Abstract
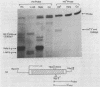
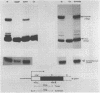
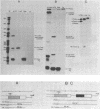
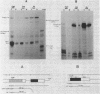
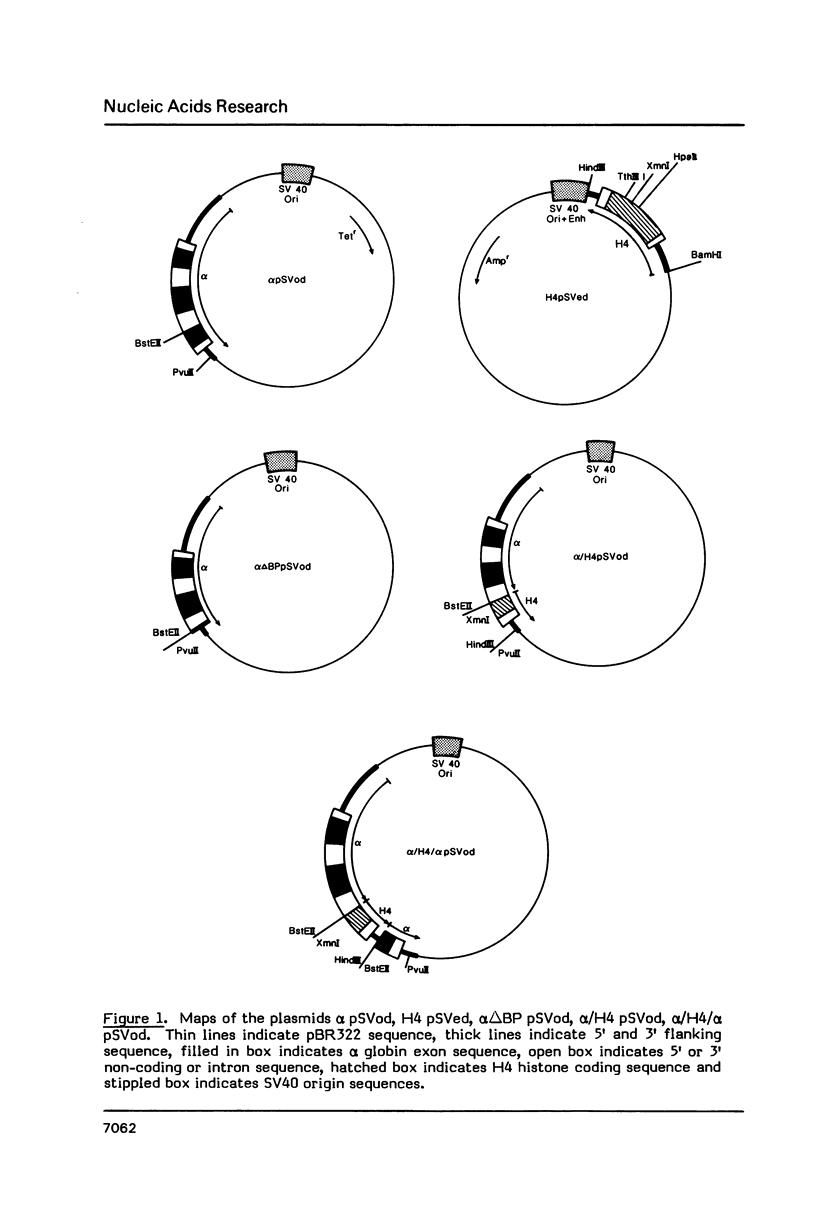
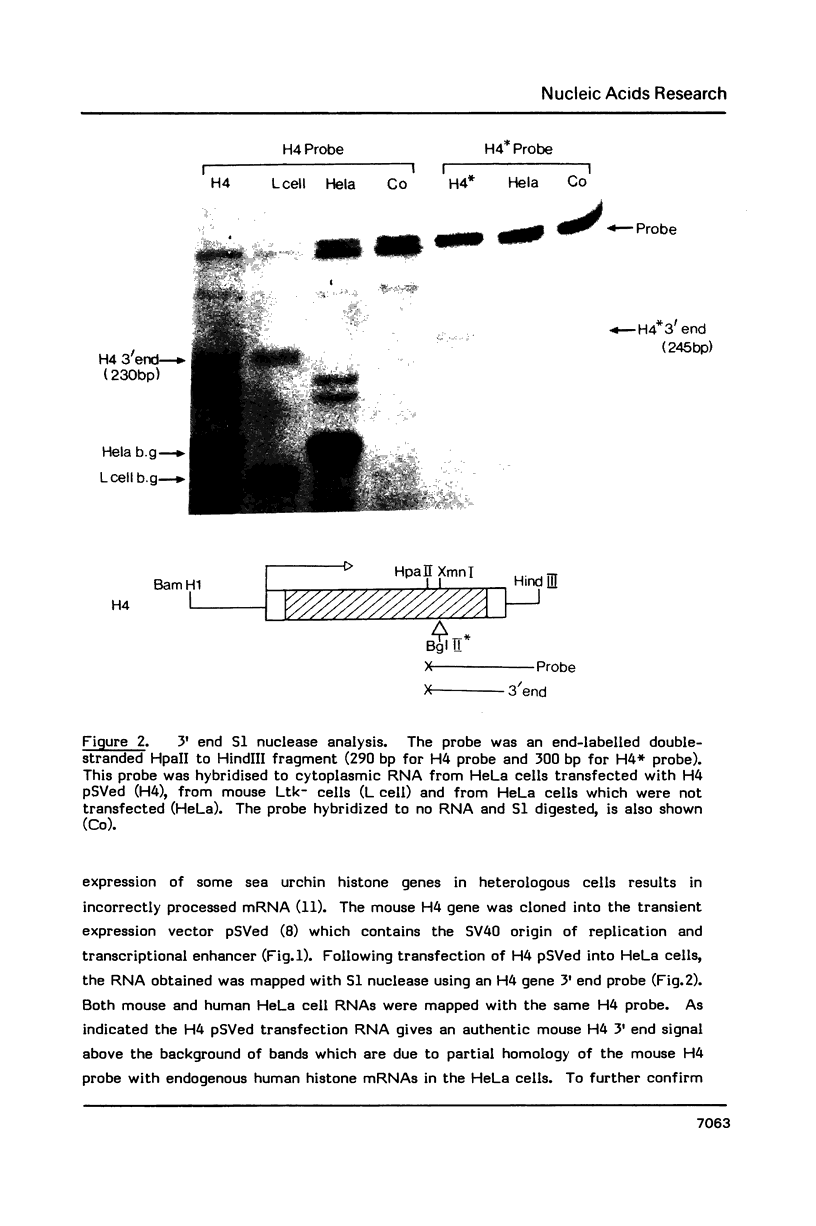
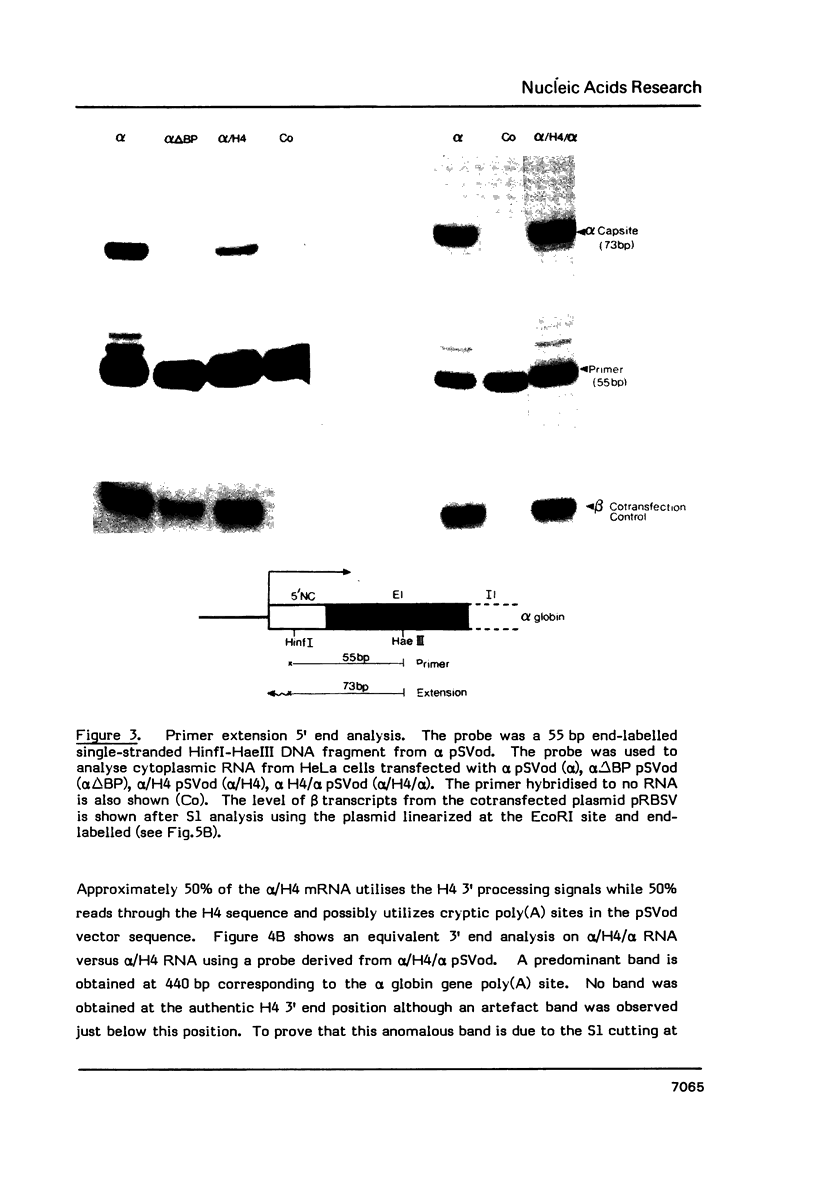
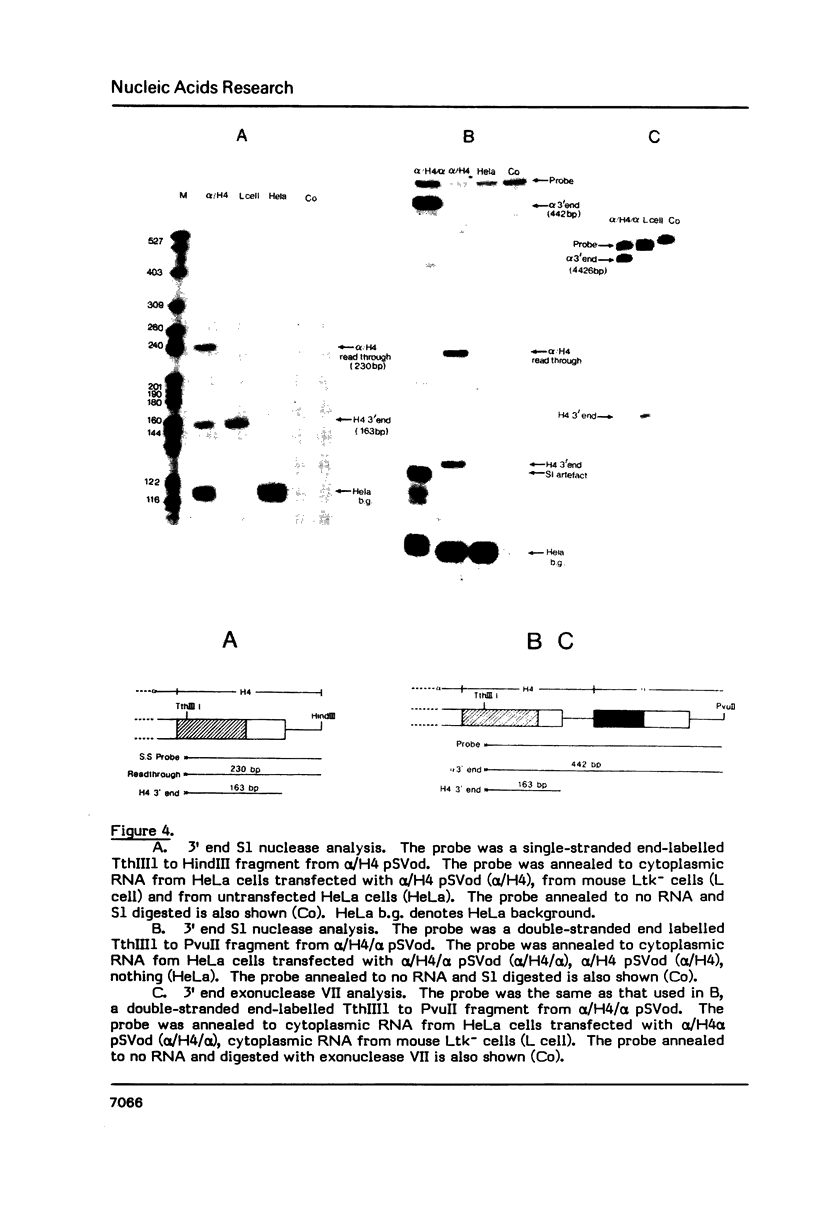
Deletion of the poly(A) site from the human alpha globin gene results in a defective gene that produces very little stable mRNA as compared to the intact gene, presumably due to the instability of the mRNA. However, if the Alpha poly(A) site is replaced by mouse histone H4 3' end processing signals, significant levels of hybrid alpha/H4 mRNA are obtained and the transcripts formed are cytoplasmic and poly(A)-. When both mouse histone 3' end processing signals and the alpha globin poly(A) site signals are placed in tandem after the alpha globin gene promoter and coding sequence, the alpha poly(A) site signals are utilized exclusively. These results show that the histone 3' end processing signals can function independently of the histone promoter and the transcripts which are normally polyadenylated (alpha globin) can be stabilized by a poly(A)- histone mRNA 3' terminus. Furthermore, these results show that the histone 3' end processing signals are less efficient than the alpha globin poly(A) site signals, if the two are placed in direct competition.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birchmeier C., Folk W., Birnstiel M. L. The terminal RNA stem-loop structure and 80 bp of spacer DNA are required for the formation of 3' termini of sea urchin H2A mRNA. Cell. 1983 Dec;35(2 Pt 1):433–440. doi: 10.1016/0092-8674(83)90176-9. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Darnell J. E., Jr Variety in the level of gene control in eukaryotic cells. Nature. 1982 Jun 3;297(5865):365–371. doi: 10.1038/297365a0. [DOI] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Mattaj I. W., De Robertis E. M. Nuclear segregation of U2 snRNA requires binding of specific snRNP proteins. Cell. 1985 Jan;40(1):111–118. doi: 10.1016/0092-8674(85)90314-9. [DOI] [PubMed] [Google Scholar]
- Nevins J. R. The pathway of eukaryotic mRNA formation. Annu Rev Biochem. 1983;52:441–466. doi: 10.1146/annurev.bi.52.070183.002301. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Rutherford T. R., Partington G. A. Transcriptional analysis of human zeta globin genes. EMBO J. 1984 Jul;3(7):1533–1540. doi: 10.1002/j.1460-2075.1984.tb02007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]