Hispanic, and White women (data not shown) yielded similar results, although significance fell to around 0.08 for the KMSK alcohol scale in adjusted models. The association of the TT genotype with KMSK cocaine and tobacco scores remained significant at p < 0.02 in the combined cohort of whites, Hispanics, and African-Americans.
Association of the TT genotype with dichotomous outcomes based on KMSK scales or on percent of visits reporting drug use
Unadjusted and adjusted odds ratios for the TT genotype in predicting the dichotomous outcome of ‘likely substance dependence’ vs. controls are shown in Table 4. In the fully adjusted model, significance falls to 0.14 for the TT genotype in predicting the outcome of ‘likely cocaine dependence’, but is significant in the simpler models. The odds ratios for the TT genotype of predicting ‘likely alcohol dependence’ are significant in all 3 models.
Table 4.
Unadjusted and adjusted odds ratios for the TT genotype of predicting the dichotomous outcomes of ‘likely substance dependence’ [based on Kreek–McHugh–Schluger–Kellogg (KMSK) cut scores] versus control subjects in African American women.
| Unadjusted [P value] | Adjusted for HIV serostatus [P value] | Adjusted for HIV, age, income and education [P value] | |
|---|---|---|---|
| ‘Likely opiate dependence’ versus controls | 1.5 (0.4–5.1) [0.53] | 1.6 (0.5–5.6) [0.46] | 1.4 (0.3–6.1) [0.65] |
| ‘Likely alcohol dependence’ versus controls | 3.7 (1.6–8.4) [0.003] | 3.6 (1.5–8.3) [0.003] | 3.0 (1.1–8.0) [0.03] |
| ‘Likely coeaine dependence’ versus controls | 2.8 (1.8–6.4) [0.014] | 2.7 (1.2–6.2) [0.02] | 2.0 (0.8–5.2) [0.14] |
The numbers in each cell are odds ratios, 95% confidence intervals and significance levels. The TT genotype was found in 8 out of 230 controls, 24 of 261 with ‘likely cocaine dependence’, 20 of 173 with ‘likely alcohol dependence’ and 4 of 79 with ‘likely opiate dependence’ (because controls exclude subjects with some substance use, the number of subjects with the TT genotype do not sum up to 42—the total number of African American participants with the TT genotype).
We used ‘frequent substance use’ as a second dichotomous outcome variable to obtain a measure independent of the KMSK scales (Table 5). Odds ratios for the TT genotype in predicting ‘frequent cocaine’ or ‘frequent excessive alcohol use’ were somewhat higher than in predicting KMSK score and all highly significant. The odds ratio for predicting ‘frequent heroin use’ were not significant.
Table 5.
Adjusted and unadjusted odds ratios for the TT genotype of predicting the dichotomous outcomes of ‘frequent substance abuse’ (based on percentage of visits reporting substance use) vs. controls in African-American women.
| Unadjusted | Adjusted for HIV-serostatus | Adjusted for HIV, age, income, and education | |
|---|---|---|---|
| Top 10% heroin use vs. controls | 1.5 (0.6-4.2), p = 0.41 | 1.6 (0.6-4.3), p = 0.41 | 2.1 (0.6-7.2), p = 0.23 |
| Top 10% excessive alcohol vs. controls | 2.9 (1.2-6.7), p = 0.015 | 3.1 (1.3-7.2), p = 0.01 | 3.6 (1.4-9.8), p = 0.01 |
| Top 10% cocaine use vs. controls | 2.7 (1.1-6.5), p = 0.03 | 2.8 (1.2-6.8), p = 0.02 | 3.4 (1.1-10.4), p = 0.03 |
The numbers in each cell are odds ratios, 95% confidence intervals, and significance levels. The TT genotype was found in 17 of 374 controls, 8 of 72 in the top 10 % of cocaine use, 9 of 77 in the top 10% alcohol percent, and 5 of 75 in top 10% of heroin use. The number of total cases for each substance are slightly greater than 70 (i.e. top ten percent of 697 participants) because of rounding off. The numbers of controls differ in tables 4 and 5 because in table 4 they were based on lifetime KMSK scales and in table 5 they were based on substance use during 15 years of follow-up in the WIHS. Many women who had used drugs at some time of their life before the study totally abstained during the study. Hence the number of controls is greater in table 5 than in table 4.
Discussion
This is largest study of the C17T polymorphism of the mu opiate receptor gene in African-American women and the only study that used a quantitative measure of substance use. In African American women, we found highly significant associations of the TT genotype with KMSK alcohol, cocaine, and tobacco scores. The association of the TT genotype with the KMSK opiate scale was not significant. When we divided subjects based on KMSK scores into those likely to have substance dependence and non-using controls, we again found significant odds ratios for the TT genotype for predicting cocaine or alcohol dependence. We confirmed this finding with an independent measure of cocaine use and of frequent excessive alcohol use based on use reported at up to 29 visits over 15 years.
Even with our large sample size, we had inadequate power to detect a significant effect of the TT genotype on opiate abuse, because of the low prevalence of use. We calculated power of 18% for detecting a significant effect of the TT genotype on the KMSK opiate scale. In contrast, we had power of 99% for detecting a significant effect on the KMSK alcohol and cocaine scales. Odds ratios for the TT genotype in predicting KMSK opiate score were similar to those for predicting KMSK alcohol, tobacco, and cocaine score, but significance levels were marginal (p = 0.07) for predicting opiate score. We suspect that the TT genotype also significantly increases risk of opiate dependence, but the sample size was too small to demonstrate this.
We could find only three previous association studies of opiates, cocaine, alcohol, or tobacco use that included at least 50 cases and 50 controls from a specific racial/ethnic group and had any cases or controls with the TT genotype. Hoehe et al. (2000) studied 158 African-American cases and 51 controls. Cases included 125 cocaine-dependent and 33 opiate-dependent subjects. The TT genotype was found in 6 cases and in no controls. Crowley et al. (2003) studied 96 African-Americans with severe opiate addiction and 99 controls. The TT genotype was found in 3 cases and in 7 controls. Kapur et al. (2007) found 13 Indo-European men with opiate addiction with the TT genotype out of 126 total cases. None of the 156 controls had the TT genotype.
Our study was not limited to African-American women, but because the TT genotype was found in only one Hispanic and no white women, we chose to perform all analyses in two ways – first confined to African-Americans and then with the entire cohort of white, Hispanic, and African-American women. Even in the latter case, we demonstrated significant odds ratios for the TT genotype for predicting KMSK cocaine, alcohol, and tobacco scores.
Racial/ethnic admixture has been a source of false positive results in case control studies of candidate genes. The marked disparity in frequency of the C17T polymorphism among the 3 racial/ethnic groups suggests that it is a marker of African ancestry. Nonetheless, there are several reasons why we do not believe genetic admixture is a cause of false positive results in our study. First, Ducci et al. (2009) recently demonstrated that African genetic heritage is not associated with opioid or cocaine addiction. Second, disease frequencies are comparable for cocaine and tobacco in the 3 ethnic/racial groups in order study. Alcohol use is more common in whites, and opiate use is less common in African-Americans. Lower frequency in the African-American women bias against finding a significant association. A spurious association might occur if we neither stratified by racial/ethnic groups nor included race/ethnicity as a covariate, but we used both methods to control for genetic admixture. Furthermore, our primary analyses utilized all of the African-Americans in the cohort rather than separating them into cases and controls. This method somewhat obviates the potential problem of uneven distribution of ancestral markers between cases and controls. Third, we used the best method available to us short of using ancestral genes to categorize racial/ethnic groups by requiring that all 4 grandparents be of the same racial/ethnic group. Fourth, possible change in function of OPRM1 is certainly a biologically plausible explanation for differences in substance dependence. Finally, our results are consistent with those found in two of three previous studies. Nonetheless, we recognize that for our dichotomous outcome analyses, we cannot rule out uneven racial/ethnic admixture between cases and controls without analyzing a complete set of ancestral genetic markers.
Does the fact that most of the study participants have HIV limit the generalizability of our findings? If a participants’ knowledge that they were HIV-seropositive influenced results, maximal substance use should have occurred after acquiring HIV. Participants reported the year when substance abuse was maximal, in most cases it was many years before they learned they were HIV-seropositive. Furthermore, when we included HIV-serostatus as a covariate in the models, our results were not changed.
What is the evidence that the TT genotype might influence function of OPRM1? The sixth position from the N-terminal of OPRM1 lies in the extra-cellular space. While the substitution of alanine for valine in the sixth amino acid is not likely to influence binding of an agonist to the receptor, it could influence signaling functions of MOR1. A recent study (Ravindranathan et al., 2009) tested A6V-MOR1A variant in the receptor activity assay, a ligand-induced efflux of intracellular calcium, and found that the EC50 for DAMGO was 13.3±0.8 compared to 35.4±4.1 for the MOR1A. Also, Emax of DAMGO differed significantly between the A6V-MOR1A variant and MOR1A (P < 0.05). However, there were no significant differences in binding affinity for DAMGO in these variants.
Lifetime prevalence of substance use and abuse was higher for all racial/ethnic groups in our study than rates reported for young adults in the Monitoring the Future study (Johnston et al., 2008). The high rates of lifetime dependence in all 3 racial/ethnic groups among both HIV-infected and HIV-uninfected participants reflect recruitment from high-risk populations.
Our original aim when this research was planned was to study the association of 6 different OPRM1 polymorphisms with KMSK scores. Our focus on the TT genotype of C17T was a natural extension of that aim. We did not correct for multiple comparisons; the best way to do so is controversial and overcorrection runs the risk of making type II errors. Our most robust results were significant at p = 0.0001, a level required for significance after correcting for 500 comparisons by the Bonferonni method. Moreover, our results are consistent for cocaine, alcohol, and tobacco and when an independent outcome measure was utilized. Nonetheless, we concede that many of our significance levels are in the p = 0.01 to 0.04 range, and might not be significant if we corrected for multiple comparisons.
This work has significant strengths and weaknesses. The large sample size of African-American participants and detailed information about the race/ethnicity of participants’ grandparents are strengths. Use of the KMSK could be viewed as both a strength and a weakness. A quantitative scale often has more power to detect differences than a qualitative scale. The KMSK is operationally defined, but it based on participants’ recall of drug use many years in the past. In studies involving participants from the United States, retrospective recall is usually the only way to obtain data about maximal drug use. Our use of dichotomous outcomes for secondary analyses may also be viewed as a strength and weakness. The dichotomous outcomes we have created are easier to understand and more consistent with analyses in the literature than the ordinal KMSK scales. On the other hand, by analyzing only the extremes of our sample, we diminish our sample size.
Social and psychological pressures for or against substance use vary over an individual's lifetime; epigenetic factors also may vary. In contrast, genetic factors influencing the use of drugs should not vary over one's lifetime. Using maximal drug use as an outcome rather than current drug use may limit the effects of psychosocial factors that cannot be completely controlled for in any analysis. Since this work was initiated, we (MJK) have been using quantitative measures of both maximal lifetime substance use and recent substance use in our analyses.
Our results raise many questions. Can the results be duplicated in women without HIV and in men with or without HIV? Substance abuse is a complex trait: what, if any, psychological traits are also associated with the TT genotype? Are there haplotypes that include the TT genotype that show even stronger associations with substance use and abuse? Such studies are being planned.
Supplementary Material
Table S1 Frequency of other OPRM1 SNPs studied and ethnic/racial group.
ACKNOWLEDGEMENTS
Supported in part by grants 1R01MH076537 (HC), 1R01MH079880 (MJK), IAID U01 318345, and NIH-NIDA P60-05130 (MJK). Data in this manuscript were collected by the Women's Interagency HIV Study (WIHS) Collaborative Study Group with centers (Principal Investigators) at New York City/Bronx Consortium (Kathryn Anastos); Brooklyn, NY (Howard Minkoff); Washington DC, Metropolitan Consortium (Mary Young); The Connie Wofsy Study Consortium of Northern California (Ruth Greenblatt); Los Angeles County/Southern California Consortium (Alexandra Levine); Chicago Consortium (Mardge Cohen); Data Coordinating Center (Stephen Gange). The WIHS is funded by the National Institute of Allergy and Infectious Diseases (UO1-AI-35004, UO1-AI-31834, UO1-AI-34994, UO1-AI-34989, UO1-AI-34993, and UO1-AI-42590) and by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (UO1-HD-32632). The study is co- funded by the National Cancer Institute, the National Institute on Drug Abuse, and the National Institute on Deafness and Other Communication Disorders. Funding is also provided by the National Center for Research Resources (UCSF-CTSI Grant Number UL1 RR024131). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
We acknowledge O Levran, V Yuferov, and D Proudnikov for useful discussions and manuscript review.
We thank the women participating in WIHS for their time, cooperation, and support.
REFERENCES
- Bacon MC, von Wyl V, et al. The Women's Interagency HIV Study: an observational cohort brings clinical sciences to the bench. Clin Diagn Lab Immunol. 2005;12(9):1013–9. doi: 10.1128/CDLI.12.9.1013-1019.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan SE, Melnick SL, Preston-Martin S, Weber K, Kalish LA, Miotti P, Young M, Greenblatt R, Sacks H, Feldman J. The women's interagency HIV study. WIHS collaborative study group. Epidemiol. 1998;9(2):117–125. [PubMed] [Google Scholar]
- Bergen AW, Kokoszka J, et al. Mu opioid receptor gene variants: lack of association with alcohol dependence. Mol Psychiatry. 1997;2(6):490–4. doi: 10.1038/sj.mp.4000331. [DOI] [PubMed] [Google Scholar]
- Berrettini WH. Human mu opioid receptor gene polynorphisms and vulnerability to substance abuse. Addiction Biology. 1997;2:303–308. doi: 10.1080/13556219772598. [DOI] [PubMed] [Google Scholar]
- Bond C, LaForge KS, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95(16):9608–13. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley JJ, Oslin DW, et al. A genetic association study of the mu opioid receptor and severe opioid dependence. Psychiatr Genet. 2003;13(3):169–73. doi: 10.1097/00041444-200309000-00006. [DOI] [PubMed] [Google Scholar]
- Dahan A, Kest B, et al. Sex-specific responses to opiates: animal and human studies. Anesth Analg. 2008;107(1):83–95. doi: 10.1213/ane.0b013e31816a66a4. [DOI] [PubMed] [Google Scholar]
- Ducci F, Roy A, et al. Association of substance use disorders with childhood trauma but not African genetic heritage in an African American cohort. Am J Psychiatry. 2009;166(9):1031–40. doi: 10.1176/appi.ajp.2009.08071068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke P, Wang T, et al. Nonreplication of association between mu-opioid-receptor gene (OPRM1) A118G polymorphism and substance dependence. Am J Med Genet. 2001;105(1):114–9. [PubMed] [Google Scholar]
- Gelernter J, Kranzler H, et al. Genetics of two mu opioid receptor gene (OPRM1) exon I polymorphisms: population studies, and allele frequencies in alcohol- and drug-dependent subjects. Mol Psychiatry. 1999;4(5):476–83. doi: 10.1038/sj.mp.4000556. [DOI] [PubMed] [Google Scholar]
- Gilbody S, Richards D, et al. Screening for depression in medical settings with the Patient Health Questionnaire (PHQ): a diagnostic meta-analysis. J Gen Intern Med. 2007;22(11):1596–602. doi: 10.1007/s11606-007-0333-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Finch SJ, Nothnagel M, Ott J. Power and sample size calculations for case-control genetic association tests when errors present: application to single nucleotide polymorphisms. Human Heredity. 2002;54:22–33. doi: 10.1159/000066696. [DOI] [PubMed] [Google Scholar]
- Hoehe MR, Kopke K, et al. Sequence variability and candidate gene analysis in complex disease: association of mu opioid receptor gene variation with substance dependence. Hum Mol Genet. 2000;9(19):2895–908. doi: 10.1093/hmg/9.19.2895. [DOI] [PubMed] [Google Scholar]
- Hurley RW, Adams MC. Sex, gender, and pain: an overview of a complex field. Anesth Analg. 2008;107(1):309–17. doi: 10.1213/01.ane.0b013e31816ba437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LD, O'Malley PM, Bachman JG, Schulenberg JE. Monitoring the Future national survey results on drug use, 1975-2007. Volume II: College students and adults ages 19-45 (NIH Publication No. 08-6418B) National Institute on Drug Abuse; Bethesda, MD: 2008. p. 319. [Google Scholar]
- Kapur S, Sharad S, et al. A118g polymorphism in mu opioid receptor gene (OPRM1): association with opiate addiction in subjects of Indian origin. J Integr Neurosci. 2007;6(4):511–22. doi: 10.1142/s0219635207001635. [DOI] [PubMed] [Google Scholar]
- Kellogg SH, McHugh PF, et al. The Kreek-McHugh-Schluger-Kellogg scale: a new, rapid method for quantifying substance abuse and its possible applications. Drug Alcohol Depend. 2003;69(2):137–50. doi: 10.1016/s0376-8716(02)00308-3. [DOI] [PubMed] [Google Scholar]
- Kreek MJ, Bart G, et al. Pharmacogenetics and human molecular genetics of opiate and cocaine addictions and their treatments. Pharmacol Rev. 2005;57(1):1–26. doi: 10.1124/pr.57.1.1. [DOI] [PubMed] [Google Scholar]
- Lacroix-Fralish ML. Sex-specific pain modulation: the growth factor, neuregulin-1, as a pro-nociceptive cytokine. Neurosci Lett. 2008;437(3):184–7. doi: 10.1016/j.neulet.2008.02.074. [DOI] [PubMed] [Google Scholar]
- Levran O, Londono D, et al. Heroin addiction in African Americans: a hypothesis-driven association study. Genes Brain Behav. 2009;8(5):531–40. doi: 10.1111/j.1601-183X.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, et al. Haplotypes at the OPRM1 locus are associated with susceptibility to substance dependence in European-Americans. Am J Med Genet B Neuropsychiatr Genet. 2003;120B(1):97–108. doi: 10.1002/ajmg.b.20034. [DOI] [PubMed] [Google Scholar]
- Luo X, Zuo L, et al. Multiple OPR genes influence personality traits in substance dependent and healthy subjects in two American populations. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(7):1028–39. doi: 10.1002/ajmg.b.30701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindranathan A, Joslyn G, et al. Functional characterization of human variants of the mu-opioid receptor gene. Proc Natl Acad Sci U S A. 2009;106(26):10811–6. doi: 10.1073/pnas.0904509106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommelspacher H, Smolka M, et al. Genetic analysis of the mu-opioid receptor in alcohol-dependent individuals. Alcohol. 2001;24(2):129–35. doi: 10.1016/s0741-8329(01)00139-2. [DOI] [PubMed] [Google Scholar]
- Spitzer RL, Kroenke K, et al. Validation and utility of a self-report version of PRIME-MD: the PHQ primary care study. Primary Care Evaluation of Mental Disorders. Patient Health Questionnaire. Jama. 1999;282(18):1737–44. doi: 10.1001/jama.282.18.1737. [DOI] [PubMed] [Google Scholar]
- Stroop J. Studies of interference in serial verbal reactions. Journal of Experimental Psychology. 1935;18:643–662. [Google Scholar]
- Tan EC, Tan CH, et al. Mu opioid receptor gene polymorphisms and heroin dependence in Asian populations. Neuroreport. 2003;14(4):569–72. doi: 10.1097/00001756-200303240-00008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Frequency of other OPRM1 SNPs studied and ethnic/racial group.