Post-translational control of critical stem cell transcription factors plays essential roles in directing cell fate determination of stem cells. Protein ubiquitylation as one of important post-translational modifications has been well demonstrated to be a crucial regulatory mechanism involved in several fundamental biological processes such as cell differentiation, development and oncogenesis. Many regulatory proteins including some key transcription factors are ubiquitylated by specific E3 ubiquitin ligases and subsequently targeted for degradation by the 26S proteasome. Similar to other reversible post-translational regulations such as phosphorylation and methylation, protein ubiquitylation can be removed by specific deubiquitylating enzymes called deubiquitylases (DUBs). Thus, critical proteins may be controlled by both ubiquitylation and deubiquitylation mechanisms in a dynamically balanced manner.
One of the most critical DUBs, the herpesvirus-associated ubiquitin-specific protease (HAUSP, also known as ubiquitin specific protease 7, USP7) was originally identified to be associated with viral proteins such as ICP0 (herpes simplex virus type 1 regulatory protein) and EBNA1 (Epstein-Barr nuclear antigen 1) during viral infection, thereby regulating ICP0 stability as well as EBNA1 transcriptional activity.1,2 A number of elegant studies demonstrated that HAUSP regulates stability and functions of several important proteins under normal and stress conditions. For example, in response to oxidative stress, HAUSP inhibits nuclear localization and transcriptional activity of FOXO4 (a forkhead box O transcription factor) by interacting with and deubiquitylating FOXO protein.3 Through a similar mechanism, HAUSP deubiquitylates tumor suppressor PTEN, and thus regulates its nuclear exclusion.4 HAUSP also regulates the stability of another crucial tumor suppressor p53 and its counterpart MDM2.5,6 During early embryonic development, HAUSP inactivation caused early embryonic lethality and the knockout mice die between E6.5 and E7.5 due to p53 activation.7 Interestingly, HAUSP and p53 double knockout mice, although extending to a more advanced embryonic development, can not survive the whole prenatal development,7 suggesting p53-independent functions of HAUSP that may be responsible for proliferation and differentiation.
Our recent studies published in Nature Cell Biology demonstrated that HAUSP prevents degradation of the repressor element 1-silencing transcription factor (REST) through deubiquitylation and thus promote maintenance of neural stem/progenitor cells (NPCs).8 REST (also known as neuron-restricted silencing factor, NRSF) is a central transcriptional repressor of neuronal differentiation. REST is highly expressed in neural tumours, in particular medulloblastomas and neuroblastomas, and aberrant REST function is associated with neurodegenerative diseases and other pathological states.9 During neuronal differentiation, REST protein is targeted for the ubiquitin-dependent proteasomal degradation by SCFβ-TrCP (an E3 ubiquitin ligase complex).10,11 Our studies revealed that REST protein can be stabilized by HAUSP that antagonizes the SCFβ-TrCP-mediated ubiquitylation.8 We have identified a critical consensus site (310-PYSS-313) on REST protein that is required for the HAUSP-mediated deubiquitylation.8 Our studies strongly suggest that deubiquitylases may play critical roles to stabilize stem cell transcription factors and promote maintenance of “stemness”. As a number of transcriptional factors (Nanog, c-Myc, Sox2 and Oct4) serving as core regulators of self-renewal and maintenance of embryonic stem cells (ESCs) and tissue stem cells have been found to be ubiquitylated by different E3 ubiquitin ligases, it is likely that each stem cell transcription factor can be deubiquitylated by a specific deubiquitylase. Identification of such deubiquitylases will be important to understand the molecular mechanisms of cell fate determination of ESCs and tissue stem cells.
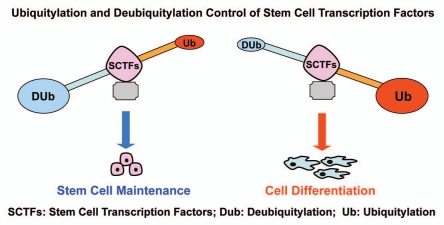
Based on our recent studies and previous studies by other groups, we propose that each stem cell transcription factor (SCTF) is regulated by both ubiquitylation and deubiquitylation at post-translational level. The protein stability of each SCTF is controlled at least by a pair of specific deubiquitylase and ubiquitin E3 ligase. The net balance of ubiquitylation and deubiquitylation of SCTFs could have a significant impact on cell fate determination of stem cells (Fig. 1). Increased ubiquitylation of SCTFs leads to degradation and induces cell differentiation, and dominant deubiquitylation of SCTFs stabilizes these transcription factors to promote maintenance of stem cells. Dissecting this paradigm of reciprocal post-translational control in stem cell regulatory networks not only advances stem cell biology but may also provide new understanding of cancer stem cells.
Figure 1.
A hypothetical model for the post-translational control of stem cell transcription factors by ubiquitylation and deubiquitylation to direct cell fate determination of stem cells.
Acknowledgements
We thank members of the Rich's laboratory for helpful scientific discussions. This work was supported by the Cleveland Clinic Foundation and a NIH R01 grant (NS070315) to S.B.
Comment on: Huang Z, et al. Nat Cell Biol. 2011;13:142–152. doi: 10.1038/ncb2153.
References
- 1.Holowaty MN, et al. J Biol Chem. 2003;278:29987–29994. doi: 10.1074/jbc.M303977200. [DOI] [PubMed] [Google Scholar]
- 2.Boutell C, et al. J Virol. 2005;79:12342–12354. doi: 10.1128/JVI.79.19.12342-12354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Horst A, et al. Nature Cell Biology. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 4.Song MS, et al. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li M, et al. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 6.Sheng Y, et al. Nat Struct Mol Biol. 2006;13:285–297. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- 7.Kon N, et al. Oncogene. 2010;29:1270–1279. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Z, et al. Nature Cell Biology. 2011;13:142–152. doi: 10.1038/ncb2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuccato C, et al. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 10.Westbrook TF, et al. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guardavaccaro D, et al. Nature. 2008;452:365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]