Abstract
Longitudinal bone growth is determined by endochondral ossification that occurs as chondrocytes in the cartilaginous growth plate undergo proliferation, hypertrophy, cell death, and osteoblastic replacement. The natriuretic peptide family consists of three structurally related endogenous ligands, atrial, brain, and C-type natriuretic peptides (ANP, BNP, and CNP), and is thought to be involved in a variety of homeostatic processes. To investigate the physiological significance of CNP in vivo, we generated mice with targeted disruption of CNP (Nppc−/− mice). The Nppc−/− mice show severe dwarfism as a result of impaired endochondral ossification. They are all viable perinatally, but less than half can survive during postnatal development. The skeletal phenotypes are histologically similar to those seen in patients with achondroplasia, the most common genetic form of human dwarfism. Targeted expression of CNP in the growth plate chondrocytes can rescue the skeletal defect of Nppc−/− mice and allow their prolonged survival. This study demonstrates that CNP acts locally as a positive regulator of endochondral ossification in vivo and suggests its pathophysiological and therapeutic implication in some forms of skeletal dysplasia.
There are two major mechanisms of bone formations, membranous and endochondral ossifications. The former occurs when mesenchymal precursor cells directly differentiate into bone-forming osteoblasts, a process by which all flat bones are formed. The latter involves the conversion of an initial cartilage template into bone and is responsible for the formation of long bones and vertebrae.
The natriuretic peptide system consists of a family of three structurally related endogenous ligands, atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP), and C-type natriuretic peptide (CNP) (1), and three membrane-bound receptors, two of which are guanylyl cyclase (GC)-coupled receptors (GC-A and GC-B) that mediate the biological actions of the ligands, and one of which is a biologically silent receptor (C-receptor) implicated in the metabolic clearance of the ligands (2, 3). ANP and BNP are cardiac hormones that are produced predominantly by the atrium and ventricle, respectively (4–6), and are thought to play important roles in the regulation of cardiovascular homeostasis, primarily through GC-A (7, 8). On the other hand, CNP occurs in a wide variety of central and peripheral tissues (9–12) and may act locally as an autocrine/paracrine regulator through GC-B (7, 8).
We have created transgenic mice overexpressing BNP under the control of the liver-specific serum amyloid P component promoter and demonstrated that they exhibit blood pressure reduction (13). They also show marked skeletal overgrowth accompanied by the activation of endochondral ossification (14). It has been also reported that C-receptor-deficient mice (Npr3−/− mice) show skeletal abnormalities similar to those seen in transgenic mice overexpressing BNP (15, 16). These observations suggest that natriuretic peptides can affect endochondral ossification. In this context, previous studies have revealed no such skeletal abnormalities in ANP (Nppa)-deficient mice (17) or BNP (Nppb)-deficient mice (18), indicating that neither ANP nor BNP is involved in endochondral ossification under physiological conditions. In this study, we generated mice with targeted disruption of CNP (Nppc−/− mice) and demonstrated that they show severe dwarfism and early death as a result of impaired endochondral ossification. Targeted expression of CNP in the growth plate chondrocytes can rescue the skeletal defect of Nppc−/− mice and allow their prolonged survival. This study demonstrates that CNP is a bona fide endogenous natriuretic peptide in the bone, where it activates endochondral ossification.
Materials and Methods
Gene Targeting and Generation of Transgenic Mice.
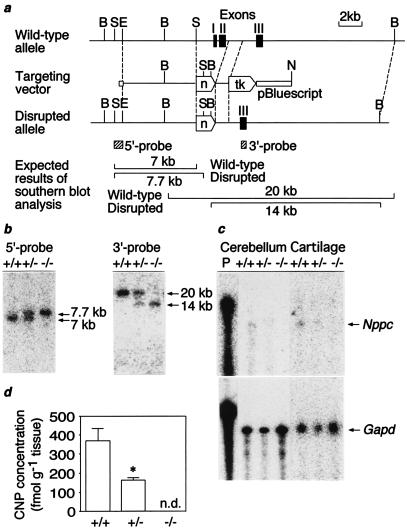
The 129/Sv mouse Nppc was isolated from a 129/Sv mouse genomic library in λFixII (Stratagene, La Jolla, CA). A targeting vector was constructed, in which exons 1 and 2 of the 129/Sv mouse Nppc that encode the entire coding sequences of mouse preproCNP were replaced by the neomycin resistance gene (Fig. 1a). The targeting vector was introduced into embryonic stem cells by electroporation (18). Double selection in G418 and ganciclovir produced seven homologously recombinant embryonic stem cell clones that were analyzed by Southern blot analysis with the 5′ and 3′ external probes indicated (Fig. 1 a and b). Male chimeras derived from two independent clones with germ-line transmission of the disrupted allele were bred to C57BL/6J or 129/SvJ females.
Figure 1.
Targeted disruption of the mouse Nppc. (a) Restriction maps of the wild-type 129/Sv mouse Nppc allele, targeting vector, and the predicted disrupted allele. Closed boxes indicate exons (I–III). Locations of 5′ and 3′ external probes are shown as hatched bars. B, BamHI; S, SphI; E, EcoRI; N, NotI; tk, Herpes simplex virus thymidine kinase gene; n, neomycin resistance gene. (b) Southern blot analysis of genomic DNAs from F2 offspring with 5′ and 3′ probes upon digestion with SphI and BamHI, respectively. (c) RNase protection analysis of Nppc and Gapd transcript in the cerebellum and tibial epiphyseal cartilage. The cerebellum and tibial cartilage are obtained from mice at 20 weeks and 7 days of age, respectively. Ten micrograms and 1 μg of total RNA were used to analyze Nppc and Gapd transcripts, respectively. (d) Cerebellar CNP concentrations at 20 weeks of age (n = 4). n.d., not detectable. *, P < 0.05 vs. Nppc+/+ mice.
Generation of transgenic mice (Tg mice) with targeted expression of CNP in the growth plate chondrocytes under the control of the mouse pro-α1(II) collagen (Col2a1) promoter [provided by B. de Crombrugghe at the M. D. Anderson Cancer Center, Houston, TX (19)] will be reported elsewhere (A.Y., Y.K., H.C., T.M., Y.O., and Kazuwa Nakao, unpublished observations). The transgene expression was detected only in the chondrocytes (19). To perform genetic rescue of Nppc−/− mice, Tg mice were mated with Nppc+/− mice, and F1 offspring heterozygous for the transgene and for the Nppc allele ablation were bred to generate Nppc−/− mice with the transgene expression (Tg/Nppc−/− mice). The care of the animals and all experiments were conducted in accordance with the institutional guidelines of Kyoto University Graduate School of Medicine.
RNA and Peptide Analysis.
Total RNA was extracted from various tissues from Nppc+/+, Nppc+/−, and Nppc−/− mice. In the cerebellum and epiphyseal cartilage, Nppc mRNA expression was assessed by RNase protection assay. Ten micrograms and 1 μg of total RNA were used to analyze Nppc and glyceraldehyde- 3-phosphate dehydrogenase (Gapd) transcripts, respectively. 32P-labeled antisense Nppc and Gapd riboprobes were generated from a 5′-rapid amplification of cDNA ends-based 129/Sv mouse Nppc cDNA fragment and mouse Gapd cDNA fragment (a gift from M. B. Prystowsky at the Albert Einstein College of Medicine), respectively. In other tissues, Nppc and Gapd mRNA expressions were assessed by reverse transcription (RT)-PCR and Southern blot analysis (Nppc: sense primer, 5′-AAAAAGGGTGACAAGACTCCAGGCAG-3′; antisense primer, 5′-GGTGTTGTGTATTGCCAGTA-3′; antisense probe, 5′-CCTTC-TTGTTGCCGCCTTTGAT-3′; Gapd: sense and antisense primers, a mouse G3PDH control amplimer set, CLONTECH; antisense probe, 5′-GCCTTGACTGTGCCGTTGAATTTGCCGTGA-3′). Cerebellar CNP concentrations were determined by a RIA for CNP (9).
Skeletal Preparation and Histology.
Skeletal preparation and histological analysis were performed as described (14). Briefly, mice were killed, skinned, eviscerated, and subjected to soft x-ray analysis (23 kVp, 5 mA for 1 min; Softron Type SRO-M5, Softron, Tokyo). Bones from 7-day-old mice were fixed in 4% paraformaldehyde in 0.01 M PBS (pH 7.4), decalcified in 10% EDTA, and embedded in paraffin. Five-micrometer-thick sections were sliced and stained with Alcian blue (pH 2.5) and hematoxylin/eosin. The slices were also analyzed for apoptosis by terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate-biotin nick end labeling assay according to the manufacturer's protocol (Takara Shuzo, Shiga, Japan).
Detection of BrdUrd-Labeled Cells.
Mice at 1 week of age were injected intraperitoneally with BrdUrd (100 μg/g body weight) and were killed 6 h later. Tibiae were fixed in 4% paraformaldehyde in 0.01 M PBS (pH 7.4), decalcified in 10% EDTA, and embedded in paraffin. Detection of BrdUrd-positive cells in the growth plate was performed by BrdUrd labeling and with a detection kit II (Roche Diagnostics).
In Situ Hybridization.
Digoxigenin-labeled antisense and sense riboprobes were obtained from reverse transcription–PCR products for Nppc, rat GC-B (nucleotides 762-1394 of rat GC-B cDNA; GenBank M26896), mouse pro-α1(I) collagen (Col1a1) (nucleotide 693-1139 of Col1a1 cDNA; GenBank M14423) and rat pro-α1(X) collagen (a gift from B. R. Olsen at Harvard Medical School, Boston, MA), mouse pro-α1(II) collagen (Col2a1) (a gift from Y. Yamada at the National Institutes of Health, Bethesda, MD), and mouse Indian hedgehog (Ihh) (a gift from K. Lee at Massachusetts General Hospital, Boston, MA) cDNA fragments with the use of a digoxigenin RNA labeling kit (Roche Diagnostics). Tibiae from 7-day-old mice were fixed in 4% paraformaldehyde, 0.5% glutaraldehyde, 1 mM CaCl2 in 0.1 M phosphate buffer (pH 7.2) for 24 h, embedded in paraffin, and sliced into 5-μm-thick sections. In situ hybridization analysis was performed as described (20).
cGMP Assay.
The tail bone cGMP concentrations in 7-day-old mice were determined by a RIA for cGMP as described (21).
Statistical Analysis.
Data are expressed as means ± SE. The statistical significance of differences in mean values was assessed by Student's t test. The difference in survival rates among genotypes was assessed by Kaplan–Meier analysis.
Results and Discussion
Generation of Nppc−/− Mice.
To investigate the physiological significance of CNP in vivo, we generated mice with a disrupted Nppc allele by gene targeting in 129/Sv mouse-derived embryonic stem cells (Fig. 1a). Male chimeras with germ-line transmission of the disrupted allele were bred to C57BL/6J and 129/SvJ females, and Nppc+/+ mice (wild type), Nppc+/− mice (heterozygous for the disrupted allele), and Nppc−/− mice (homozygous for the disrupted allele) were obtained (Fig. 1b). The Nppc mRNA levels were decreased by ≈50% in the cerebellum and tibial epiphyseal cartilage from Nppc−/− mice relative to those of Nppc+/+ mice (Fig. 1c). Cerebellar CNP concentrations were also decreased in Nppc+/− mice relative to Nppc+/+ mice (P < 0.05, n = 4) (Fig. 1d). No Nppc mRNA or CNP was detected in the cerebellum or cartilage from Nppc−/− mice (Fig. 1 c and d). The Nppc transcript was also detected in other tissues, including the cerebrum, pituitary, heart, kidney, ovary, and uterus, in Nppc+/+ mice but not in Nppc−/− mice (data not shown).
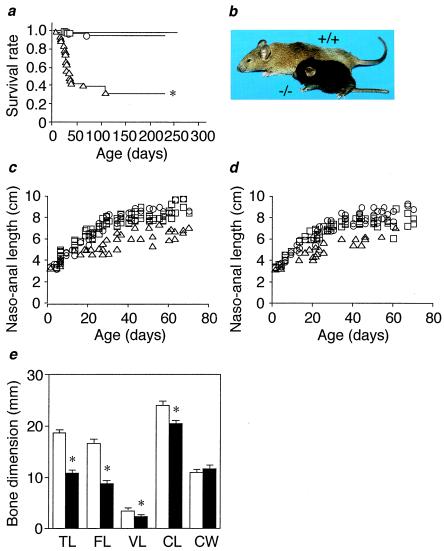
Analysis of 96 intercrosses between 129/B6 hybrids heterozygous for the disrupted allele revealed that the genotype ratio of +/+:+/−:−/− at weaning (at 4 weeks of age) is 1.00:2.13:0.37 (n = 648), indicating a deviation from the expected Mendelian proportions (P < 0.001 by χ2 test). On the other hand, the genotype ratio of +/+:+/−:−/− at 16.5 days post coitus was 1.0:1.5:1.4 (n = 65), suggesting that Nppc−/− mice are not embryonically lethal. More than half of Nppc−/− mice died before weaning, and only 30% survived at 100 days of age and thereafter (Fig. 2a). No appreciable differences in the genotype ratio and survival rate were noted between the 129/B6 hybrid and 129/Sv pure backgrounds.
Figure 2.
Dwarfism and mortality in Nppc−/− mice. (a) Survival curves of Nppc+/+ (○), Nppc+/− (□), and Nppc−/− (▵) mice. (b) Gross appearance of Nppc+/+ and Nppc−/−mice at 10 weeks of age. (c and d) Growth curves of Nppc+/+ (○), Nppc+/− (□), and Nppc−/− (▵) mice. (c) Males. (d) Females. (e) Soft x-ray analysis of Nppc+/+ and Nppc−/− mice at 10 weeks of age. Bone dimensions in e are of Nppc+/+ (open bars) and Nppc−/− (closed bars) mice (n = 4). TL, tibial length; FL, femoral length; VL, fifth lumbar vertebral length; CL, naso-occipital length of the calvarium; CW, maximal interparietal distance of the calvarium. *, P < 0.05 vs. Nppc+/+ mice.
At birth, Nppc−/− pups had a body weight and a naso-anal length about 90% of those of Nppc+/+ pups. In Nppc−/− mice, dwarfism with short tails and extremities became prominent as they grew (Fig. 2b). The naso-anal lengths of male and female Nppc−/− mice were 60–70% of those of Nppc+/+ mice at 4–10 weeks of age (Fig. 2 c and d). Body weights of Nppc−/− mice were about 70% of those of Nppc+/+ mice, and no significant differences in visceral organ/body weight ratios were noted between genotypes at 20 weeks of age. No other gross abnormalities were found in Nppc−/− mice.
Soft x-ray analysis revealed that the longitudinal growth of vertebrae and tail and limb bones is affected in Nppc−/− mice. The longitudinal lengths of femurs, tibiae, and vertebrae in Nppc−/− mice were 50–80% of those in Nppc+/+ mice (Fig. 2e). The naso-occipital length of the calvarium, which depends on the growth of occipital and sphenoidal bones formed through endochondral ossification, was also reduced significantly in Nppc−/− mice relative to Nppc+/+ mice (n = 4, P < 0.05) (Fig. 2e). On the other hand, in Nppc−/− mice, there were no appreciable changes in the shape and interparietal width of the skull vault, which are formed by membranous ossification. These observations indicate that loss of CNP affects endochondral ossification but not membranous ossification in vivo.
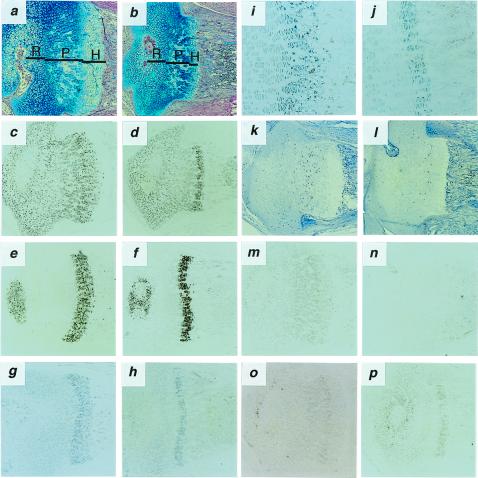
Longitudinal bone growth is determined by endochondral ossification in the cartilaginous growth plate, which is located at both ends of vertebrae and long bones (22). The cartilaginous growth plate consists of the resting, proliferative, and hypertrophic zones of chondrocytes. Histologically, Nppc−/− mice displayed striking narrowing of the growth plate of vertebrae and long bones compared with Nppc+/+ mice at 7 days of age (Fig. 3 a and b). The heights of the proliferative and hypertrophic zones were markedly reduced in Nppc−/− mice, whereas no significant differences in the resting zone were noted between genotypes. In situ hybridization analysis revealed no appreciable difference in the intensity of Col2a1 and type X collagen (Col10a1) mRNA expression between genotypes (Fig. 3 c–f). No ectopic expression of Col1a1 mRNA was found in the growth plate from Nppc−/− mice (data not shown). These findings suggest that chondrocyte precursors are capable of differentiating into hypertrophic chondrocytes in the growth plate from Nppc−/− mice. However, the band width of Ihh-expressing cells [cells committing to differentiation into hypertrophic chondrocytes (23)] was narrowed in Nppc−/− mice relative to Nppc+/+ mice (Fig. 3 g–j), although the intensity of Ihh expression was not remarkably different between genotypes. Notably, the ratio of the height of the hypertrophic zone to the height of the proliferative zone was decreased by ≈50% in Nppc−/− mice compared with Nppc+/+ mice (Fig. 3 a and b). These observations suggest that the rate of cell differentiation into hypertrophic chondrocytes is reduced in Nppc−/− mice. The number of hypertrophic chondrocytes positive for terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate-biotin nick end labeling was not significantly different between genotypes (data not shown), suggesting that chondrocyte apoptosis is unaffected in Nppc−/− mice.
Figure 3.
Histological analysis of proximal epiphyseal cartilages of the tibia from 7-day-old Nppc+/+ and Nppc−/− mice. (a and b) Alcian blue-hematoxylin/eosin staining of tibial epiphyseal cartilages from Nppc+/+ (a) and Nppc−/− (b) mice. Resting (R), proliferating (P), and hypertrophic (H) zones are indicated. (c–j, m–p) In situ hybridization analysis of tibial epiphyseal cartilages from Nppc+/+ (c, e, g, i, m, o) and Nppc−/− (d, f, h, j, n, p) mice showing expression of mRNA for Col2a1 (c, d), Col10a1 (e, f), Ihh (g-j), Nppc (m, n), or Npr2 (o, p). (k and l) Immunohistochemical detection of BrdUrd-labeled chondrocytes in the tibial growth plate from Nppc+/+ (k) and Nppc−/− (l) mice. (Magnification: a–h, k–p, ×40; i, j, ×100.)
We also assessed the consequence of Nppc ablation on the proliferation of the growth plate chondrocytes. There was a significant reduction of BrdUrd-labeled cells in Nppc−/− mice relative to Nppc+/+ mice (Fig. 3 k and l), demonstrating that CNP promotes chondrocyte proliferation in vivo.
In Nppc+/+ mice, Nppc mRNA was detected in proliferative and prehypertrophic chondrocytes in the growth plate but not detected in Nppc−/− mice (Fig. 3 m and n). Expression of mRNA for GC-B (Npr2) was detected predominantly in the proliferative and prehypertrophic chondrocytes in both Nppc−/− and Nppc+/+ mice (Fig. 3 o and p). The tail bone cGMP concentrations in Nppc−/− mice were markedly smaller than those in Nppc+/+ mice at 7 days of age (9.7 ± 2.2 vs. 25.6 ± 2.7 pmol/g tissue, n = 6, P < 0.01), suggesting that CNP is a major determinant of cGMP in the bone. Evidence has indicated that CNP increases the height of the proliferative and hypertrophic zones of chondrocytes and cartilage matrix production in the organ cultures of fetal mouse and rat long bones via cGMP production (21, 24). It is therefore likely that CNP acts primarily on proliferative and prehypertrophic chondrocytes expressing GC-B, thus stimulating growth plate chondrocyte proliferation and differentiation in vivo through cGMP-mediated pathway. Histology of other organs was not remarkable in Nppc−/− mice at 20 weeks of age (data not shown).
Transgenic Rescue of Nppc−/− Mice.
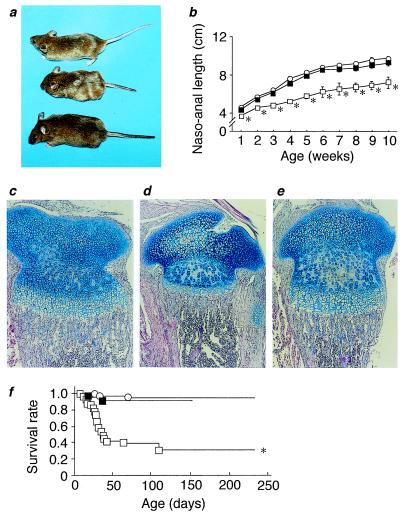
To determine whether local expression of CNP in the bone can rescue the dwarfism of Nppc−/− mice in vivo, they were crossed with mice with transgenic expression of CNP in the growth plate chondrocytes (Tg mice). The genetically “rescued” animals [or Nppc−/− mice with the transgene expression (Tg/Nppc−/− mice)] were of normal appearance (Fig. 4a), and their skeletons were indistinguishable from those of Nppc+/+ mice. During postnatal development, no significant difference in the naso-anal length was observed between Tg/Nppc−/− and Nppc+/+ mice (Fig. 4b). Body weights of Tg/Nppc−/− mice were not different from those of Nppc+/+ mice at 10 weeks of age (24.3 ± 1.9 vs. 21.9 ± 1.1 g, n = 6). Histologically, no appreciable differences in tibial growth plate cartilage were noted between Tg/Nppc−/− and Nppc+/+ mice (Fig. 4 c–e). The tail bone cGMP concentrations in Tg/Nppc−/− mice (17.2 ± 4.9 pmol/g tissue, n = 6) were also roughly comparable to those in Nppc+/+ mice (25.6 ± 2.7 pmol/g tissue, n = 6, P > 0.05). These findings demonstrate that CNP, when expressed locally in the growth plate chondrocytes, can rescue the skeletal defect of Nppc−/− mice via the cGMP-mediated mechanism in vivo. Although CNP is expressed in a variety of central and peripheral tissues, the data of this study indicate that CNP acts locally as a positive regulator of endochondral ossification. Targeted expression of CNP in the growth plate chondrocytes also resulted in prolonged survival of Nppc−/− mice; most Tg/Nppc−/− mice examined (11 of 13 mice) survived up to 20 weeks of age (Fig. 4f). Therefore, Nppc−/− mice are short-lived because of their skeletal abnormalities.
Figure 4.
Genetic rescue of Nppc−/− mice by crosses with transgenic mice with targeted expression of CNP in the growth plate chondrocytes. (a) Gross appearance of Nppc+/+ mice, Nppc−/− mice without the transgene expression, and Nppc−/−mice with the transgene expression (Tg/Nppc−/− mice) at 20 weeks of age. (From Top to Bottom) Nppc+/+, Nppc−/−, and Tg/Nppc−/− mice. (b) Growth curves of Nppc+/+ (○), Nppc−/− (□), and Tg/Nppc−/− (■) mice. *P < 0.05 vs. Nppc+/+ mice. (c–e) Alcian blue-hematoxylin/eosin stainings of tibial epiphyseal cartilages from 7-day-old Nppc+/+ (c), Nppc−/− (d), and Tg/Nppc−/− (e) mice. (f) Survival curves of Nppc+/+ (○), Nppc−/− (□), and Tg/Nppc−/− mice (■). *, P < 0.05 vs. Nppc+/+ mice.
Transgenic mice with elevated plasma BNP concentrations show skeletal overgrowth due to increased endochondral ossification (13, 14), whereas mice with targeted disruption of BNP exhibit no skeletal abnormalities (18). Thus, BNP, a hormone derived from the cardiac ventricle, is not involved in endochondral ossification under physiological conditions. This study has established CNP as an endogenous ligand to activate the cGMP production in the bone, thereby regulating endochondral ossification. In this context, mice with targeted disruption of cGMP-dependent protein kinase II, an intracellular mediator of cGMP that is expressed in late proliferative and early hypertrophic chondrocytes in the growth plate, show dwarfism due to impaired endochondral ossification (25). We postulate that the CNP/GC-B/cGMP-dependent protein kinase II signaling cascade plays a critical role in endochondral ossification. Recently, Npr3−/− mice have been reported to show skeletal overgrowth and increased endochondral ossification (15, 16). It is likely that the metabolic clearance of CNP produced locally is delayed in Npr3−/− mice, thereby activating endochondral ossification.
The Nppc−/− mice exhibit gross phenotypes and histologic features similar to those found in patients with achondroplasia (26) and mouse models for achondroplasia (27, 28). Several gain-of-function mutations in the fibroblast growth factor receptor 3 gene have been found in most achondroplastic patients and those with two other distinct skeletal dysplasias, hypochondroplasia and thanatophoric dysplasia (29–32). Our data suggest that CNP may be one of the causative genes for such skeletal dysplasias of unknown origin and may be useful for treatment of achondroplasia.
Acknowledgments
We thank T. Shiroishi for the 129/SvJ mouse strain; K. Katsuki, G. Katsuura, K. Watanabe, K. Okada, H. Hiratani, and M. Nagamoto for technical assistance; and Y. Isa and Y. Nakajima for secretarial assistance. This work is supported by grants from Research for the Future of the Japan Society for the Promotion of Science (JSPS-RFTF 96100204 and 98L00801); the Japanese Ministry of Education, Science, Sports, and Culture; the Japanese Ministry of Health and Welfare; the Kanae Foundation for Life and Sociomedical Science; the Study Group of Molecular Cardiology; the Mochida Memorial Foundation for Medical and Pharmaceutical Research; the Tanabe Medical Frontier Conference; and the Cell Science Research Foundation.
Abbreviations
- ANP
atrial natriuretic peptide
- BNP
brain natriuretic peptide
- CNP
C-type natriuretic peptide
- GC-B
guanylyl cyclase-B
- Tg
transgenic
- Nppc, Npr2, Col2a1, Gapd
mouse genes for CNP, GC-B, pro-α1(II) collagen, and glyceraldehyde-3-phosphate dehydrogenase, respectively
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rosenzweig A, Seidman C E. Annu Rev Biochem. 1991;60:229–255. doi: 10.1146/annurev.bi.60.070191.001305. [DOI] [PubMed] [Google Scholar]
- 2.Chinkers M, Garbers D L. Annu Rev Biochem. 1991;60:553–575. doi: 10.1146/annurev.bi.60.070191.003005. [DOI] [PubMed] [Google Scholar]
- 3.Maack T, Suzuki M, Almeida F A, Nussenzveig D, Scarborough R M, McEnroe G A, Lewicki J A. Science. 1987;238:675–678. doi: 10.1126/science.2823385. [DOI] [PubMed] [Google Scholar]
- 4.Mukoyama M, Nakao K, Hosoda K, Suga S, Saito Y, Ogawa Y, Shirakami G, Jougasaki M, Obata K, Yasue H, et al. J Clin Invest. 1991;87:1402–1412. doi: 10.1172/JCI115146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dagnino L, Drouin J, Nemer M. Mol Endocrinol. 1991;5:1292–1300. doi: 10.1210/mend-5-9-1292. [DOI] [PubMed] [Google Scholar]
- 6.Tamura N, Ogawa Y, Yasoda A, Itoh H, Saito Y, Nakao K. J Mol Cell Cardiol. 1996;28:1811–185. doi: 10.1006/jmcc.1996.0170. [DOI] [PubMed] [Google Scholar]
- 7.Koller K J, Lowe D G, Bennett G L, Minamino N, Kangawa K, Matsuo H, Goeddel D V. Science. 1991;252:120–123. doi: 10.1126/science.1672777. [DOI] [PubMed] [Google Scholar]
- 8.Suga S, Nakao K, Hosoda K, Mukoyama M, Ogawa Y, Shirakami G, Arai H, Saito Y, Kambayashi Y, Inouye K, et al. Endocrinology. 1992;130:229–239. doi: 10.1210/endo.130.1.1309330. [DOI] [PubMed] [Google Scholar]
- 9.Komatsu Y, Nakao K, Suga S, Ogawa Y, Mukoyama M, Arai H, Shirakami G, Hosoda K, Nakagawa O, Hama N, et al. Endocrinology. 1991;129:1104–1106. doi: 10.1210/endo-129-2-1104. [DOI] [PubMed] [Google Scholar]
- 10.Suga S, Nakao K, Itoh H, Komatsu Y, Ogawa Y, Hama N, Imura H. J Clin Invest. 1992;90:1145–1149. doi: 10.1172/JCI115933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chrisman T D, Schulz S, Potter L R, Garbers D L. J Biol Chem. 1993;268:3698–3703. [PubMed] [Google Scholar]
- 12.Vollmar A M, Gerbes A L, Nemer M, Schulz R. Endocrinology. 1993;132:1872–1874. doi: 10.1210/endo.132.4.8462485. [DOI] [PubMed] [Google Scholar]
- 13.Ogawa Y, Itoh H, Tamura N, Suga S, Yoshimasa T, Uehira M, Matsuda S, Shiono S, Nishimoto H, Nakao K. J Clin Invest. 1994;93:1911–1921. doi: 10.1172/JCI117182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suda M, Ogawa Y, Tanaka K, Tamura N, Yasoda A, Takigawa T, Uehira M, Nishimoto H, Itoh H, Saito Y, et al. Proc Natl Acad Sci USA. 1998;95:2337–2342. doi: 10.1073/pnas.95.5.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsukawa N, Grzesik W J, Takahashi N, Pandey K N, Pang S, Yamauchi M, Smithies O. Proc Natl Acad Sci USA. 1999;96:7403–7408. doi: 10.1073/pnas.96.13.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaubert J, Jaubert F, Martin N, Washburn L L, Lee B K, Eicher E M, Guenet J L. Proc Natl Acad Sci USA. 1999;96:10278–10283. doi: 10.1073/pnas.96.18.10278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.John S W, Krege J H, Oliver P M, Hagaman J R, Hodgin J B, Pang S C, Flynn T G, Smithies O. Science. 1995;267:679–681. doi: 10.1126/science.7839143. [DOI] [PubMed] [Google Scholar]
- 18.Tamura N, Ogawa Y, Chusho H, Nakamura K, Nakao K, Suda M, Kasahara M, Hashimoto R, Katsuura G, Mukoyama M, et al. Proc Natl Acad Sci USA. 2000;97:4239–4244. doi: 10.1073/pnas.070371497. . (First Published March 28, 2000; 10.1073/pnas.070371497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou G, Garofalo S, Mukhopadhyay K, Lefebvre V, Smith C N, Eberspaecher H, de Crombrugghe B. J Cell Sci. 1995;108:3677–3684. doi: 10.1242/jcs.108.12.3677. [DOI] [PubMed] [Google Scholar]
- 20.Mori K, Ogawa Y, Ebihara K, Tamura N, Tashiro K, Kuwahara T, Mukoyama M, Sugawara A, Ozaki S, Tanaka I, et al. J Biol Chem. 1999;274:15701–15705. doi: 10.1074/jbc.274.22.15701. [DOI] [PubMed] [Google Scholar]
- 21.Yasoda A, Ogawa Y, Suda M, Tamura N, Mori K, Sakuma Y, Chusho H, Shiota K, Tanaka K, Nakao K. J Biol Chem. 1998;273:11695–11700. doi: 10.1074/jbc.273.19.11695. [DOI] [PubMed] [Google Scholar]
- 22.Howell D S, Dean D D. Disorders of Bone and Mineral Metabolism. New York: Raven; 1992. [Google Scholar]
- 23.Vortkamp A, Lee K, Lanske B, Segre G V, Kronenberg H M, Tabin C J. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 24.Mericq V, Uyeda J A, Barnes K M, De Luca F, Baron J. Pediatr Res. 2000;47:189–193. doi: 10.1203/00006450-200002000-00007. [DOI] [PubMed] [Google Scholar]
- 25.Pfeifer A, Aszodi A, Seidler U, Ruth P, Hofmann F, Fassler R. Science. 1996;274:2082–2086. doi: 10.1126/science.274.5295.2082. [DOI] [PubMed] [Google Scholar]
- 26.Oberklaid F, Danks D M, Jensen F, Stace L, Rosshandler S. J Med Genet. 1979;16:140–146. doi: 10.1136/jmg.16.2.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naski M C, Colvin J S, Coffin J D, Ornitz D M. Development (Cambridge, UK) 1998;125:4977–4988. doi: 10.1242/dev.125.24.4977. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Spatz M K, Kannan K, Hayk H, Avivi A, Gorivodsky M, Pines M, Yayon A, Lonai P, Givol D. Proc Natl Acad Sci USA. 1999;96:4455–4460. doi: 10.1073/pnas.96.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiang R, Thompson L M, Zhu Y Z, Church D M, Fielder T J, Bocian M, Winokur S T, Wasmuth J J. Cell. 1994;78:335–342. doi: 10.1016/0092-8674(94)90302-6. [DOI] [PubMed] [Google Scholar]
- 30.Rousseau F, Bonaventure J, Legeai-Mallet L, Pelet A, Rozet J M, Maroteaux P, Le Merrer M, Munnich A. Nature (London) 1994;371:252–254. doi: 10.1038/371252a0. [DOI] [PubMed] [Google Scholar]
- 31.Tavormina P L, Shiang R, Thompson L M, Zhu Y Z, Wilkin D J, Lachman R S, Wilcox W R, Rimoin D L, Cohn D H, Wasmuth J J. Nat Genet. 1995;9:321–328. doi: 10.1038/ng0395-321. [DOI] [PubMed] [Google Scholar]
- 32.Rousseau F, Saugier P, Le Merrer M, Munnich A, Delezoide A L, Maroteaux P, Bonaventure J, Narcy F, Sanak M. Nat Genet. 1995;10:11–12. doi: 10.1038/ng0595-11. [DOI] [PubMed] [Google Scholar]