Abstract
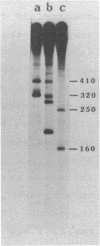
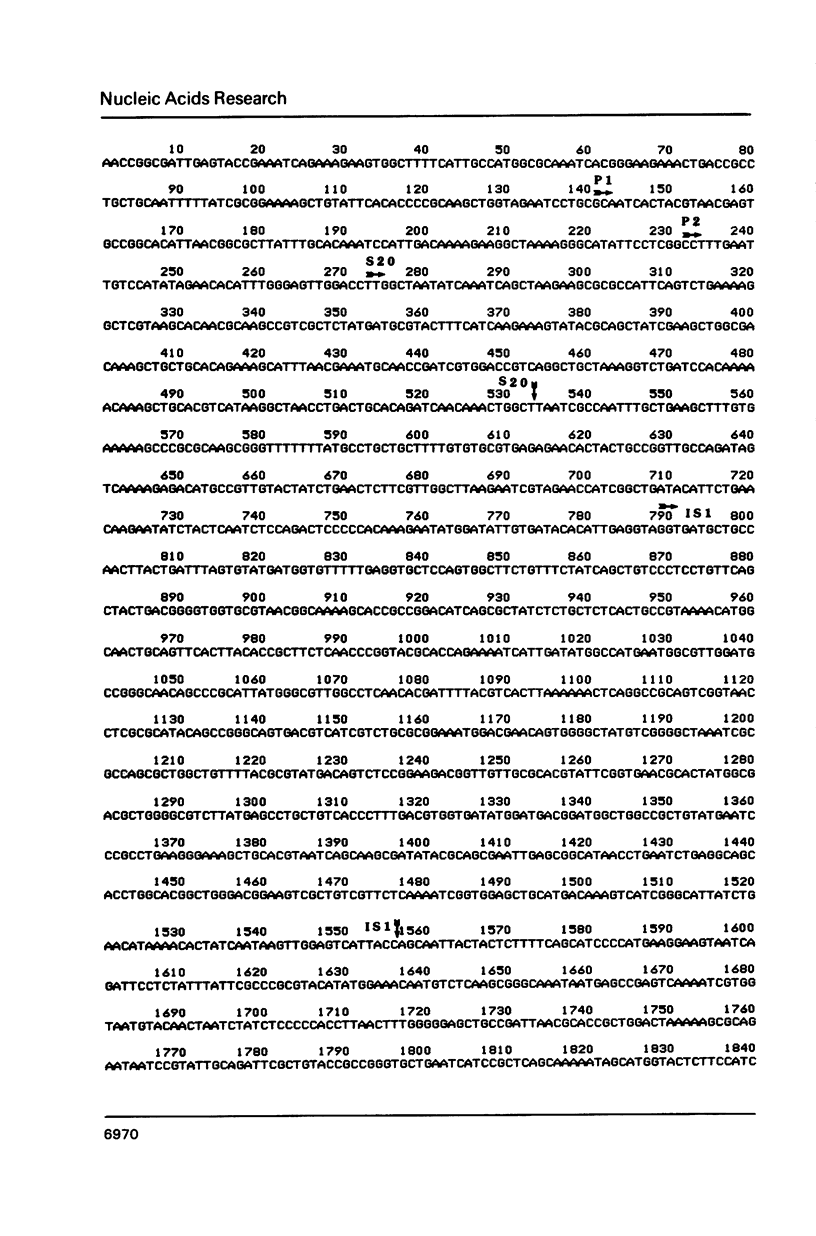
The sequence of nucleotides extending over 2.3 kb distal to the gene for ribosomal protein S20 of E. coli has been determined. Included in the sequence is an efficient rho-independent terminator 50 b.p. distal to the coding sequence for S20, a complete copy of IS1 which lacks, however, flanking direct repeats, and finally, an open reading frame capable of encoding a 28 kDa polypeptide of unknown function. Several lines of evidence suggest that the IS1 sequence described here must represent one of the copies resident in the bacterial chromosome rather than a newly transposed copy. Northern blotting experiments show that the gene for S20 is functionally monocistronic under all conditions tested in several genetic backgrounds. Thus it seems unlikely that the distal copy of IS1 plays any role in the termination or stability of mRNA transcribed from the gene for S20.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Apirion D., Watson N. Mapping and characterization of a mutation in Escherichia coli that reduces the level of ribonuclease III specific for double-stranded ribonucleic acid. J Bacteriol. 1975 Oct;124(1):317–324. doi: 10.1128/jb.124.1.317-324.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biel S. W., Adelt G., Berg D. E. Transcriptional control of IS1 transposition in Escherichia coli. J Mol Biol. 1984 Apr 5;174(2):251–264. doi: 10.1016/0022-2836(84)90337-1. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk G. R. E. coli ribosomal protein operons: the case of the misplaced genes. Cell. 1985 Aug;42(1):7–8. doi: 10.1016/s0092-8674(85)80093-3. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Burton Z., Burgess R. R., Lin J., Moore D., Holder S., Gross C. A. The nucleotide sequence of the cloned rpoD gene for the RNA polymerase sigma subunit from E coli K12. Nucleic Acids Res. 1981 Jun 25;9(12):2889–2903. doi: 10.1093/nar/9.12.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Johnsrud L., Miller J. H. DNA sequence at the integration sites of the insertion element IS1. Cell. 1978 Mar;13(3):411–418. doi: 10.1016/0092-8674(78)90315-x. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Christie G. E., Farnham P. J., Platt T. Synthetic sites for transcription termination and a functional comparison with tryptophan operon termination sites in vitro. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4180–4184. doi: 10.1073/pnas.78.7.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Court D., Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dennis P. P., Nomura M. Regulation of the expression of ribosomal protein genes in Escherichia coli. J Mol Biol. 1975 Sep 5;97(1):61–76. doi: 10.1016/s0022-2836(75)80022-2. [DOI] [PubMed] [Google Scholar]
- Farnham P. J., Greenblatt J., Platt T. Effects of NusA protein on transcription termination in the tryptophan operon of Escherichia coli. Cell. 1982 Jul;29(3):945–951. doi: 10.1016/0092-8674(82)90457-3. [DOI] [PubMed] [Google Scholar]
- Friesen J. D., Parker J., Watson R. J., Fiil N. P., Pedersen S. Isolation of transducing phage carrying rps T, the structural gene for ribosomal protein S20. Mol Gen Genet. 1976 Feb 27;144(1):115–118. doi: 10.1007/BF00277314. [DOI] [PubMed] [Google Scholar]
- Grindley N. D. IS1 insertion generates duplication of a nine base pair sequence at its target site. Cell. 1978 Mar;13(3):419–426. doi: 10.1016/0092-8674(78)90316-1. [DOI] [PubMed] [Google Scholar]
- Holmes W. M., Platt T., Rosenberg M. Termination of transcription in E. coli. Cell. 1983 Apr;32(4):1029–1032. doi: 10.1016/0092-8674(83)90287-8. [DOI] [PubMed] [Google Scholar]
- Hu M., Deonier R. C. Comparison of IS1, IS2 and IS3 copy number in Escherichia coli strains K-12, B and C. Gene. 1981 Dec;16(1-3):161–170. doi: 10.1016/0378-1119(81)90072-x. [DOI] [PubMed] [Google Scholar]
- Hu M., Deonier R. C. Mapping of IS1 elements flanking the argF gene region on the Escherichia coli K-12 chromosome. Mol Gen Genet. 1981;181(2):222–229. doi: 10.1007/BF00268430. [DOI] [PubMed] [Google Scholar]
- Johnsrud L. DNA sequence of the transposable element IS1. Mol Gen Genet. 1979 Jan 31;169(2):213–218. doi: 10.1007/BF00271673. [DOI] [PubMed] [Google Scholar]
- Kamio Y., Lin C. K., Regue M., Wu H. C. Characterization of the ileS-lsp operon in Escherichia coli. Identification of an open reading frame upstream of the ileS gene and potential promoter(s) for the ileS-lsp operon. J Biol Chem. 1985 May 10;260(9):5616–5620. [PubMed] [Google Scholar]
- Liebke H., Hatfull G. The sequence of the distal end of the E. coli ribosomal RNA rrnE operon indicates conserved features are shared by rrn operons. Nucleic Acids Res. 1985 Aug 12;13(15):5515–5525. doi: 10.1093/nar/13.15.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L., Zengel J. M. Expression of ribosomal genes in bacteria. Adv Genet. 1982;21:53–121. doi: 10.1016/s0065-2660(08)60297-7. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Mackie G. A. Cloning of fragments of lambda dapB2 DNA and identification of the dapB gene product. J Biol Chem. 1980 Sep 25;255(18):8928–8935. [PubMed] [Google Scholar]
- Mackie G. A. Nucleotide sequence of the gene for ribosomal protein S20 and its flanking regions. J Biol Chem. 1981 Aug 10;256(15):8177–8182. [PubMed] [Google Scholar]
- Mackie G. A., Parsons G. D. Identification of gene products from cloned fragments of the left arm of lambda dapB2. Can J Biochem. 1982 Mar;60(3):338–346. doi: 10.1139/o82-040. [DOI] [PubMed] [Google Scholar]
- Mackie G. A., Parsons G. D. Tandem promoters in the gene for ribosomal protein S20. J Biol Chem. 1983 Jun 25;258(12):7840–7846. [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Nomura M., Gourse R., Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Nyman K., Nakamura K., Ohtsubo H., Ohtsubo E. Distribution of the insertion sequence IS1 in gram-negative bacteria. Nature. 1981 Feb 12;289(5798):609–612. doi: 10.1038/289609a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci U S A. 1978 Feb;75(2):615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postle K., Good R. F. A bidirectional rho-independent transcription terminator between the E. coli tonB gene and an opposing gene. Cell. 1985 Jun;41(2):577–585. doi: 10.1016/s0092-8674(85)80030-1. [DOI] [PubMed] [Google Scholar]
- Read H. A., Das Sarma S., Jaskunas S. R. Fate of donor insertion sequence IS1 during transposition. Proc Natl Acad Sci U S A. 1980 May;77(5):2514–2518. doi: 10.1073/pnas.77.5.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regue M., Remenick J., Tokunaga M., Mackie G. A., Wu H. C. Mapping of the lipoprotein signal peptidase gene (lsp). J Bacteriol. 1984 May;158(2):632–635. doi: 10.1128/jb.158.2.632-635.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif H. J., Saedler H. IS1 is involved in deletion formation in the gal region of E. coli K12. Mol Gen Genet. 1975;137(1):17–28. doi: 10.1007/BF00332538. [DOI] [PubMed] [Google Scholar]
- Smiley B. L., Lupski J. R., Svec P. S., McMacken R., Godson G. N. Sequences of the Escherichia coli dnaG primase gene and regulation of its expression. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4550–4554. doi: 10.1073/pnas.79.15.4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M., Loranger J. M., Wu H. C. Isolation and characterization of an Escherichia coli clone overproducing prolipoprotein signal peptidase. J Biol Chem. 1983 Oct 25;258(20):12102–12105. [PubMed] [Google Scholar]
- Webster T., Tsai H., Kula M., Mackie G. A., Schimmel P. Specific sequence homology and three-dimensional structure of an aminoacyl transfer RNA synthetase. Science. 1984 Dec 14;226(4680):1315–1317. doi: 10.1126/science.6390679. [DOI] [PubMed] [Google Scholar]
- Yamagata H., Daishima K., Mizushima S. Cloning and expression of a gene coding for the prolipoprotein signal peptidase of Escherichia coli. FEBS Lett. 1983 Jul 25;158(2):301–304. doi: 10.1016/0014-5793(83)80600-0. [DOI] [PubMed] [Google Scholar]
- York M. K., Stodolsky M. Characterization of P1argF derivatives from Escherichia coli K12 transduction. I. IS1 elements flank the argF gene segment. Mol Gen Genet. 1981;181(2):230–240. doi: 10.1007/BF00268431. [DOI] [PubMed] [Google Scholar]