Abstract
The etiology of depression is still poorly understood, but two major causative hypotheses have been put forth: the monoamine deficiency and the stress hypotheses of depression. We evaluate these hypotheses using animal models of endogenous depression and chronic stress. The endogenously depressed rat and its control strain were developed by bidirectional selective breeding from the Wistar–Kyoto (WKY) rat, an accepted model of major depressive disorder (MDD). The WKY More Immobile (WMI) substrain shows high immobility/despair-like behavior in the forced swim test (FST), while the control substrain, WKY Less Immobile (WLI), shows no depressive behavior in the FST. Chronic stress responses were investigated by using Brown Norway, Fischer 344, Lewis and WKY, genetically and behaviorally distinct strains of rats. Animals were either not stressed (NS) or exposed to chronic restraint stress (CRS). Genome-wide microarray analyses identified differentially expressed genes in hippocampi and amygdalae of the endogenous depression and the chronic stress models. No significant difference was observed in the expression of monoaminergic transmission-related genes in either model. Furthermore, very few genes showed overlapping changes in the WMI vs WLI and CRS vs NS comparisons, strongly suggesting divergence between endogenous depressive behavior- and chronic stress-related molecular mechanisms. Taken together, these results posit that although chronic stress may induce depressive behavior, its molecular underpinnings differ from those of endogenous depression in animals and possibly in humans, suggesting the need for different treatments. The identification of novel endogenous depression-related and chronic stress response genes suggests that unexplored molecular mechanisms could be targeted for the development of novel therapeutic agents.
Keywords: animal models, depression, microarray, selective breeding, Wistar Kyoto rat
Introduction
Major depressive disorder (MDD) is the third most prevalent and costly disease worldwide, and it is projected to become number one by 2030 (WHO, GBD report, 2004). In addition to increasing the risk for suicide, MDD presents frequent comorbidity with other psychiatric disorders, and it has been identified as a risk factor for many illnesses, including obesity, cardiovascular, neurodegenerative and other diseases.1, 2, 3, 4 Understanding the pathophysiology of MDD, and providing better treatments, would improve mental health and quality of life of the largest psychiatric population in the world.5
There are two major hypotheses with regard to the etiology of MDD: the monoamine deficiency and stress hypotheses of depression. The monoamine deficiency hypothesis proposes that MDD can be etiologically explained by a deficiency in the monoamine neurotransmitters serotonin, norepinephrine and dopamine. Although this hypothesis is still prominent and is the basis of most antidepressant development to date, current opinion is that monoamine deficiency only partially explains MDD.6, 7, 8 On the basis of observations in MDD, the stress hypothesis relates pathological alterations in the stress-responsive hypothalamic-pituitary-adrenal (HPA) axis to causality of depression.9, 10, 11, 12, 13, 14, 15, 16, 17 Despite a large body of data supporting a role for stress in MDD, the nature and degree of its involvement remains uncertain.18 Knowing whether stress-induced depression shares any or all molecular mechanisms with that of endogenous depression may guide the development of different treatment alternatives.
Animal models that reproduce key symptoms of MDD offer a unique opportunity for experimental exploration unfeasible in human studies.13, 14, 17, 19, 20, 21, 22 Selective breeding has been used to generate animal models of depression with extreme traits.23, 24, 25, 26, 27, 28 We used this approach to create two substrains of the Wistar Kyoto (WKY) rat, an accepted model of MDD that replicates many behavioral and physiological endophenotypes present in major depression.29, 30, 31, 32 The two substrains of the WKY, identified as WKY More Immobile (WMI) and WKY Less Immobile (WLI), were generated by bidirectional selective breeding from the WKY based on depressive behavior in the forced swim test (FST) of the naïve, unstressed animals; therefore, WMIs represent an endogenous depression model.33 The chronic stress model employed four phylogenetically, physiologically and behaviorally different rat strains34, 35 to identify general, strain-independent molecular characteristics of the chronic stress response. We chose a chronic restraint stress (CRS) paradigm that has been shown to increase depression-like behaviors36, 37, 38, 39, 40, 41, 42 and reduce hippocampal volume.43, 44, 45
The hippocampus and amygdala were selected to investigate gene expression profiles, as these brain regions have been strongly implicated in the cause and consequences of both depression and chronic stress.46, 47, 48, 49, 50, 51 Our goal was to examine both the monoamine and stress hypotheses of depression, the former by identifying the contribution of genes implicated by the monoamine hypothesis to the two depression model expression profiles, and the latter by defining the overlap between the expression profiles of endogenous depression and chronic stress response. Our present results found no evidence for the involvement of monoamine neurotransmission-related genes in the expression profile of the endogenous depression model and found divergent expression profiles in endogenous depression and chronic stress models. However, novel target genes and pathways have emerged from this study, which have the potential to advance our knowledge with regard to the etiology of depression and its treatment.
Methods
Animal models
All procedures were approved by the Institutional Animal Care and Use Committee of Northwestern University. The WMI-WLI selective breeding commenced as described previously.33 Animals showing the most extreme FST behavior within each line were selected for breeding, specifically avoiding sibling mating until the G5 generation. Adult WMI and WLI male animals from the 13th generation of selective breeding were employed in this study. The experimental design included daily administration of desipramine (10 mg kg−1) or saline, subcutaneously for 14 days, to both WMI and WLI animals (n=9 per treatment per strain). Animals were killed by fast decapitation on the 15th day immediately after removal from the home cage.
For the CRS experiment, adult male Fischer 344 (F344), Brown-Norway (BN), Lewis (LEW) and WKY rats were obtained from Harlan Laboratories (Indianapolis, IN, USA) (n=9 per strain per treatment). Rats were either exposed to CRS in a breathable decapicone for 2 h per day for a 2-week period or remained in their home cage (no stress, NS). Body weight was monitored throughout the experiment. On the 15th day, both groups of rats were tested in the elevated plus maze test in parallel and killed by decapitation immediately following the 5-min test. Animals were ∼100 days old at the time of sample collections. Blood samples were collected for the determination of plasma corticosterone (CORT) levels and adrenal weight was also determined. The elevated plus maze test of anxiety was used as a confirmation of strain differences in anxiety measures basally (NS group) and after CRS: the results are not shown since the purpose of this study is to identify chronic stress-induced changes in gene expression independently of strain effects.
Behavioral testing
FST was performed as described previously.30, 52 Briefly, on day 1, rats were individually placed in the water tank (water temperature 22–24 °C) for 15 min. After 24 h, rats were once again placed in the tank for a 5-min test session. The test session was videotaped and scored by a trained observer, using the scoring system developed by Detke et al.53 The open-field test was performed as described previously,54 but the animals' movements were followed and analyzed by the TSE Videomot 2 version 5.75 software.
Radioimmunoassay for plasma CORT
Assays were carried out in duplicate, as described previously,29 using the CORT RIA kit (MP Biomedicals, Solon, OH, USA). The assay sensitivity was 2–4 pg per tube. The intra- and interassay coefficients of variation were 3.5 and 8%, respectively.
Microarray experiments
Brain regions were dissected immediately after decapitation, as described previously,55 and stored at −80 °C in RNAlater (Ambion, Austin, TX, USA). Individual hippocampi and amygdalae were homogenized in TRIzol (Invitrogen, Carlsbad, CA, USA), and RNA was isolated following the manufacturer's protocol. All RNA samples were treated with DNase1 (Qiagen, Valencia, CA, USA) according to the manufacturer's methods.
Total RNA isolated from WMI and WLI brain regions was reverse-transcribed, and-double stranded cDNA was synthesized with the GeneChip® Expression 3-Amplification One-cycle kit (Affymetrix, Santa Clara, CA, USA. In an in vitro reaction with T7 RNA polymerase, the cDNA was linearly amplified and labeled with biotinylated nucleotides (Affymetrix). Ten micrograms of biotin-labeled and fragmented cRNA was then hybridized onto Rat Genome 230 2.0 GeneChip arrays (Affymetrix).
Total RNA from the chronic stress experiment was reverse transcribed, followed by second-strand cDNA synthesis. For each sample, an in vitro transcription reaction was carried out incorporating biotinylated nucleotides according to the manufacturer's protocol for Illumina® Totalprep RNA amplification kit (Ambion). Biotin-labeled cRNA, 1.5 μg, was then hybridized onto RatRef-12 Expression BeadChips (Illumina, San Diego, CA, USA).
Statistical analysis for WMI-WLI experiment
Probe intensity data from Rat 230v2 Affymetrix GeneChip arrays were read into the R software environment (http://www.R-project.org) directly from CEL files using the R/affy package.56 Affymetrix data was normalized using the robust multiarray average method for probe set data.57 Data quality was assessed using histograms of signal intensities, scatter plots and hierarchical clustering of samples.
Analysis of variance (ANOVA) methods, performed with the R/maanova package,58, 59 were used to statistically resolve gene expression differences. The experimental design included chronic administration of desipramine or saline to both WMI and WLI animals, of which three animals per strain per treatment were randomly selected for the microarray analyses. As we were only interested in the strain effect in this experiment, treatment (desipramine or saline) was a covariate. For each probe set g, strain i, covariate j and replicate k, a linear model for the log-transformed expression measure, yijkg, can be formulated as a sum of components that contribute to the overall intensity value:
where μg is the mean intensity over all 12 samples, αig is the effect of strain i (i=1, 2), βjg is the additive effect of covariate j (j=1, 2), (αβ)ijg is the interaction effect of strain i, covariate j and probe set g and ɛijkg is the residual error for strain i, covariate j, replicate k and probe set g, respectively. To identify transcripts with a high probability of being differentially expressed between strains, statistical tests for the null hypothesis H0: αig=0 vs the alternative H0: αig≠0 were performed. We excluded transcripts in which the strain effect was not consistent between different covariate levels, as identified by a statistical test for the null hypothesis H0: (αβ)ijg=0 vs the alternative H0: (αβ)ijg≠0. Both these tests were performed using Fs, a modified F statistic incorporating shrinkage estimates of residual variance.60 P-values were calculated by permuting model residuals 1000 times. Unless otherwise noted, transcripts with differences between strains were identified as those with P-values, or estimated false-positive rates, less than 0.01 for the hypothesis H0: αig=0 and greater than 0.01 for the hypothesis H0: (αβ)ijg=0. As an adjustment for multiple testing, we used the q-value transformation of the P-values to estimate false discovery rate.61 To relate probe sets to genes, probe set IDs were mapped to symbols using NetAffx (http://www.affymetrix.com/analysis/netaffx/). When reporting fold changes, we consider WLI to be the reference strain. If the measured expression level is higher in the WMI strain than in the WLI strain, the fold change is positive, and if it is lower in WMI than in WLI, the fold change is negative.
For each experiment, we also applied models of combined data from both tissues to measure the differences in transcript abundance between tissue. We used the model
which is equivalent to model (1), except that there is an additional variable (γkg) for tissue with indices k=1,2 for the amygdala and hippocampus. Because we applied the normalization separately by tissue, we mean centered the normalized samples before applying this model.
Statistical analysis for stress experiment
Probe intensity data from Illumina Chip arrays were read directly into the R software environment from bead summary files produced by BeadStudio using the R/beadarray package.62 Quantile normalization was applied to the Illumina bead summary data using the R/preprocessCore package.63 Data quality was assessed as described for the WMI-WLI experiment.
Analysis of variance methods were similarly applied for this experiment. Twelve rats were exposed to chronic stress conditions, and 12 rats were not exposed to chronic stress. The animals for each stress condition consisted of three rats each from four different strains (BN-SS, F344, LEW and WKY). The linear model of equation (1) was applied for each probe set g. In this case, αig, is the effect of condition i (i=1, 2), βjg is the additive effect of strain j (j=1, 2, 3, 4), which is a covariate. To identify transcripts exhibiting consistent differential expression owing to condition across strains, we performed and applied statistical tests for the null hypotheses H0: αig=0 vs H0: αig≠0 and H0: (αβ)ijg=0 vs H0: (αβ)ijg≠0 that are analogous to those described above for the WLI-WMI comparison. Probe set IDs were mapped to symbols using Illumina's annotation resource (http://www.illumina.com/support/annotation_files.ilmn). When reporting fold changes, we consider the NS to be the reference condition. If the measured expression in the CRS condition is greater than in the NS condition, the fold change is positive. If it is less under CRS than under NS, the fold change is negative. To facilitate across-experiment comparison by gene, probes were mapped to gene symbols using current probe annotation files provided by the array manufacturers. The suffixes ‘_predicted' and ‘_mapped' were truncated from the Illumina symbol assignments. Where there were multiple probes for the same symbol, probes with the largest F statistic for the relevant hypothesis test were chosen to give a one-to-one mapping across experiments for 10 112 genes. The linear model of equation (2) was applied analogously for this experiment to obtain the distribution of estimated tissue effects.
Real-time RT-PCR
Real-time reverse transcription-polymerase chain reaction (RT-PCR) was used to confirm microarray results for a subset of randomly selected genes from those fulfilling the criteria of significant (P<0.01) expression differences. Reverse transcription for 2 μg of each sample was performed with Invitrogen's Superscript® III First-Strand kit (18080-051) according to the manufacturer's protocol. Primers were designed to amplify 80–150 bp regions and to contain a maximum amount of microarray probe sequence using default settings of ABI's Primer Express software (version 3.0, PE Applied Biosystems, Carlsbad, CA, USA). Primer sequences are listed in Supplementary Table S1. Forty nanograms of cDNA was amplified in 20 μl reactions (1 × SYBR green reaction mix (ABI, Carlsbad, CA, USA), 250 μ primers) in the ABI 7900HT PCR machine using the relative quantification (–ddCt) method, with 18s RNA as the internal control.
Results
Selective breeding and characterization of WMI and WLI strains
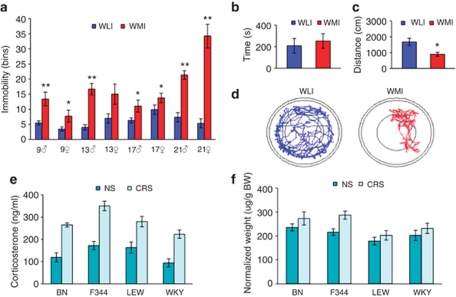
The WMI and WLI rat strains consistently maintained significant, dichotomous FST phenotypes throughout 21 generations of selective breeding (Figure 1a), with WMIs always exhibiting greater immobility scores than WLIs. In the open-field test of exploration/anxiety, WMI and WLI male animals showed similar level of exploration of the inner circle (Figure 1b), suggesting no differences in anxiety-related behaviors. In contrast, WMIs explored the arena significantly less than WLIs (Figure 1c), and the activity traces (Figure 1d) illustrate freezing-like behavior exhibited by the WMIs, a behavioral pattern very similarly to psychomotor retardation. There were no differences in the two strains' basal plasma thyroxine and CORT levels or their CORT responses to acute restraint stress (Supplementary Table S2). Furthermore, no additional behavioral differences were observed between the two strains, including anxiety-like behaviors as assessed by the elevated plus maze and defensive burying tests, confirming the results of the open-field test, and spatial learning and memory as measured in the Morris water maze (Supplementary Table S2). These data verify that the behavioral differences between the two substrains of WKYs are not fear or anxiety driven, but rather related to depressive state.
Figure 1.
The endogenous depression model, the Wistar–Kyoto More Immobile (WMI) strain, shows depressive behavior not linked to fear/anxiety (a–d). Chronic restraint stress (CRS) increases adrenocortical function consistently in all four strains (e and f). (a) In the forced swim test (FST), immobility scores of the WMI and WKY Less Immobile (WLI) animals differ significantly across generations. (b) Time spent in the inner circle of the open-field test, (c) total distance traveled and (d) movement traces of representative WMI and WLI animals. (e) Plasma corticosterone levels were consistently elevated after CRS in all four strains and (f) adrenal weights were consistently greater in the CRS group of all strains. *P<0.01, **P<0.001.
CRS decreased body weight gain, increased plasma CORT levels and adrenal weight
Genetic polymorphisms between the strains range from 25.9% between F344 and LEW to 66% between BN and WKY, representing a substantial interstrain variation. However, CRS affected all strain of rats. Specifically, CRS resulted in a lower body weight gain compared with NS rats (F[1,65]=212.9, P<0.001), but there was no weight loss in response to the CRS paradigm (body weight on day 1: 239±5 g; on day 14 of CRS: 244±5 g). Plasma CORT levels were elevated in all rats exposed to the chronic stress procedure compared with those not subjected to stress (Figure 1e; strain: (F[1,65]=105.37, P<0.001) and those subjected to stress (F[3,65]=6.75, P<0.001; strain, stress interaction NS). As expected, chronic stress increased adrenal weight in all four rat strains (Figure 1f; strain: F[3,41]=1.34, NS; stress: F[1,41]=4.77, P=0.035). Taken together, these data established that all CRS rats were undergoing expected physiological changes associated with CRS exposure.
Differential gene expression profile in the amygdala and hippocampus of WMI and WLI male rats
Six hundred and thirty-eight genes in the amygdala and 463 in the hippocampus were differentially expressed between the WMI and WLI animals (P<0.01). Genes that additionally had fold changes above 1.4 (40% increase or decrease) are listed in Table 1. The complete data set is shown in Supplementary Tables S3 (amygdala) and S4 (hippocampus). Twenty-seven genes were differentially expressed in both brain regions between WMI and WLI in the same direction.
Table 1. Differentially expressed depression genes (top) and stress genes (bottom) in the amygdala and hippocampus.
| Probe set | Gene name | Rat/human symbol | Cytoband | Fold change | P-value |
|---|---|---|---|---|---|
| Amygdala | |||||
| 1370215_at | Complement component 1, q subcomponent, beta polypeptide | C1qb/C1QB | 5q36 | −1.60 | 9.96E−03 |
| 1376198_at | Adipocyte-specific adhesion molecule | Asam/ASAM | 8q22 | −1.60 | 9.55E−06 |
| 1380497_at | Ash1 (absent, small or homeotic)-like (Drosophila) | Ash1l/ASH1L | 2q34 | 1.58 | 1.84E−04 |
| 1390835_at | Solute carrier family 47, member 1 | Slc47a1/SLC47A1 | 10q22–q23 | −1.55 | 5.16E−04 |
| 1387029_at | Complement factor H | Cfh/CFH | 13q13 | −1.54 | 2.26E−03 |
| 1378518_at | Ewing sarcoma breakpoint region 1 | Ewsr1/EWSR1 | 14q21 | 1.54 | 8.50E−05 |
| 1394940_at | Family with sequence similarity 46, member A | Fam46a/FAM46A | 8q31 | −1.53 | 3.78E−03 |
| 1398522_at | Ankyrin repeat and LEM domain containing 2 | Ankle2/ANKLE2 | 12q16 | 1.50 | 8.22E−06 |
| 1379917_at | Adaptor-related protein complex 3, sigma 1 subunit | Ap3s1 /AP3S1 | 18q11 | −1.49 | 3.11E−03 |
| 1379264_at | Zinc- and ring finger 1 | Znrf1/ZNRF1 | 4q24 | −1.49 | 7.88E−04 |
| 1376175_at | Glioblastoma amplified sequence | Gbas/GBAS | 12q13 | 1.48 | 1.47E−03 |
| 1394964_at | BAT2 domain containing 1 | Bat2d1/BAT2D1 | 13q22 | 1.48 | 8.48E−06 |
| 1370989_at | Ret proto-oncogene | Ret/RET | 4q42 | −1.47 | 1.15E−03 |
| 1367749_at | Lumican | Lum/LUM | 7q13 | −1.46 | 5.74E−03 |
| 1394814_at | Translocated promoter region | Tpr/TPR | 13q21 | 1.46 | 2.37E−04 |
| 1383997_at | Mitogen-activated protein kinase 1 | Mapk1/MAPK1 | 11q23 | −1.45 | 3.46E−04 |
| 1391524_at | Similar to WD repeat domain 11 protein | RGD1564964/PHIP | 8q31 | 1.44 | 5.17E−05 |
| 1369742_at | Hect (homologous to the E6-AP (UBE3A) carboxyl-terminus) domain and RCC1 (CHC1)-like domain (RLD) 1 | Herc1/HERC1 | 8q24 | 1.44 | 4.21E−04 |
| 1385101_a_at | Coiled-coil domain containing 127 | Ccdc127/CCDC127 | 1p11 | 1.43 | 9.94E−05 |
| 1391788_at | Sin3-associated polypeptide, 18 kDa | Sap18/SAP18 | 15p12 | 1.43 | 6.33E−04 |
| 1378520_at | B-cell CLL/lymphoma 11B (zinc-finger protein) | Bcl11b/BCL11B | 6q32 | 1.42 | 2.68E−04 |
| 1390112_at | EGF-containing fibulin-like extracellular matrix protein 1 | Efemp1/EFEMP1 | 14q22 | −1.42 | 2.45E−04 |
| 1384000_at | SRY (sex determining region Y)-box 4 | Sox4/SOX4 | 17p12 | 1.42 | 6.48E−03 |
| 1370891_at | Cd48 molecule | Cd48/CD48 | 13q24 | −1.41 | 8.88E−03 |
| 1381829_at | Zinc-finger protein 318 | Zfp318/ZNF318 | 9q12 | 1.41 | 3.40E−03 |
| 1370122_at | RAB27B, member RAS oncogene family | Rab27b/RAB27B | 18q12.1 | 1.40 | 6.89E−04 |
| 1380569_at | Ring finger protein 41 | Rnf41/RNF41 | 7q11 | −1.40 | 7.40E−05 |
| Hippocampus | |||||
| 1392948_at | Chloride intracellular channel 6 | Clic6/CLIC6 | 11q11 | −3.35 | 4.25E−05 |
| 1379281_at | Sclerostin domain containing 1 | Sostdc1/SOSTDC1 | 6q16 | −3.30 | 2.20E−05 |
| 1374320_at | Coagulation factor V (proaccelerin, labile factor) | F5/F5 | 13q22 | −3.12 | 1.04E−04 |
| 1378365_at | Solute carrier family 4, sodium bicarbonate cotransporter, member 5 | Slc4a5/SLC4A5 | 4q34 | −2.83 | 1.39E−04 |
| 1370384_a_at | Prolactin receptor | Prlr/PRLR | 2q16 | −2.61 | 2.09E−04 |
| 1367598_at | Transthyretin | Ttr/TTR | 18p | −2.60 | 5.93E−03 |
| 1377434_at | Membrane frizzled-related protein | Mfrp/MFRP | 8q22 | −2.55 | 2.20E−04 |
| 1375465_at | Orthodenticle homolog 2 (Drosophila) | Otx2/OTX2 | 15p14 | −2.52 | 4.78E−05 |
| 1376944_at | Prolactin receptor | Prlr/PRLR | 2q16 | −2.25 | 4.54E−04 |
| 1382083_at | Coagulation factor C homolog, cochlin (Limulus polyphemus) | Coch/COCH | 6q22 | −1.80 | 8.12E−06 |
| 1368606_at | Solute carrier organic anion transporter family, member 1a5 | Slco1a5/SLCO1A2 | 4q44 | −1.79 | 1.01E−03 |
| 1372299_at | Cyclin-dependent kinase inhibitor 1C (P57) | Cdkn1c/CDKN1C | 1q42 | −1.77 | 2.67E−05 |
| 1367700_at | Fibromodulin | Fmod/FMOD | 13q13 | −1.70 | 1.16E−05 |
| 1371849_at | 5′-Nucleotidase domain containing 2 | Nt5dc2/NT5DC2 | 16p16 | −1.63 | 2.72E−04 |
| 1369625_at | Aquaporin 1 | Aqp1/AQP1 | 4q24 | −1.61 | 1.29E−03 |
| 1367682_at | Midkine | Mdk/MDK | 3q24 | −1.61 | 5.17E−04 |
| 1368536_at | Ectonucleotide pyrophosphatase/phosphodiesterase 2 | Enpp2/ENPP2 | 7q31 | −1.60 | 3.55E−04 |
| 1391211_at | Atpase, class VI, type 11C | Atp11c/ATP11C | Xq36 | −1.58 | 1.36E−03 |
| 1376728_at | Hypothetical RNA binding protein RGD1359713 | RGD1359713/RBM47 | 14p11 | −1.55 | 1.24E−03 |
| 1375026_at | Calmodulin-like 4 | Calml4/CALML4 | 8q24 | −1.54 | 1.40E−03 |
| 1387791_at | Angiotensin I-converting enzyme (peptidyl-dipeptidase A) 1 | Ace/ACE | 10q32.1 | −1.49 | 5.35E−03 |
| 1368046_at | Solute carrier family 31 (copper transporters), member 1 | Slc31a1/SLC31A1 | 5q24 | −1.49 | 3.36E−03 |
| 1370068_at | Phospholipase A2, group V | Pla2g5/PLA2G5 | 5q36 | −1.48 | 3.94E−03 |
| 1376285_at | GULP, engulfment adaptor PTB domain containing 1 | Gulp1/GULP1 | 9q22 | −1.48 | 6.20E−03 |
| 1386770_x_at | Potassium voltage-gated channel, Isk-related subfamily, gene 2 | Kcne2/KCNE2 | 11q11 | −1.48 | 1.69E−03 |
| 1374139_at | Cerebellar degeneration-related 2 | Cdr2/CDR2 | 1q35-q36 | −1.46 | 5.40E−04 |
| 1368202_a_at | Disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | Dab2/DAB2 | 2q16 | −1.44 | 2.68E−05 |
| 1369798_at | ATPase, Na+/K+ transporting, beta 2 polypeptide | Atp1b2/ATP1B2 | 10q24 | −1.42 | 5.24E−03 |
| Amygdala | |||||
| 4610273 | Hemoglobin, beta | MGC72973/HBB | 1q32 | −1.61 | 1.91E−03 |
| 3140088 | Ribosomal protein L30 | Rpl30/RPL30 | 7q22 | −1.58 | 2.07E−03 |
| 6650278 | Hemoglobin alpha, adult chain 2 | Hba-a2/HBA1 | 10q12 | −1.57 | 1.93E−06 |
| 4210725 | Transthyretin (prealbumin, amyloidosis type I) | Ttr/TTR | 18p12 | −1.55 | 3.47E−03 |
| 2510524 | Hemoglobin, beta | Hbb/HBB | 1q32 | −1.53 | 1.93E−-06 |
| Hippocampus | |||||
| 6650278 | Hemoglobin alpha, adult chain 2 | Hba-a2/HBA1 | 10q12 | −1.80 | 2.90E−06 |
| 4780059 | Insulin-like growth factor 2 (somatomedin A) | Igf2/IGF2 | 1q41 | −1.79 | 7.16E−05 |
| 2510524 | Hemoglobin, beta | Hbb/HBB | 1q32 | −1.75 | 4.84E−06 |
| 4610273 | Hemoglobin, beta | MGC72973/HBB | 1q32 | −1.64 | 9.67E−05 |
| 3840156 | Collagen, type I, alpha 2 | Col1a2/COL1A2 | 4q13 | −1.46 | 5.32E−03 |
Gene expression changes in response to chronic stress in the amygdala and hippocampus of four strains of male rats
To eliminate strain-specific effects in our selection attributable to differences in stress reactivity or potential cognitive differences between strains, genes were selected based on consistent, significant expression level differences in all four strains. We found 125 genes in the amygdala and 126 genes in the hippocampus that were differentially expressed between the CRS and NS conditions (P<0.01) (Supplementary Tables S5 and S6). From these, the 10 amygdalar and hippocampal genes with absolute fold changes above 1.4 are shown in Table 1. Twelve of the genes showed the same directional changes in both brain regions.
Minimal overlap between depression and stress response
Both the WMI-WLI and CRS-NS gene sets were examined for brain region-specific overlap using all known aliases to account for possible differences between microarray platform target nomenclatures. Only two genes in the amygdala (Kcnj14 and Mprl9) and four in the hippocampus (Chi3l1, collagen type I alpha 1 (Col1a1), matrix metallopeptidase 14 (Mmp14) and RGD1305680) showed differential expression in both the chronic stress response and endogenous depression comparisons. On the basis of the stress hypothesis of depression, transcripts with lowered expression levels in WMI animals should show similarly decreased expression in CRS animals, and increased expression in WMIs should correspond to increased expression in CRS. When we examined the directionality of fold changes for the six overlapping genes, only three genes (Col1a1, Mmp14 and RGD1305680), all in hippocampal tissue, showed these hypothesized directional changes. This provides suggestive evidence that the molecular signature of chronic stress and endogenous depressive behavior in these brain regions are not similar. However, since comparison of overlap between small sets of selected genes can be sensitive to thresholding, we also compared the differences between model effects for CRS and NS with the differences for WMI and WLI.
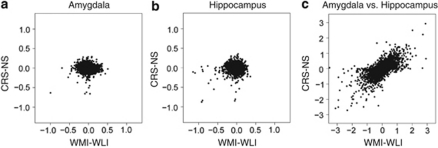
Expression differences in both models were relatively small (Figures 2a and b). If there is an underlying relationship between expression differences in response to CRS and endogenous depressive behavior for many genes, we should see significant positive correlation between the differences in gene expression across the two experiments. In particular, the differences between WMI and WLI model effects and differences between CRS and NS model effects ((α2g−α1g) in equation (1)) should show positive correlation. The Spearman's correlation (ρ) for these comparisons of differences were not significantly different than zero in either tissue (ρ=−0.12, amygdala; ρ=−0.02, hippocampus; Figures 2a and b). This lack of observed correlation could be explained by the different platforms or by the chronologies of the experiments. To address this concern, equation (2), which combined data of both brain regions and estimates the tissue effects, was employed. The tissue (interbrain region) effects showed significant positive correlation between experiments (ρ=0.48, P<0.001; Figure 2c). We also measured correlation for the subset of genes for which the magnitude of tissue effects were comparable to the CRS-NS and WMI-WLI effects, specifically those having a magnitude below 0.4 on both platforms. Within this subset, we still found significant positive correlation (ρ=0.31, P<0.001). We conclude that we should have observed correlation between the CRS-NS and WMI-WLI effects if it were present despite the platform differences. This suggests that, on a global scale, the effects of chronic stress and endogenous depression on the amygdalar and hippocampal transcriptome are dissimilar.
Figure 2.
An across-experiment, gene-to-gene mapping was created for 10 112 gene symbols. Scatter plots are shown for observed effects obtained from the chronic restraint stress-no stress (CRS-NS) microarray experiment (vertical axis) and the Wistar–Kyoto More Immobile-WKY Less Immobile (WMI-WLI) microarray experiment (horizontal axis) by applying the linear models of Equation (1) and (2). The statistics are differences in condition effects, α2g−α1g, for the amygdala (a) and hippocampus (b), and differences in brain region-related effects, γ2g−γ1g (c).
Confirmation of the expression differences by real-time RT-PCR
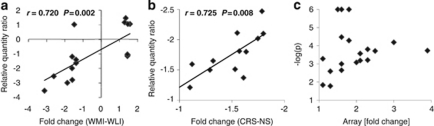
We carried out quantitative RT-PCR confirmation of the microarray data using total RNA samples from both experiments and primer pairs described in Supplementary Table S1. Pearson's correlation of fold change from the Affymetrix microarray for 16 genes with relative quantification ratios from qPCR (RQ ratio) showed significant concurrence of data (r=0.720, P=0.002; Figure 3a). Pearson's correlation of fold change from the Illumina microarray for nine genes (three were confirmed in both brain regions) with relative quantification ratios from qPCR showed similarly significant correlation (r=0.725, P=0.008; Figure 3b). Positive and negative fold changes are defined in Methods.
Figure 3.
(a) Validation of genes differentially expressed in Wistar–Kyoto More Immobile (WMI) and WKY Less Immobile (WLI) hippocampi or amygdala (n=6 per strain) by real-time reverse transcription-polymerase chain reaction (RT-PCR). The correlation between fold change in the Affymetrix microarray experiment and real-time RT-PCR determination of relative quantification ratios are shown (Pearson's correlation, r=0.720, P=0.002). (b) Validation of genes differentially expressed in chronic restraint stress (CRS) vs no stress (NS) (n=12 per treatment) hippocampi or amygdala by real-time RT-PCR. The correlation between fold change in the Illumina array experiment and real-time RT-PCR determination of relative quantification ratios are shown (Pearson's correlation, r=0.725, P=0.008). (c) Lack of correlation between absolute fold change and significant P-value (–log P) from the microarray analyses for genes with expression changes validated by real-time RT-PCR.
When we examined the validated transcript fold change in the microarrays in relation to their respective P-value, we found no significant correlation (r=0.113, P>0.05; Figure 3c). These data confirm that significance and fold change criteria cannot be used interchangeably, but that expression changes can be confirmed by real-time RT-PCR, even if the fold change is small.
The molecular pathways of depression and stress response genes
The lists of hippocampal and amygdalar genes with significant expression differences were combined into single gene lists for each of our two models, and the rat gene symbols were converted to their human orthologs to access a larger body of literature. These master ortholog lists were subjected to functional annotation using the PANTHER Classification System.64 In addition, we used the PANTHER binomial statistics tool to compare our annotated ortholog lists to the NCBI Homo sapiens reference gene list to determine significant over- or under-representation of molecular function, biological process or pathway classification terms.65
Table 2 shows significantly over- and under-represented terms from both gene sets. A group of genes identified to belong to the integrin signaling pathway (including Araf, Arf1, Arf4, Arhgap10, Col12a1, Col15a1, Col1a1, Col4a1, Col5a2, Col6a6, Crkl, Elmo1, Fnlb, Grb2, Itga11, Itga6, Itgae, Itgb6, Itgb8, Itgbl1, Mapk1, Pik3r1, Pitk2b, Rac1, Rhoa and Tln2) were most overly represented in the WMI-WLI comparison. From the CRS-NS comparison, the Huntington disease pathway showed the only significantly clustered group of genes (Capn9, Gapdh, Rhog, Tnfaip8, Tnfaip8l3 and tumor suppressor protein 53).
Table 2. Over- and under-represented gene ontology (GO) groups determined by the Panther software analysis.
| NCBI (19 911)a | STR (206)b | EXP | +/− | P-value | DEP (1065)c | EXP | +/− | P-value | |
|---|---|---|---|---|---|---|---|---|---|
| Pathway | |||||||||
| Huntington disease | 167 | 6 | 1.73 | + | 8.30E−03 | 10 | 9 | + | 4.04E−01 |
| Integrin signaling pathway | 189 | 3 | 1.96 | + | 3.11E−01 | 26 | 10 | + | 1.88E−05 |
| Ras pathway | 79 | 0 | 0.82 | − | 4.41E−01 | 14 | 4 | + | 1.28E−04 |
| Heterotrimeric G-protein signaling pathway– Gq alpha- and Go alpha-mediated pathway | 134 | 0 | 1.39 | − | 2.49E−01 | 16 | 7 | + | 2.91E−03 |
| Angiogenesis | 191 | 0 | 1.98 | − | 1.37E−01 | 19 | 10 | + | 8.55E−03 |
| Axon guidance mediated by semaphorins | 43 | 1 | 0.44 | + | 3.59E−01 | 7 | 2 | + | 9.28E−03 |
| Biological process | |||||||||
| Primary metabolic process | 7959 | 95 | 82.34 | + | 4.26E−02 | 533 | 426 | + | 1.88E−11 |
| Metabolic process | 8276 | 101 | 85.62 | + | 1.82E−02 | 548 | 443 | + | 4.98E−11 |
| Cellular process | 6309 | 83 | 65.27 | + | 5.60E−03 | 433 | 337 | + | 4.35E−10 |
| Cell communication | 4408 | 65 | 45.61 | + | 1.13E−03 | 313 | 236 | + | 2.13E−08 |
| Protein metabolic process | 3247 | 41 | 33.59 | + | 9.86E−02 | 240 | 174 | + | 8.45E−08 |
| Signal transduction | 4234 | 61 | 43.81 | + | 3.02E−03 | 296 | 226 | + | 2.73E−07 |
| Transport | 2857 | 42 | 29.56 | + | 1.13E−02 | 213 | 153 | + | 3.10E−07 |
| Neurological system process | 1995 | 29 | 20.64 | + | 3.89E−02 | 152 | 107 | + | 7.07E−06 |
| Vesicle-mediated transport | 1160 | 13 | 12 | + | 4.24E−01 | 98 | 62 | + | 7.86E−06 |
| Intracellular signaling cascade | 1609 | 17 | 16.65 | + | 5.01E−01 | 127 | 86 | + | 9.00E−06 |
| System process | 2264 | 33 | 23.42 | + | 2.73E−02 | 167 | 121 | + | 1.41E−05 |
| Endocytosis | 575 | 8 | 5.95 | + | 2.47E−01 | 56 | 31 | + | 2.03E−05 |
| Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 3827 | 39 | 39.59 | − | 5.01E−01 | 257 | 205 | + | 4.43E−05 |
| Synaptic transmission | 635 | 10 | 6.57 | + | 1.25E−01 | 59 | 34 | + | 4.53E−05 |
| Protein transport | 1646 | 18 | 17.03 | + | 4.39E−01 | 124 | 88 | + | 8.73E−05 |
| Intracellular protein transport | 1646 | 18 | 17.03 | + | 4.39E−01 | 124 | 88 | + | 8.73E−05 |
| Cellular amino-acid and derivative metabolic process | 367 | 3 | 3.8 | − | 4.73E−01 | 38 | 20 | + | 1.29E−04 |
| Amino-acid transport | 77 | 1 | 0.8 | + | 5.50E−01 | 13 | 4 | + | 3.47E−04 |
| Carbohydrate metabolic process | 952 | 17 | 9.85 | + | 2.10E−02 | 76 | 51 | + | 4.44E−04 |
| Cell–cell signaling | 1374 | 22 | 14.22 | + | 2.82E−02 | 102 | 73 | + | 6.19E−04 |
| Developmental process | 3052 | 40 | 31.58 | + | 6.60E−02 | 202 | 163 | + | 7.65E−04 |
| Cellular component organization | 1492 | 18 | 15.44 | + | 2.84E−01 | 108 | 80 | + | 1.00E−03 |
| Response to stress | 500 | 9 | 5.17 | + | 7.74E−02 | 44 | 27 | + | 1.18E−03 |
| Mitosis | 635 | 7 | 6.57 | + | 4.86E−01 | 52 | 34 | + | 2.04E−03 |
| Vitamin transport | 95 | 2 | 0.98 | + | 2.58E−01 | 13 | 5 | + | 2.25E−03 |
| Ectoderm development | 1469 | 24 | 15.2 | + | 1.81E−02 | 104 | 79 | + | 2.46E−03 |
| Lipid metabolic process | 1119 | 14 | 11.58 | + | 2.70E−01 | 82 | 60 | + | 2.92E−03 |
| Cell cycle | 1882 | 20 | 19.47 | + | 4.84E−01 | 128 | 101 | + | 3.23E−03 |
| Cellular component morphogenesis | 1170 | 15 | 12.1 | + | 2.32E−01 | 84 | 63 | + | 4.42E−03 |
| Anatomical structure morphogenesis | 1170 | 15 | 12.1 | + | 2.32E−01 | 84 | 63 | + | 4.42E−03 |
| Visual perception | 412 | 5 | 4.26 | + | 4.23E−01 | 35 | 22 | + | 6.00E−03 |
| Carbohydrate transport | 187 | 6 | 1.93 | + | 1.39E−02 | 19 | 10 | + | 6.94E−03 |
| Cell surface receptor linked signal transduction | 2235 | 33 | 23.12 | + | 2.32E−02 | 146 | 120 | + | 6.98E−03 |
| Response to stimulus | 1798 | 34 | 18.6 | + | 4.49E−04 | 113 | 96 | + | 4.29E−02 |
| Immune system process | 2630 | 43 | 27.21 | + | 1.50E−03 | 156 | 141 | + | 9.12E−02 |
| Immune response | 756 | 16 | 7.82 | + | 5.73E−03 | 40 | 40 | − | 5.14E−01 |
| Macrophage activation | 305 | 9 | 3.16 | + | 4.86E−03 | 16 | 16 | − | 5.35E−01 |
| Molecular function | |||||||||
| Binding | 6794 | 76 | 70.29 | + | 2.21E−01 | 467 | 363 | + | 3.01E−11 |
| Protein binding | 3200 | 36 | 33.11 | + | 3.19E−01 | 247 | 171 | + | 1.10E−09 |
| Catalytic activity | 5336 | 65 | 55.21 | + | 7.36E−02 | 363 | 285 | + | 9.65E−08 |
| Transporter activity | 942 | 11 | 9.75 | + | 3.84E−01 | 87 | 50 | + | 9.21E−07 |
| Enzyme regulator activity | 1187 | 9 | 12.28 | − | 2.11E−01 | 103 | 63 | + | 1.46E−06 |
| Transmembrane transporter activity | 897 | 11 | 9.28 | + | 3.26E−01 | 81 | 48 | + | 5.02E−06 |
| Transferase activity | 1593 | 16 | 16.48 | − | 5.16E−01 | 121 | 85 | + | 7.65E−05 |
| Kinase regulator activity | 320 | 0 | 3.31 | − | 3.55E−02 | 35 | 17 | + | 8.46E−05 |
| Translation factor activity, nucleic acid binding | 107 | 0 | 1.11 | − | 3.30E−01 | 17 | 6 | + | 9.55E−05 |
| Translation initiation factor activity | 80 | 0 | 0.83 | − | 4.36E−01 | 14 | 4 | + | 1.46E−04 |
| Nucleic acid binding | 3863 | 43 | 39.97 | + | 3.23E−01 | 253 | 207 | + | 2.58E−04 |
| Small GTPase regulator activity | 495 | 4 | 5.12 | − | 4.17E−01 | 45 | 26 | + | 5.46E−04 |
| RNA binding | 530 | 3 | 5.48 | − | 2.00E−01 | 47 | 28 | + | 6.97E−04 |
| Translation regulator activity | 105 | 0 | 1.09 | − | 3.36E−01 | 15 | 6 | + | 7.08E−04 |
| Hydrolase activity | 2236 | 34 | 23.13 | + | 1.43E−02 | 154 | 120 | + | 7.34E−04 |
| Guanyl-nucleotide exchange factor activity | 160 | 0 | 1.66 | − | 1.90E−01 | 19 | 9 | + | 1.33E−03 |
| Lipid transporter activity | 92 | 0 | 0.95 | − | 3.85E−01 | 13 | 5 | + | 1.71E−03 |
| Receptor binding | 1235 | 17 | 12.78 | + | 1.42E−01 | 89 | 66 | + | 3.11E−03 |
| RNA splicing factor activity, transesterification mechanism | 268 | 1 | 2.77 | − | 2.34E−01 | 26 | 14 | + | 3.31E−03 |
| Helicase activity | 159 | 1 | 1.65 | − | 5.10E−01 | 17 | 9 | + | 6.43E−03 |
| Kinase inhibitor activity | 122 | 0 | 1.26 | − | 2.82E−01 | 14 | 7 | + | 7.15E−03 |
| Amino-acid transmembrane transporter activity | 98 | 0 | 1.01 | − | 3.62E−01 | 12 | 5 | + | 7.60E−03 |
| Deaminase activity | 43 | 0 | 0.44 | − | 6.41E−01 | 7 | 2 | + | 9.28E−03 |
| Acyltransferase activity | 193 | 0 | 2 | − | 1.34E−01 | 19 | 10 | + | 9.46E−03 |
| Extracellular matrix structural constituent | 147 | 7 | 1.52 | + | 9.41E−04 | 15 | 8 | + | 1.47E−02 |
Abbreviations: DEP, depression genes; EXP, expected (rounded to nearest whole number); NCBI, National Center for Biotechnology Information; STR, chronic stress response genes.
Total number of genes referenced from NCBI.
Stress response genes (P<0.01) per GO term out of 194 total.
Depression genes (P<0.01) per GO term out of 998 total.
P<0.01 values are bolded.
The biological process terms ‘Cellular Process', ‘Cell Communication' and ‘Signal Transduction' were significantly over-represented in both the CRS-NS and the WMI-WLI gene lists. It is of further interest that metabolic process terms are the most over-represented in the WMI-WLI comparison, while cellular process terms, mainly immune system-related, are over-represented in the CRS-NS gene list. There was no overlap in over- or under-represented molecular function terms between the stress response and endogenous depression gene lists.
Discussion
This study identified novel molecular signatures of chronic stress and of endogenous depressive behavior in animal models. However, absence of direct association between the gene expression profiles suggests that independent molecular mechanisms regulate these two states.
Among genes showing significantly altered transcription levels in the amygdala and hippocampus in the endogenous depression model, there was a complete omission of serotoninergic, adrenergic and dopaminergic neurotransmission-related genes. This finding, although surprising, is congruent with gene expression studies of human postmortem MDD brains.66, 67, 68 We did find that multiple newer antidepressant targets, including Grm5,69 and a number of phosphodiesterases,70 were differentially expressed between WMIs and WLIs. In addition to these previously described targets, we identified several novel depressive behavior-related genes and pathways. Physiologically intriguing findings include the decreased mRNA levels of prolactin receptor and angiotensin I-converting enzyme in the WMI hippocampus; these genes have known peripheral function, but they have not been associated with depressive behavior as yet. Other intriguing examples of novel implications for endogenous depression include the solute carrier family 4, sodium bicarbonate co-transporter and member 5 gene (Slc4a5), which has recently been associated with metabolic phenotypes.71 Interestingly, metabolic processes were the most over-represented biological process terms in the WMI-WLI gene ontology. The integrin signaling pathway, which was over-represented in the endogenous depression gene set, contains different members of the collagen family, some of which are also implicated in the chronic stress response. Finally, mRNA levels of dehydrodolichyl diphosphate synthase are increased in both the hippocampus and the amygdala of WMI. Dehydrodolichyl diphosphate synthase is responsible for synthesizing dolichol, which accumulates in the neuropathological human brain.72
The set of genes whose hippocampal or amygdalar expression patterns were altered by chronic stress in all four rat strains represent a generalizable molecular response to chronic stress. Many of these genes have shown matching directional changes by unpredictable chronic mild stress, and these include: Col1a1, tissue plasminogen activator, insulin-like growth factor binding protein 2, amyloid beta precursor protein and transthyretin in the amygdala and/or hippocampus.39, 73 In addition, stress has previously been found to increase the expression of amyloid beta precursor protein74, 75 and neuropeptide Y.76, 77, 78 Our findings, however, also implicate a number of novel ‘stress genes'. Among these, the decreased mRNA levels of tumor suppressor protein 53, insulin-like growth factor 2 and hemoglobin beta and alpha a1 are of particular interest. Tumor suppressor protein 53 has been linked to the pathology of Huntington disease,79 a finding that is connected to the over-representation of our chronic stress response genes involved in the Huntington pathway. Insulin-like growth factor 2 is an imprinted gene, described as having both neuroprotective and neurodegenerative properties,80 as well as involvement in metabolic disorders. Recently, hemoglobin beta and alpha transcripts and proteins have been found in cortical and hippocampal astrocytes, mature oligodendrocytes and a subpopulation of dopaminergic neurons.81 If the decreased expression of hemoglobin beta and alpha a1 is a manifestation of chronic stress-induced decreases in oxygen storage, these findings would implicate chronic stress in neurodegenerative processes. If, however, they are related to the neurotoxicity of hemoglobins,82 the decreased expression of hemoglobin beta and alpha a1 and of tumor suppressor protein 53 would propose the heretical idea of chronic stress being neuroprotective.
If depression in general is a result of chronic stress and stress-related factors, these genes should be shared with the endogenous ‘depression transcriptome', as defined by WMI-WLI differences. However, a surprisingly small number of genes were shared between WMI-WLI and NS-CRS differentially expressed genes in the amygdala and hippocampus. Genes showing parallel directional expression changes in the hippocampus between the ‘endogenous depression' and the ‘chronic stress' gene sets include: Col1a1, Mmp14 and RGD1305680 (homologous to the KIAA0240 gene). Mmp14 (also known as MT1-MMP) degrades endogenous beta amyloids83 and is implicated in the pathogenesis of central nervous system inflammatory disorders. It is also a tethered membrane collagenase, which, together with the involvement of Col1a1, implies the significance of extracellular matrix remodeling in depressive behavior and chronic stress. Ontological analysis further and more broadly confirmed the dissimilarity between the endogenous depressive behavior and generic stress response expression profiles. Functional annotation of the genes involved in both states revealed few common biological processes and no common molecular functions between the two gene sets.
This study using animal models contends that endogenous depression and chronic stress response are regulated by independent molecular pathways. In humans, an inference of this finding would be that stress-induced and endogenous subtypes of depression are etiologically distinct. There are both human and animal studies supporting this assumption. For example, only a subset of depressive phenotypes can be causally attributed to stressful life events, and these phenotypes indicate only a moderately increased risk of developing depression.84, 85 In addition, although the chronic mild stress model is thought of as a model of depression, some animals are resistant to the anhedonic effect of stress.73, 86, 87, 88 Similarly, some animals are resilient to learned helplessness in the congenital learned helplessness model of depression,25, 89 suggesting that differences in vulnerability to stress may determine the development of depressive phenotype in these models. Thus, it is possible that stress-susceptible individuals could develop a subtype of depression distinct from endogenous depression; this subtype may show greater comorbidity with other stress-related disorders. The existence of different subtypes of depression is in further agreement with the findings of Krishnan and Nestler,84 which state that there is no ‘unified theory' of depression. Should our findings translate to MDD, they suggest that different treatment strategies, dependent on the patient's depressive subtype, could be beneficial. Furthermore, as most animal models of depression are based on stress, the above findings predict that using various genetic animal models of depression could lead to novel drug targets that may achieve successful treatment of patients in which current treatment methods fail.
Acknowledgments
This work was supported the Davee Foundation, the RD Foundation, and NIH grant MH077234. The authors also wish to thank Laura Sittig, Elif Tunc-Ozcan, and Timothy Ullmann for helpful suggestions on the manuscript and Drs. Frasier Aird and Claire Will for early work on the endogenous depression model.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Rumsfeld JS, Ho PM. Depression and cardiovascular disease: a call for recognition. Circulation. 2005;111:250–253. doi: 10.1161/01.CIR.0000154573.62822.89. [DOI] [PubMed] [Google Scholar]
- Swaab DF, Bao AM, Lucassen PJ. The stress system in the human brain in depression and neurodegeneration. Ageing Res Rev. 2005;4:141–194. doi: 10.1016/j.arr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Bornstein SR, Schuppenies A, Wong ML, Licinio J. Approaching the shared biology of obesity and depression: the stress axis as the locus of gene–environment interactions. Mol Psychiatry. 2006;11:892–902. doi: 10.1038/sj.mp.4001873. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Ormel J, Demler O, Stang PE. Comorbid mental disorders account for the role impairment of commonly occurring chronic physical disorders: results from the National Comorbidity Survey. J Occup Environ Med. 2003;45:1257–1266. doi: 10.1097/01.jom.0000100000.70011.bb. [DOI] [PubMed] [Google Scholar]
- Altar CA, Vawter MP, Ginsberg SD. Target identification for CNS diseases by transcriptional profiling. Neuropsychopharmacology. 2009;34:18–54. doi: 10.1038/npp.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado PL. Depression: the case for a monoamine deficiency. J Clin Psychiatry. 2000;61 (Suppl 6:7–11. [PubMed] [Google Scholar]
- Ruhe HG, Mason NS, Schene AH. Mood is indirectly related to serotonin, norepinephrine and dopamine levels in humans: a meta-analysis of monoamine depletion studies. Mol Psychiatry. 2007;12:331–359. doi: 10.1038/sj.mp.4001949. [DOI] [PubMed] [Google Scholar]
- Fava M, Kendler KS. Major depressive disorder. Neuron. 2000;28:335–341. doi: 10.1016/s0896-6273(00)00112-4. [DOI] [PubMed] [Google Scholar]
- Holsboer F. The stress hormone system is back on the map. Curr Psychiatry Rep. 2000;2:454–456. doi: 10.1007/s11920-000-0001-y. [DOI] [PubMed] [Google Scholar]
- Wong ML, Licinio J. Research and treatment approaches to depression. Nat Rev Neurosci. 2001;2:343–351. doi: 10.1038/35072566. [DOI] [PubMed] [Google Scholar]
- Gold PW, Chrousos GP. Organization of the stress system and its dysregulation in melancholic and atypical depression: high vs low CRH/NE states. Mol Psychiatry. 2002;7:254–275. doi: 10.1038/sj.mp.4001032. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Barrot M, DiLeone RJ, Eisch AJ, Gold SJ, Monteggia LM. Neurobiology of depression. Neuron. 2002;34:13–25. doi: 10.1016/s0896-6273(02)00653-0. [DOI] [PubMed] [Google Scholar]
- Hasler G, Drevets WC, Manji HK, Charney DS. Discovering endophenotypes for major depression. Neuropsychopharmacology. 2004;29:1765–1781. doi: 10.1038/sj.npp.1300506. [DOI] [PubMed] [Google Scholar]
- Cryan JF, Holmes A. The ascent of mouse: advances in modelling human depression and anxiety. Nat Rev Drug Discov. 2005;4:775–790. doi: 10.1038/nrd1825. [DOI] [PubMed] [Google Scholar]
- de Kloet ER, Joels M, Holsboer F. Stress and the brain: from adaptation to disease. Nat Rev Neurosci. 2005;6:463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- Bale TL. Stress sensitivity and the development of affective disorders. Horm Behav. 2006;50:529–533. doi: 10.1016/j.yhbeh.2006.06.033. [DOI] [PubMed] [Google Scholar]
- Muller MB, Holsboer F. Mice with mutations in the HPA-system as models for symptoms of depression. Biol Psychiatry. 2006;59:1104–1115. doi: 10.1016/j.biopsych.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Monroe SM, Reid MW. Gene–environment interactions in depression research: genetic polymorphisms and life-stress polyprocedures. Psychol Sci. 2008;19:947–956. doi: 10.1111/j.1467-9280.2008.02181.x. [DOI] [PubMed] [Google Scholar]
- Jacobson LH, Cryan JF. Feeling strained? Influence of genetic background on depression-related behavior in mice: a review. Behav Genet. 2007;37:171–213. doi: 10.1007/s10519-006-9106-3. [DOI] [PubMed] [Google Scholar]
- Kas MJ, Fernandes C, Schalkwyk LC, Collier DA. Genetics of behavioural domains across the neuropsychiatric spectrum; of mice and men. Mol Psychiatry. 2007;12:324–330. doi: 10.1038/sj.mp.4001979. [DOI] [PubMed] [Google Scholar]
- Urani A, Chourbaji S, Gass P. Mutant mouse models of depression: candidate genes and current mouse lines. Neurosci Biobehav Rev. 2005;29:805–828. doi: 10.1016/j.neubiorev.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Tecott LH. The genes and brains of mice and men. Am J Psychiatry. 2003;160:646–656. doi: 10.1176/appi.ajp.160.4.646. [DOI] [PubMed] [Google Scholar]
- Scott PA, Cierpal MA, Kilts CD, Weiss JM. Susceptibility and resistance of rats to stress-induced decreases in swim-test activity: a selective breeding study. Brain Res. 1996;725:217–230. doi: 10.1016/0006-8993(96)00093-5. [DOI] [PubMed] [Google Scholar]
- Steimer T, Driscoll P. Divergent stress responses and coping styles in psychogenetically selected Roman high-(RHA) and low-(RLA) avoidance rats: behavioural, neuroendocrine and developmental aspects. Stress. 2003;6:87–100. doi: 10.1080/1025389031000111320. [DOI] [PubMed] [Google Scholar]
- Henn FA, Vollmayr B. Stress models of depression: forming genetically vulnerable strains. Neurosci Biobehav Rev. 2005;29:799–804. doi: 10.1016/j.neubiorev.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Weiss JM, Cierpial MA, West CHK. Selective breeding of rats for high and low motor activity in a swim test: toward a new animal model of depression. Pharmacol Biochem Behav. 1998;61:49–66. doi: 10.1016/s0091-3057(98)00075-6. [DOI] [PubMed] [Google Scholar]
- Weiss JM, West CH, Emery MS, Bonsall RW, Moore JP, Boss-Williams KA. Rats selectively-bred for behavior related to affective disorders: proclivity for intake of alcohol and drugs of abuse, and measures of brain monoamines. Biochem Pharmacol. 2008;75:134–159. doi: 10.1016/j.bcp.2007.09.027. [DOI] [PubMed] [Google Scholar]
- El Yacoubi M, Bouali S, Popa D, Naudon L, Leroux-Nicollet I, Hamon M, et al. Behavioral, neurochemical, and electrophysiological characterization of a genetic mouse model of depression. Proc Natl Acad Sci USA. 2003;100:6227–6232. doi: 10.1073/pnas.1034823100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redei E, Pare WP, Aird F, Kluczynski J. Strain differences in hypothalamic-pituitary-adrenal activity and stress ulcer. Am J Physiol. 1994;266 (Part 2:R353–R360. doi: 10.1152/ajpregu.1994.266.2.R353. [DOI] [PubMed] [Google Scholar]
- Redei EE, Ahmadiyeh N, Baum A, Sasso D, Slone J, Solberg LC, et al. Novel animal models of affective disordersIn: Tucker GJ, McKinney W (eds).Seminars in Clinical Neuropsychiatryvol. 6.WB Saunders Company: Philadelphia; 2001 [DOI] [PubMed] [Google Scholar]
- Solberg LC, Ahmadiyeh N, Baum AE, Vitaterna MH, Takahashi JS, Turek FW, et al. Depressive-like behavior and stress reactivity are independent traits in a Wistar Kyoto × Fisher 344 cross. Mol Psychiatry. 2003;8:423–433. doi: 10.1038/sj.mp.4001255. [DOI] [PubMed] [Google Scholar]
- Rittenhouse PA, Lopez-Rubalcava C, Stanwood GD, Lucki I. Amplified behavioral and endocrine responses to forced swim stress in the Wistar–Kyoto rat. Psychoneuroendocrinology. 2002;27:303–318. doi: 10.1016/s0306-4530(01)00052-x. [DOI] [PubMed] [Google Scholar]
- Will CC, Aird F, Redei EE. Selectively bred Wistar–Kyoto rats: an animal model of depression and hyper-responsiveness to antidepressants. Mol Psychiatry. 2003;8:925–932. doi: 10.1038/sj.mp.4001345. [DOI] [PubMed] [Google Scholar]
- Thomas MA, Chen CF, Jensen-Seaman MI, Tonellato PJ, Twigger SN. Phylogenetics of rat inbred strains. Mamm Genome. 2003;14:61–64. doi: 10.1007/s00335-002-2204-5. [DOI] [PubMed] [Google Scholar]
- Gomez F, Lahmame A, de Kloet ER, Armario A. Hypothalamic-pituitary-adrenal response to chronic stress in five inbred rat strains: differential responses are mainly located at the adrenocortical level. Neuroendocrinology. 1996;63:327–337. doi: 10.1159/000126973. [DOI] [PubMed] [Google Scholar]
- Wood GE, Norris EH, Waters E, Stoldt JT, McEwen BS. Chronic immobilization stress alters aspects of emotionality and associative learning in the rat. Behav Neurosci. 2008;122:282–292. doi: 10.1037/0735-7044.122.2.282. [DOI] [PubMed] [Google Scholar]
- Bravo JA, Diaz-Veliz G, Mora S, Ulloa JL, Berthoud VM, Morales P, et al. Desipramine prevents stress-induced changes in depressive-like behavior and hippocampal markers of neuroprotection. Behav Pharmacol. 2009;20:273–285. doi: 10.1097/FBP.0b013e32832c70d9. [DOI] [PubMed] [Google Scholar]
- Gil M, Marti J, Armario A. Inhibition of catecholamine synthesis depresses behavior of rats in the holeboard and forced swim tests: influence of previous chronic stress. Pharmacol Biochem Behav. 1992;43:597–601. doi: 10.1016/0091-3057(92)90196-m. [DOI] [PubMed] [Google Scholar]
- Joo Y, Choi KM, Lee YH, Kim G, Lee DH, Roh GS, et al. Chronic immobilization stress induces anxiety- and depression-like behaviors and decreases transthyretin in the mouse cortex. Neurosci Lett. 2009;461:121–125. doi: 10.1016/j.neulet.2009.06.025. [DOI] [PubMed] [Google Scholar]
- Kim KS, Han PL. Optimization of chronic stress paradigms using anxiety- and depression-like behavioral parameters. J Neurosci Res. 2006;83:497–507. doi: 10.1002/jnr.20754. [DOI] [PubMed] [Google Scholar]
- Haenisch B, Bilkei-Gorzo A, Caron MG, Bonisch H. Knockout of the norepinephrine transporter and pharmacologically diverse antidepressants prevent behavioral and brain neurotrophin alterations in two chronic stress models of depression. J Neurochem. 2009;111:403–416. doi: 10.1111/j.1471-4159.2009.06345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veena J, Srikumar BN, Raju TR, Shankaranarayana Rao BS. Exposure to enriched environment restores the survival and differentiation of new born cells in the hippocampus and ameliorates depressive symptoms in chronically stressed rats. Neurosci Lett. 2009;455:178–182. doi: 10.1016/j.neulet.2009.03.059. [DOI] [PubMed] [Google Scholar]
- Lee T, Jarome T, Li SJ, Kim JJ, Helmstetter FJ. Chronic stress selectively reduces hippocampal volume in rats: a longitudinal magnetic resonance imaging study. Neuroreport. 2009;20:1554–1558. doi: 10.1097/WNR.0b013e328332bb09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas A, Pillai AG, Chattarji S. Recovery after chronic stress fails to reverse amygdaloid neuronal hypertrophy and enhanced anxiety-like behavior. Neuroscience. 2004;128:667–673. doi: 10.1016/j.neuroscience.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Vyas A, Bernal S, Chattarji S. Effects of chronic stress on dendritic arborization in the central and extended amygdala. Brain Res. 2003;965:290–294. doi: 10.1016/s0006-8993(02)04162-8. [DOI] [PubMed] [Google Scholar]
- Bremner JD, Narayan M, Anderson ER, Staib LH, Miller HL, Charney DS. Hippocampal volume reduction in major depression. Am J Psychiatry. 2000;157:115–118. doi: 10.1176/ajp.157.1.115. [DOI] [PubMed] [Google Scholar]
- Sheline YI, Wang PW, Gado MH, Csernansky JG, Vannier MW. Hippocampal atrophy in recurrent major depression. Proc Natl Acad Sci USA. 1996;93:3908–3913. doi: 10.1073/pnas.93.9.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline YI, Gado MH, Price JL. Amygdala core nuclei volumes are decreased in recurrent major depression. NeuroReport. 1998;9:2023–2028. doi: 10.1097/00001756-199806220-00021. [DOI] [PubMed] [Google Scholar]
- Sheline YI, Sanghavi M, Mintun MA, Gado MH. Depression duration but not age predicts hippocampal volume loss in medically healthy women with recurrent major depression. J Neurosci. 1999;19:5034–5043. doi: 10.1523/JNEUROSCI.19-12-05034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings RS, Parsey RV, Oquendo MA, Arango V, Mann JJ. Volumetric analysis of the prefrontal cortex, amygdala, and hippocampus in major depression. Neuropsychopharmacology. 2004;29:952–959. doi: 10.1038/sj.npp.1300371. [DOI] [PubMed] [Google Scholar]
- Kim JS, Shukla SD. Acute in vivo effect of ethanol (binge drinking) on histone H3 modifications in rat tissues. Alcohol Alcohol. 2006;41:126–132. doi: 10.1093/alcalc/agh248. [DOI] [PubMed] [Google Scholar]
- Solberg LC, Baum AE, Ahmadiyeh N, Shimomura K, Li R, Turek FW, et al. Sex- and lineage-specific inheritance of depression-like behavior in the rat. Mamm Genome. 2004;15:648–662. doi: 10.1007/s00335-004-2326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detke MJ, Rickels M, Lucki I. Active behaviors in the rat forced swimming test differentially produced by serotonergic and noradrenergic antidepressants. Psychopharmacology. 1995;121:66–72. doi: 10.1007/BF02245592. [DOI] [PubMed] [Google Scholar]
- Nosek K, Dennis K, Andrus BM, Ahmadiyeh N, Baum AE, Woods LC, et al. Context and strain-dependent behavioral response to stress. Behav Brain Funct. 2008;4:23. doi: 10.1186/1744-9081-4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcoxon JS, Kuo AG, Disterhoft JF, Redei EE. Behavioral deficits associated with fetal alcohol exposure are reversed by prenatal thyroid hormone treatment: a role for maternal thyroid hormone deficiency in FAE. Mol Psychiatry. 2005;10:961–971. doi: 10.1038/sj.mp.4001694. [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Churchill GA. Using ANOVA to analyze microarray data. Biotechniques. 2004;37:173–175. doi: 10.2144/04372TE01. [DOI] [PubMed] [Google Scholar]
- Wu X, Pang ST, Sahlin L, Blanck A, Norstedt G, Flores-Morales A. Gene expression profiling of the effects of castration and estrogen treatment in the rat uterus. Biol Reprod. 2003;69:1308–1317. doi: 10.1095/biolreprod.103.015420. [DOI] [PubMed] [Google Scholar]
- Cui X, Hwang JT, Qiu J, Blades NJ, Churchill GA. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics. 2005;6:59–75. doi: 10.1093/biostatistics/kxh018. [DOI] [PubMed] [Google Scholar]
- Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc Natl Acad Sci USA. 2005;102:12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning MJ, Smith ML, Ritchie ME, Tavare S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, et al. PANTHER: a browseable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, Campbell MJ. Transcription, genomes, function. Trends Genet. 2000;16:409–415. doi: 10.1016/s0168-9525(00)02065-5. [DOI] [PubMed] [Google Scholar]
- Sibille E, Arango V, Galfalvy HC, Pavlidis P, Erraji-Benchekroun L, Ellis SP, et al. Gene expression profiling of depression and suicide in human prefrontal cortex. Neuropsychopharmacology. 2004;29:351–361. doi: 10.1038/sj.npp.1300335. [DOI] [PubMed] [Google Scholar]
- Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V, et al. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PLoS One. 2009;4:e6585. doi: 10.1371/journal.pone.0006585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira A, Turecki G. Genome wide gene expression studies in mood disorders. OMICS. 2006;10:444–454. doi: 10.1089/omi.2006.10.444. [DOI] [PubMed] [Google Scholar]
- Covington HE, III, Vialou V, Nestler EJ. From synapse to nucleus: novel targets for treating depression. Neuropharmacology. 2010;58:683–693. doi: 10.1016/j.neuropharm.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito K, Reierson GW, Luo HR, Wu GS, Licinio J, Wong ML. Phosphodiesterase genes and antidepressant treatment response: a review. Ann Med. 2009;41:177–185. doi: 10.1080/07853890802441169. [DOI] [PubMed] [Google Scholar]
- Stutz AM, Teran-Garcia M, Rao DC, Rice T, Bouchard C, Rankinen T. Functional identification of the promoter of SLC4A5, a gene associated with cardiovascular and metabolic phenotypes in the HERITAGE Family Study. Eur J Hum Genet. 2009;17:1481–1489. doi: 10.1038/ejhg.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S, Zhang YW, Takahashi S, Koyama T. Identification of human dehydrodolichyl diphosphate synthase gene. Biochim Biophys Acta. 2003;1625:291–295. doi: 10.1016/s0167-4781(02)00628-0. [DOI] [PubMed] [Google Scholar]
- Bergstrom A, Jayatissa MN, Thykjaer T, Wiborg O. Molecular pathways associated with stress resilience and drug resistance in the chronic mild stress rat model of depression: a gene expression study. J Mol Neurosci. 2007;33:201–215. doi: 10.1007/s12031-007-0065-9. [DOI] [PubMed] [Google Scholar]
- Dong H, Csernansky JG. Effects of stress and stress hormones on amyloid-beta protein and plaque deposition. J Alzheimers Dis. 2009;18:459–469. doi: 10.3233/JAD-2009-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Yu DH, Lee YS. Gene expression profiling of rat cerebral cortex development using cDNA microarrays. Neurochem Res. 2009;34:1030–1038. doi: 10.1007/s11064-008-9867-6. [DOI] [PubMed] [Google Scholar]
- Zambello E, Fuchs E, Abumaria N, Rygula R, Domenici E, Caberlotto L. Chronic psychosocial stress alters NPY system: different effects in rat and tree shrew. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:122–130. doi: 10.1016/j.pnpbp.2009.10.011. [DOI] [PubMed] [Google Scholar]
- Thorsell A, Slawecki CJ, El Khoury A, Mathe AA, Ehlers CL. The effects of social isolation on neuropeptide Y levels, exploratory and anxiety-related behaviors in rats. Pharmacol Biochem Behav. 2006;83:28–34. doi: 10.1016/j.pbb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Heilig M, Widerlov E. Neurobiology and clinical aspects of neuropeptide Y. Crit Rev Neurobiol. 1995;9:115–136. [PubMed] [Google Scholar]
- Feng Z, Jin S, Zupnick A, Hoh J, de Stanchina E, Lowe S, et al. p53 tumor suppressor protein regulates the levels of huntingtin gene expression. Oncogene. 2006;25:1–7. doi: 10.1038/sj.onc.1209021. [DOI] [PubMed] [Google Scholar]
- Dikkes P, D BJ, Guo WH, Chao C, Hemond P, Yoon K, et al. IGF2 knockout mice are resistant to kainic acid-induced seizures and neurodegeneration. Brain Res. 2007;1175:85–95. doi: 10.1016/j.brainres.2007.05.068. [DOI] [PubMed] [Google Scholar]
- Biagioli M, Pinto M, Cesselli D, Zaninello M, Lazarevic D, Roncaglia P, et al. Unexpected expression of alpha- and beta-globin in mesencephalic dopaminergic neurons and glial cells. Proc Natl Acad Sci USA. 2009;106:15454–15459. doi: 10.1073/pnas.0813216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Mori T, Sumii T, Lo EH. Hemoglobin-induced cytotoxicity in rat cerebral cortical neurons: caspase activation and oxidative stress. Stroke. 2002;33:1882–1888. doi: 10.1161/01.str.0000020121.41527.5d. [DOI] [PubMed] [Google Scholar]
- Liao MC, Van Nostrand WE. Degradation of soluble and fibrillar amyloid beta-protein by matrix metalloproteinase (MT1-MMP) in vitro. Biochemistry. 2010;49:1127–1136. doi: 10.1021/bi901994d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V, Nestler EJ. The molecular neurobiology of depression. Nature. 2008;455:894–902. doi: 10.1038/nature07455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Karkowski LM, Prescott CA. Causal relationship between stressful life events and the onset of major depression. Am J Psychiatry. 1999;156:837–841. doi: 10.1176/ajp.156.6.837. [DOI] [PubMed] [Google Scholar]
- Strekalova T, Spanagel R, Bartsch D, Henn FA, Gass P. Stress-induced anhedonia in mice is associated with deficits in forced swimming and exploration. Neuropsychopharmacology. 2004;29:2007–2017. doi: 10.1038/sj.npp.1300532. [DOI] [PubMed] [Google Scholar]
- Jayatissa MN, Bisgaard C, Tingstrom A, Papp M, Wiborg O. Hippocampal cytogenesis correlates to escitalopram-mediated recovery in a chronic mild stress rat model of depression. Neuropsychopharmacology. 2006;31:2395–2404. doi: 10.1038/sj.npp.1301041. [DOI] [PubMed] [Google Scholar]
- Bisgaard CF, Jayatissa MN, Enghild JJ, Sanchez C, Artemychyn R, Wiborg O. Proteomic investigation of the ventral rat hippocampus links DRP-2 to escitalopram treatment resistance and SNAP to stress resilience in the chronic mild stress model of depression. J Mol Neurosci. 2007;32:132–144. doi: 10.1007/s12031-007-0025-4. [DOI] [PubMed] [Google Scholar]
- Schulz D, Mirrione MM, Henn FA. Cognitive aspects of congenital learned helplessness and its reversal by the monoamine oxidase (MAO)-B inhibitor deprenyl. Neurobiol Learn Mem. 2010;93:291–301. doi: 10.1016/j.nlm.2009.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.