Abstract
Amyloid diseases, including Alzheimer's disease, are characterized by aggregation of normally functioning proteins or peptides into ordered, β-sheet rich fibrils. Most of the theories on amyloid toxicity focus on the nuclei or oligomers in the fibril formation process. The nuclei and oligomers are transient species, making their full characterization difficult. We have isolated toxic protein species that act like an oligomer and may provide the first evidence of a stable reactive species created by disaggregation of amyloid fibrils. This reactive species was isolated by dissolving amyloid fibrils at high pH and it has a mass >100 kDa and a diameter of 48 ± 15 nm. It seeds the formation of fibrils in a dose dependent manner, but using circular dichroism and deep ultraviolet resonance Raman spectroscopy, the reactive species was found to not have a β-sheet rich structure. We hypothesize that the reactive species does not decompose at high pH and maintains its structure in solution. The remaining disaggregated insulin, excluding the toxic reactive species that elongated the fibrils, returned to native structured insulin. This is the first time, to our knowledge, that a stable reactive species of an amyloid reaction has been separated and characterized by disaggregation of amyloid fibrils.
Introduction
Amyloid diseases are characterized by the transformation of proteins and peptides into ordered β-sheet containing fibrils. The formation of the fibrils is suspected to proceed through a nucleation-elongation pathway that is characterized by a lag phase, where nuclei form and proceed to oligomer formation, culminating in fibril elongation and growth (1). Researchers are continuing to search for oligomers and the nuclei that initiate fibril formation because these entities may be the cause of amyloid toxicity and disease. Small Aβ oligomers have been shown to impair synaptic plasticity (2), create holes in cells to cause calcium leakage (3), and disrupt cellular communication (4). Yet to the contrary, amyloid fibrils have been shown to pierce cellular membranes causing cell death (5), demonstrating conflicting information on the oligomer toxicity debate. This debate will not be satisfied until an amyloid nucleus and oligomers can be stably formed, isolated, and studied to better understand their structure, function, reactivity, and toxicity.
The structure and function of amyloid oligomers has eluded us, due to their transient and heterogeneous nature, but we do have evidence of oligomers. Multiple amyloid structures that are believed to be oligomers have been imaged with atomic force microscopy (AFM) and they appear to have a common round shape with a hollow center (6). Two different groups have identified the nucleus of insulin amyloid formation to contain six monomers (7,8), although the actual shape of nucleus is still under debate. Recent evidence shows that hexamer nuclei are common in nature, but the preferred arrangement of the hexamers is not circular or a compact equilibrium structure, but a more irregular structure (9). Electrospray mobility analysis demonstrated that insulin jumps from a trimer to fibrils without a detectable hexamer intermediate (10). Recent in silico predictions confirm this possibility using phase-diagram analysis (11). The manipulation of solution pH, ionic strength, and temperature has been shown to create metastable oligomers of Aβ (12,13), but according to Dobson and others, it is unclear if metastable oligomers formed in this manner lead to fibril formation (14).
Without direct evidence of nuclei and oligomers, indirect methods have given some insight into the structure of oligomeric species from different peptides and proteins. Glabe and co-workers have found antibodies that bind preferentially to Aβ oligomers over fibrils or monomers (15). Toxic oligomers of HypF-N amyloid peptide have more solvent-accessible hydrophobic domains than do nontoxic oligomers (16), as has also been shown for Aβ (17) and superoxide dismutase protein (SOD1) (18), found in amyotrophic lateral sclerosis.
Amyloid fibrils are known to be mechanically strong and stable. They are known to give structural integrity to biofilms (19) and have the strength of steel and the stiffness of silk (20). Fibril can be cleaved and fibril breakage has been suspected as a method of fibril renewal and propagation (21) and recently, sonication has been used to create uniform fibrils for study (22). Yet only a few studies have reported dissolving amyloid fibrils, and with only harsh conditions like guanidine HCl (23), DMSO (24), and ionic liquids (25). The stability of insulin fibrils was recently explored and demonstrated to be pH dependent, with a disaggregation of fibrils occurring above a pH of 8 (26). We confirm here that high pH can dissolve insulin amyloid fibrils. Native protein is released from the fibril, but a small species remains in a conformation that seeds insulin fibril formation, is toxic to cells, and has not been fully characterized.
In this study, we used the amyloid protein insulin. It is a commonly used in vitro amyloid system that in harsh conditions (65°C and pH 1.6) creates long, linear amyloid fibrils (1,27). Insulin is also a known component of amyloid plaques in injection-localized insulin amyloidosis (28). Insulin fibrils have been studied to understand their structure and the critical region of the protein involved in the cross β-sheet core of the fibrils (29). Knowing the core amino acids aides in the understanding of the nucleus before fibril formation, but it does not give firm evidence of the nucleus or oligomers. We have formed and recovered a reactive and toxic species from insulin fibrils by disaggregating the fibrils using high pH and removing a large amount of insulin that refolded into normally folded insulin. This reactive species could be a building block of the fibril formation, it could be created during dissolution, or it could be carried by the fibril during the fibril formation/dissolution process. The reactive species has no-detectable β-sheet content and is not affected by the high pH environment. The remaining insulin monomers/dimers, that exhibit near-native state behavior, can be separated from the reactive species. We speculate that the reactive species exists during the formation of insulin fibrils and has a distinct structure and stability compared with the native insulin that elongates the fibril.
Materials and Methods
Materials
The insulin was a generous gift from Novo Nordisk (Gentofte, Denmark) and the membranes used for diafiltration were a generous gift from Millipore (Billerica, MA). All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise mentioned and all solutions were filtered with a 0.22 μm filter before use.
Aggregation kinetics
Before insulin aggregation, the insulin was pretreated by pH cycling (30). This reduced the amount of preformed aggregates in the insulin solution and improved reproducibility. The insulin was then diluted to 2 mg/ml in insulin buffer (100 mM NaCl, 25 mM HCl, pH 1.6) and the pH adjusted with 10 M HCl and heated to 65°C. Analysis of fibril formation was performed on a Hitachi U-2000 UV-Vis Spectrophotometer (Hitachi High Technologies America, San Jose, CA) by measurement of the absorbance at 600 nm. The data were empirically fit to a sigmoidal curve to determine the lag time and apparent rate constant (1,31).
Fibril disaggregation
Fibrils were washed with insulin buffer and concentrated to 20 mg/ml. Sodium hydroxide (1 M) was added until the pH reached 11.0 (±0.2). The solution remained at 22°C overnight until the solution was clear. The disaggregated fibrils were centrifuged at 12,500 rpm in an accuSpin Micro centrifuge (Fisher Scientific, Waltham, MA) for 15 min to remove any remaining fibrils.
Diafiltration
Toxic insulin reactive species were separated by diafiltration using an Amicon stirred filtration cell equipped with a solvent reservoir and regenerated cellulose membranes with a 100 kDa MWCO from Millipore. Briefly, solutions of disaggregated fibrils were taken to a pH of 3 with 10 M HCl. We preferred to perform the diafiltration using insulin buffer, but a pH of 1.6 dissolved the regenerated cellulose membranes, therefore a pH of 3 was chosen. Diafiltration buffer (100 mM NaCl, pH 3) was added to the solvent reservoir and the filtration proceeded at a transmembrane pressure of 10–15 psi for a total of 10 diavolumes. The concentration of the remaining solutions was measured by the absorbance at 280 nm and all insulin samples were assumed to have the same extinction coefficient of 4,785 M−1 cm−1 (32).
Circular dichroism
Circular dichroism (CD) spectra were obtained from a Jasco (Tokyo, Japan) 815 spectrometer at room temperature using four accumulations of spectra and then smoothing by averaging five consecutive data points. All samples contained 100 mM NaCl at the specified pH. The CD spectra were deconvoluted with DichroWeb (33) using the CDSSTR algorithm (34) and the average of reference data sets 7 and SP175 (35). The melt curves were obtained every 0.5°C from 25 to 90°C with a ramp rate of 1°C/min. The retentate melt curve was obtained at 4°C/min due to aggregation occurring at a ramp rate of 1°C/min. The melt temperature for the sigmoidal pH 11 samples was calculated as the midpoint of the transition from the lower temperature baseline to the upper temperature asymptote.
Deep ultraviolet resonance Raman
The details of Deep ultraviolet resonance Raman (DUVRR) spectroscopic measurements have been reported elsewhere (36). Briefly, a 197-nm laser beam was focused into a spinning Suprasil NMR tube containing 100 μL of sample solution. Scattered radiation was collected in backscattering geometry, dispersed using a home-built double monochromator, and detected with a liquid nitrogen-cooled CCD camera (Roper Scientific, Tucson, AZ). The spectrum was recorded for 10 min. GRAMS/AI (7.01) software (Thermo Fisher Scientific, Pittsburgh, PA) was used for Raman spectroscopic data processing. The contribution of Suprasil, solvent, and buffer was quantitatively subtracted.
Toxicity
Rat neuroendocrine cells (PC-12 adherent, ATCC) were cultured in high-glucose Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 5% fetal bovine serum, 10% horse serum, and 1% penicillin-streptomycin (12). The cells were scrapped from the flask and re-suspended at a concentration of 1.1 × 105 cells/ml and 90 μl of cell suspension was added to each well of a 96-well plate (CellBIND, Corning, Corning, NY) to adhere for 24 h. Afterward, 10 μl of sample for toxicity was added and allowed to incubate at 37°C, 5% CO2, and 100% humidity for 48 h. A solution of 5 mg/ml of thiazolyl blue tetrazolium bromide (MTT) was added at a volume of 10 μl/well and returned to the incubator for 4–6 h. The solubilization buffer (10% SDS, 0.01M HCl in water) was added at 100 μl/well and incubated for 4–16 h (37). The absorbance at 562 nm was read on a Tecan (Raleigh, N.C.) Safire2 plate reader and normalized to the pH 1.6 buffer controls. MTT reduction was defined as the absorbance of the sample divided by the absorbance of the control multiplied by 100% for a percent of the control.
Small angle neutron scattering
Small angle neutron scattering (SANS) was performed on the CG-3 beam line at Oakridge National Laboratory (Oak Ridge, TN) at room temperature in deuterated solvents. Detector to sample distances were 1.7 and 6 m. All spectra were analyzed using the Guinier analysis (7,38), as detailed in the Methods in the Supporting Material.
Results
Properties of disaggregated fibrils
Once insulin fibrils are dissolved using high pH, we hypothesized that the insulin that dissolved from fibrils would be structurally different from insulin that was not subjected to the β-sheet rich fibril state. To test this hypothesis, we examined the structural configurations of different insulin states. We used native insulin as a control and cycled it through pH 11 (Fig. 1 A) to determine if the extreme pH would change the insulin structure and ability to form amyloid fibrils. In the same manner, insulin fibrils were formed by a standard method of low pH and increased temperature for 4 h (1,31), and then followed the same procedure of pH cycling (Fig. 1 B). A fibrils solution had a >90% loss in turbidity (as measured by the absorbance at 600 nm) within 10 min of the pH increase and the presence of 100 mM NaCl increased this time to 30 min (Fig. 2 A). It is likely that the combined hydrophobic and electrostatic interactions hold the fibril structure together (29,39). The shielding of electrostatic repulsion by salt in the aggregation buffer has been shown experimentally (13) and calculated (11) to promote formation of fibrils for other amyloid proteins. This is likely due to the reduction of the electrostatic free energy of the amyloid species and delays fibril disaggregation in salt solutions.
Figure 1.
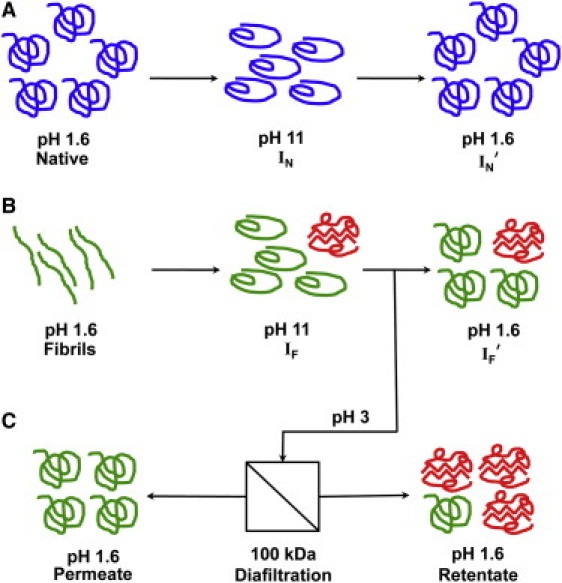
Disaggregation of insulin fibrils. (A) Native insulin was cycled to pH 11 (IN) for 1–2 min and returned to pH 1.6 (IN′) to serve as a control for the fibril disaggregation. (B) For fibril disaggregation, insulin fibrils were formed after ∼4 h at pH 1.6 and 65°C and a concentration of 2 mg/ml insulin. The fibrils were washed and the pH increased to 11(for 16 h). The fibrils were fully disaggregated and any remaining fibrils were removed by centrifugation (IF), they were returned to pH 1.6 (IF′) and stored for several days at this condition. (C) Fibrils that were disaggregated were separated at pH 3 with diafiltration using a 100 kDa synthetic membrane to removed native insulin (permeate) and concentrate the oligomers (retentate).
Figure 2.
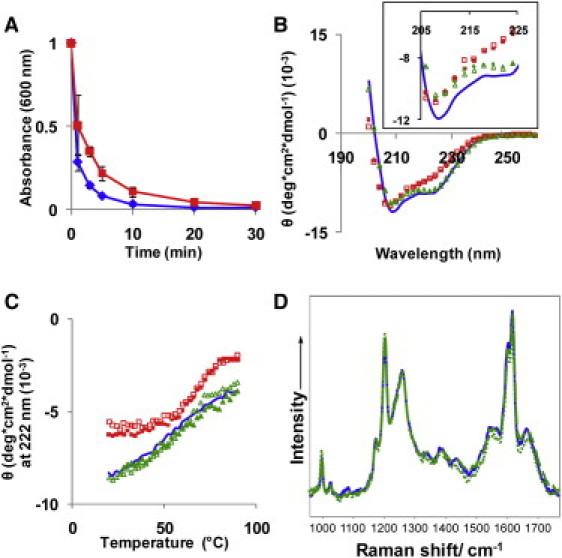
Disaggregated insulin fibrils (IF and IF′) cannot be distinguished from native insulin that had been pH-cycled (IN and IN′). (A) The disaggregation of insulin fibrils takes ∼10 min and is slower (30 min) in the presence of 100 mM NaCl (red squares) compared with no salt present (blue diamonds). The secondary structure of insulin that had been in fibrils (IF′) was compared with native insulin that had not previously been in fibrils (IN′) using B and C CD and D DUVRR. The inset in B shows an enlarged view of the range that is critical for α-helix content, 208 and 223 nm (41). By subjecting insulin to pH 11, there was some loss of native insulin structure, but this appears to be only due to the high pH since insulin that had (IF) and had not (IN) previously been in a fibril contained the same minor loss of structure. Key: (B and C) Native insulin pH 1.6 (blue solid line), IN (red solid squares), IF (red open squares), IN′ (green solid triangles), and IF′ (green open triangles). (D) Native insulin pH 1.6 (blue solid line), IN′ (green long dashes), and IF′ (green dots). Please refer to Fig. 1 for details.
By subjecting native insulin to a pH of 11, there was an 18% (±7%) reduction in α-helix content and a 12% (±7%) reduction in β-sheet structure between the native insulin and the insulin that had cycled through pH 11 and returned to pH 1.6, which was determined by deconvoluting the CD curve (Fig. 2 B) using DichroWeb (33). This reduction in α-helix and β-sheet content was reflected in an increase in disordered structure. Similar to this pH effect, we have previously shown that hen egg lysozyme, an α-helix-rich protein like insulin, loses α-helix content to random coil (disordered) content on exposure to hydrophobic interfaces in less than a few seconds (40). There was a distinct CD spectrum for insulin samples at pH 11 and those at pH 1.6, regardless of whether the samples had been previously in the β-sheet rich fibrils. This change in structure of insulin with pH is confirmed by Pocker and Biswas (41) and shown in the inset in Fig. 2 B. Insulin has been shown to have great structural flexibility within a pH range of 1–7 (39) and this most likely continues until a pH of 12, where insulin is chemically denatured and does not return to folded insulin when the pH is lowered. The gradual increase of molar ellipticity with temperature of the pH 1.6 insulin (Fig. 2 C) suggests a quaternary structure change in the insulin from hexamer to dimer to monomer (42,43). For pH 11 insulin, the protein is most likely already a dimer, and it retains a classic sigmoidal melt curve with a melt temperature of 67°C (±2°C). The melt temperature is the same whether the insulin had been in fibrils (IF) or not (IN). The structure of the insulin regardless if it had previously been in fibrils was also predicted to be similar with DUVRR spectroscopy (Fig. 2 D and Fig. S1 in the Supporting Material), which is a sensitive measure of β-sheet content (44,45).
We attempted to disrupt the disulfide bond interactions and intermolecular noncovalent bonds within the disaggregated insulin to better understand the source of stability of the seeding structure. Neither the addition of 5 mM DTT, 5 mM DTT with 10 min heating at 60°C, nor 3 M guanidine HCl was able to change the seeding behavior of the disaggregated insulin fibrils (data not shown). This suggests that either strong hydrogen bonding or strong hydrophobic interactions stabilized these aggregates and required >3 M guanidine HCl to disrupt the seeding action of the stable reactive species from the disaggregated fibrils.
Although the structure of the insulin appeared to be similar whether the protein had previously been in fibrils or not, there was evidence that something had changed in the insulin that had been in fibrils. Insulin that had previously been in fibrils (IF and IF′) had a Thioflavin-T (ThT) fluorescence, whereas insulin that had been pH-cycled (IN and IN′) did not (Fig. 3 A). A non-β-sheet rich structure of an Aβ mutant, as shown by CD, has also been shown to have a ThT signal (17). Some of the difference in IF and IF′ is due to the fluorescence intensity difference in ThT with pH (Fig. S2), but it is likely that a change in secondary structure, as shown by the CD in Fig. 2 B, is also responsible for the intensity difference. The samples with a ThT fluorescence intensity >1 decreased the lag time of a new insulin aggregation run in a dose-dependent manner (Fig. 3 B and Fig. S3), whereas the samples with a ThT intensity <1 did not change the lag time of a new insulin run (Fig. S3). This evidence supports the hypothesis that a part of the fibril, or a reactive species associated with the fibril, did not dissolve at high pH and remained in solution. We pursued the separation and analysis of this entity that was causing both a ThT signal and a decrease in lag time to produce fibrils.
Figure 3.
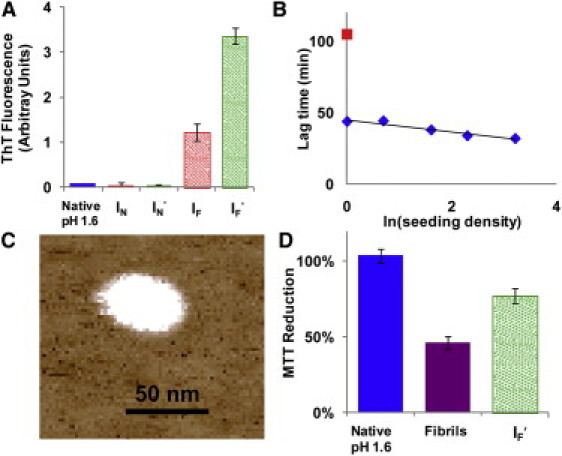
Amyloid characteristics of disaggregated insulin. (A) Thioflavin-T (ThT) fluorescence of insulin samples shows that disaggregated insulin (IF and IF′) has a ThT signal, whereas native insulin (IN and IN′) does not. (B) Seeding of IF′ into a new kinetic run reduces the lag time in a dose-dependent fashion (blue diamonds). For reference, the control without seeding is shown as a red square. (C) AFM of IF shows large aggregates 48 ± 15 nm in diameter at pH 11. (D) Fibrils and disaggregated fibrils (IF′) demonstrate cellular toxicity by an MTT reduction using PC-12 cells, whereas insulin that has not been in fibrils (Native insulin) does not reduce MTT. All MTT samples were added at pH 1.6, but the cell media buffered the samples to pH 7.4.
Large aggregates from disaggregated insulin were discovered using AFM imaging (Fig. 3 C). The aggregates were 48 ± 15 nm in diameter and fairly symmetrical in nature. They were also flat, with an average height of 3 ± 1 nm, compared to fibrils with a height of 3–4 nm. These same shaped aggregates have also been found in solutions of native insulin at pH 1.6 that had been at 65°C for 90 min (just before the onset of fibril formation) along with small, thin protofibrils (Fig. S4). This suggests that the aggregates found in the disaggregated insulin are also found in the lag time samples, at least as related to size of the aggregates. The size of the aggregates appears to be very large, but these large structures have been seen by others (46,47). It is possible that the core of these large aggregates contains the core of the fibril and the aggregate rearranges to form fibrils. Another possibility is that fibrils use the surface of the aggregate as a nucleation site and then break away from the aggregate as they elongate (47,48). Whatever the mechanism, these aggregates are not found in solutions before heating to 65°C, and therefore must be formed from the protein during the lag phase of fibril formation.
The insulin solution after disaggregation of fibrils is toxic, as determined by the MTT cell proliferation assay, using rat adrenal gland cells (PC-12 adherent), but does not reduce MTT as much as insulin fibrils (Fig. 3 D). There are many reports that amyloid fibrils are not toxic to cells (12,49,50), whereas different cell lines demonstrate toxicity toward fibrils (5,51,52). The disaggregated insulin is also toxic to cells, whereas insulin that has not been in fibrils, or exposed to 65°C for extended periods is not toxic. Oligomers of amyloid formation are known to be toxic to cells (53,54), and so this supports the hypothesis that the reactive species from the disaggregated insulin fibrils are early formed species involved in fibril formation.
Removal of insulin seeding activity
Because the aggregates in the disaggregated insulin were 48 ± 15 nm in size, we removed them with a 0.22 μm filter. This treatment removed all seeding activity from the disaggregated insulin (Fig. 4 A). The aggregates imaged were smaller than the 220 nm nominal mean pore size of the filter, but the filter likely acted as a depth filter and the aggregates adhered to the pore walls of the filter, thus removing them from solution. The absorbance at 280 nm before and after filtration did not change measurably; therefore, no perceptible amount of dissolved protein was removed from solution. To better quantify the removal of the aggregates from solution, the average size of the particles in solution was measured with SANS (Fig. 4, B and C). The disaggregated samples (IF and IF′) had an average hydrated radius of gyration (Rg) of 3.93 ± 0.21 and 3.12 ± 0.23 nm, respectively. This is much smaller than the 50–100 nm aggregates observed with AFM (Fig. 3 C) and this is likely because the aggregates are very low in concentration, as observed by no perceptible change in protein concentration when they are removed with a 0.22 μm filter. When the solution was filtered (0.22 μm), the value of Rg was reduced to 1.07 ± 0.05 nm, which is comparable with the native insulin at 1.27 ± 0.01 nm and the theoretical value of 1.4 nm (7). The Rg and molecular mass were calculated using the Guinier analysis, which is detailed in the SI Text, in Fig. S4 and summarized in Fig. 4 C.
Figure 4.
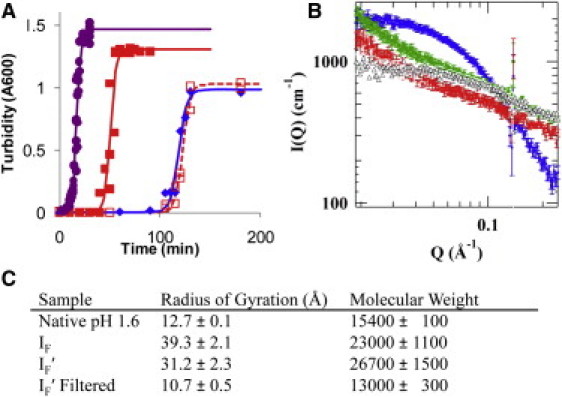
Removal of seeding behavior by 0.22 μm filtration of disaggregated insulin. (A) The kinetics of fibril formation of 2 mg/ml insulin, pH 1.6 and 65°C, with 1% fibrils (purple circles), 1% disaggregated insulin (IF′) (red squares), 1% of 0.22 μm filtered disaggregated insulin (open squares) and no seed control (blue diamonds). Empirical line fits were used to determine the lag time and kinetic elongation rate (1,31). Filtration with a 0.22 μm filter was able to remove any seeding behavior from the disaggregated insulin (IF′) (B) SANS spectra for IF (red), IF′ (green), IF′ filtered with a 0.22 μm filter (open black), and native insulin (blue). (C) Results from SANS spectra that were analyzed with the Guinier analysis, which only examines the low Q range.
Separation and analysis of insulin seeding structure
The removal of the entity that seeds the kinetic reaction with filtration led us to the hypothesis that we could separate this seeding structure for further analysis. Diafiltration was performed with a 100 kDa molecular mass cut-off membrane (Fig. 1 C); any protein smaller than 100 kDa would pass through the membrane and anything larger would be withheld by the membrane. We hypothesized that any insulin that was in dimer (11.6 kDa) to hexamer (34.8 kDa) form would pass through the membrane, whereas larger reactive species would be retained. This was a method to concentrate and purify the reactive species for further characterization. After passing 10 diavolumes of insulin buffer at pH 3 through the membrane (one diavolume is defined as the volume of liquid in the system at the start), the two fractions were characterized (Fig. 5). The retentate, or the fraction that was unable to pass through the 100 kDa membrane, maintained the seeding ability of the disaggregated insulin (Fig. 5 A) and was measured as 1% of the total protein mass by UV/vis spectroscopy, assuming the same absorption coefficient as native insulin. The measurable protein content shows that the retentate was not pure insulin reactive species and most likely contained native insulin. When samples were subjected to >10 diavolumes, the protein content continued to drop and CD and DUVRR spectra could not be obtained. The permeate, or the fraction that passed through the membrane, did not seed the kinetic reaction and had the same lag time as the control insulin that had not been in fibrils or pH cycled to pH 11. Through this purification process of removing native insulin from the sample, we were able to observe a difference in the CD spectra (Fig. 5 B) of the retentate and the permeate, but only a small rise in the Amide I, Amide II, and Cα-H bands in the DUVRR spectra (55) (Fig. 5 C). The CD spectrum of the retentate showed a loss of 35% (±10%) of the α-helix and within error there was not a change in β-sheet structure, with the α-helix loss mainly being compensated by an increase in unordered structure (34), which was determined by deconvoluting the CD curve using DichrowWeb (25). The latter conclusion was supported by the DUVRR spectroscopy because the small changes evident in Raman spectra (Fig. 5 B) were indicative of α-helix melting and formation of unordered structures, which might include turns (45). The important observation was the lack of any noticeable contribution from a newly formed β-sheet (45). Noteworthy here, CD and Raman spectroscopes are complimentary techniques because CD is most sensitive to α-helix structure and Raman to β-sheet. High specificity and selectivity of DUVRR spectroscopy to β-sheet conformation has allowed for monitoring directly and characterizing the nucleation kinetics during lysozyme fibrillation (56). As a result, we concluded that the seeding structure was more unordered than native insulin and contained little newly formed β-sheet, certainly not more than a few percent relative to the overall secondary structure compassion (the detection limit of DUVRR spectroscopy).
Figure 5.
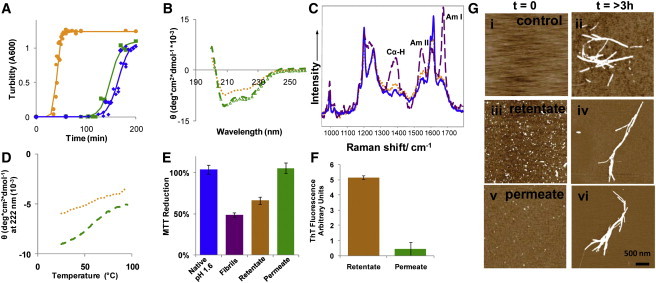
Separation of seeding behavior using diafiltration. Insulin was separated by diafiltration as shown in Fig. 1C. (A) The kinetics of fibril formation as described in Fig. 4 with 1% retentate (orange circles), 1% permeate (green squares), and no seed control (blue diamonds). (B) CD spectra show the retentate (orange dots) looses α-helix and permeate (green dashes) have the same structure as the disaggregated insulin (IF′) (open green triangles) at pH 1.6. (C) DUVRR shows little change in the β-sheet structure of the retentate (orange dots) as compared to native insulin (blue solid line). Insulin fibrils (purple dashes) have a large amount of β-sheet structure shown by the increase in the Amide I, Amide II, and Cα-H intensity. (D) Both the retentate (orange dots) and permeate (green dashes) at pH 1.6 have a continuous melt curve, as seen by native insulin at pH 1.6 in Fig. 2C. (E) The retentate reduces MTT, whereas the permeate does not and the retentate has a strong ThT signal, whereas (F) the permeate does not. All samples produce insulin fibrils with similar morphology (i.e., a height of 3–4 nm and a length of 1–3 μm) after 4 h of incubation (G ii, iv, and vi). The time zero samples (i, iii, and v) showed aggregates in iii the retentate and v the permeate samples but not (i) the control sample.
Permeate continued to display qualities of native insulin, including no toxicity (i.e., no reduction in MTT signal) (Fig. 5 E) and a minimal ThT signal (Fig. 5 F), whereas the retentate displayed all of the qualities of an insulin fibril oligomer (Fig. 5, E and F and Fig. S6). Even though the time to formation was different, the fibrils formed from the native insulin, the seeded retentate, and the seeded permeate all had similar fibril thickness (3–4 nm) and a length between 1 and 3 μm. The only differences were the amount of measurable material in the zero time images (Fig. 5 G). We have shown that regardless of the time it takes for insulin fibrils to form, the same size and molecular mass oligomer or nucleus is involved in the process (7), which may explain that although the oligomers are present at different times during the reaction, the same species is responsible for fibril formation and leads to the same fibril structure.
Discussion
Some reported methods of dissolving amyloid fibrils have used guanidine HCl (23), which is a strong protein denaturant, and destabilizing osmolyte, DMSO (24), and ionic liquids (25). Lipids have also been shown to break down Aβ fibrils into toxic protofibrils (57), as well as high pH (26). Only in the case of the ionic liquid was protein activity recovered. However, for the study of lysozyme amyloid fibrils dissolution with ionic liquids, the focus of the study was on the large amount of lysozyme that remained active, but there may have been a small amount of lysozyme that did not refold into normally active lysozyme. All of the solvents, with exception to DMSO (58), greatly disrupt ionic or charge interactions of proteins (59), including high pH. High pH was able to disrupt most of the fibril interactions and release close to native folded insulin, demonstrating that the fibril interactions are strongly electrostatic (60). The model by Eisenberg and co-workers show two different interactions dominate the insulin fibril core, a dry steric zipper that is dominated by hydrophobic interactions and a water containing layer that is likely hydrogen bonding (29). By disruption of the hydrogen-bonding layer, the fibril appears to dissociate and release insulin. Yet from our study, there appears to be a reactive species that is not disturbed under high pH conditions.
Using pH to dissolve insulin fibrils led to two populations of insulin: one that seeded the reaction and decreased the lag time and one that did not affect the reaction. The majority of the insulin (>99%) is in the later population and had no noticeable effect on the seeding reaction. The release of active peptides from amyloid fibrils has been demonstrated for many different hormones (52), but the activity of the nonnatively folded peptides was not examined, if they existed. Here, we focused on the small population that does seed the insulin fibril formation reaction, not the released insulin. It appears that the seeding phenomenon is caused by a large structure of insulin that is ∼48 ± 15 nm in diameter (Fig. 3 C). We hypothesize that this large structure is a reactive species and is stable at pH 11. The origin of this species is likely from one of three possibilities, as shown schematically in Fig. 6, either the fibrils rearrange during dissolution and form the reactive species, the reactive species is an integral part of the fibrils, or the reactive species is present in solution and associated with the fibrils. This reactive species is not β-sheet rich, but contains rather disordered protein structure. Recently, stable oligomers of Aβ have been formed with either the addition of surfactants (61) or by the stabilization of a β-hairpin using engineered disulfide bonds (14). Both of these structures contain some form of β-sheet structure, yet they were formed in a different manner than we have formed here. Dobson and co-workers have shown that the β-hairpin constrained formation does not form fibrils and state that it is possible for high molecular mass aggregates, which are 19–25 nm in diameter and bind to the A11 antibody, to form fibrils (14). We may have produced what Dobson and co-workers term high molecular weight aggregates in our experiments by high pH dissolution of insulin fibrils. Both species have minimal β-sheet content and are considered unstructured. Similar high molecular weight aggregates have also been found in vivo (15), leading us to suggest that these aggregates may be oligomers that are on the pathway to fibril formation and incorporated into or adsorbed onto the fibrils. In the past, we have used cooling experiments to demonstrate that insulin oligomers, when removed from the fibrillation reaction and cooled to room temperature for 1–28 days, continue to form nuclei that are unable to from fibrils at the colder temperature, but easily do so when seeded at the original high temperature (31). We have also shown through seeding experiments that fibrils prefer to grow asymmetrically and usually at a constant rate (32). This suggests that the reactive species isolated here from fibril disaggregation are formed during the lag time, but have not been previously studied because a β-sheet rich structure are often being sought for study, not the structure reported here. As stated by others, many different aggregates are formed from amyloid proteins and not all form fibrils (14,62,63). This reverse-engineering approach of dissolving fibrils created structures that either preexist in or on the fibril or are created during the process of fibril dissolution and seed the reaction to form additional fibrils.
Figure 6.

Cartoon of suggested insulin fibril formation and disaggregation. Native insulin at pH 1.6 and 65°C undergoes a molecular rearrangement to reactive species during the fibril formation lag time, shown in the upper right. The process continues at 65°C until some event causes the fibrils to elongate. Finally, an asymptotic phase is reached when no free insulin is left in solution to elongate the fibrils. When the fibrils are placed at pH 11 and 20°C, there appears to create a solution that is similar, if not the same, as the reactive species formation phase found during the lag time. We reach this conclusion by the same sized aggregates that are imaged in prefibrillar and disaggregated samples (Fig. S4B and Fig. 3C) and that the disaggregated solution is toxic (Fig. 3D). We hypothesize that either i), the fibrils are uniform or the reactive species are refolded during the disaggregation process, ii), the fibrils contain the reactive species and the reactive species are released during high pH disaggregation, or iii), the reactive species are associated with the fibrils and are stable and do not dissolve at high pH.
Acknowledgments
We thank Arne Staby, Novo Nordisk, for the generous gift of insulin and Millipore for the gift of membranes. We also thank Marimar Lopez for assistance with the CD, Peter Tessier, Jonathan Dordick, Ali Reza A. Ladiwala, and Jason C. Lin for assistance with the toxicity and ThT assays, and Shuo Qian, Qui Zhang, and Sai Venkatesh Pingali at Oakridge National Laboratory for assistance with the SANS experiments. The SANS measurements conducted at Oak Ridge National Laboratory's High Flux Isotope Reactor were sponsored by the Scientific User Facilities Division, Office of Basic Energy Sciences, U. S. Department of Energy.
This work was funded in part by the Department of Energy (DE-FE02-90ER14114 and DE-FE02-001ER46429) (G.B), National Science Foundation (CTS-94-00610) (G.B), and National Institutes of Health (R01AG033719) (I.K.L).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Aging or the National Institutes of Health.
Footnotes
Caryn L. Heldt's present address is Department of Chemical Engineering, Michigan Technological University, Houghton, MI.
Supporting Material
References
- 1.Nielsen L., Khurana R., Fink A.L. Effect of environmental factors on the kinetics of insulin fibril formation: elucidation of the molecular mechanism. Biochemistry. 2001;40:6036–6046. doi: 10.1021/bi002555c. [DOI] [PubMed] [Google Scholar]
- 2.Shankar G.M., Li S.M., Selkoe D.J. Amyloid-beta protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat. Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LaFerla F.M. Calcium dyshomeostasis and intracellular signalling in Alzheimer's disease. Nat. Rev. Neurosci. 2002;3:862–872. doi: 10.1038/nrn960. [DOI] [PubMed] [Google Scholar]
- 4.Pigino G., Morfini G., Brady S. Disruption of fast axonal transport is a pathogenic mechanism for intraneuronal amyloid beta. Proc. Natl. Acad. Sci. USA. 2009;106:5907–5912. doi: 10.1073/pnas.0901229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedrich R.P., Tepper K., Fändrich M. Mechanism of amyloid plaque formation suggests an intracellular basis of Abeta pathogenicity. Proc. Natl. Acad. Sci. USA. 2010;107:1942–1947. doi: 10.1073/pnas.0904532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quist A., Doudevski I., Lal R. Amyloid ion channels: a common structural link for protein-misfolding disease. Proc. Natl. Acad. Sci. USA. 2005;102:10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nayak A., Sorci M., Belfort G. A universal pathway for amyloid nucleus and precursor formation for insulin. Proteins. 2009;74:556–565. doi: 10.1002/prot.22169. [DOI] [PubMed] [Google Scholar]
- 8.Vestergaard B., Groenning M., Svergun D.I. A helical structural nucleus is the primary elongating unit of insulin amyloid fibrils. PLoS Biol. 2007;5:e134. doi: 10.1371/journal.pbio.0050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng G., Arkus N., Manoharan V.N. The free-energy landscape of clusters of attractive hard spheres. Science. 2010;327:560–563. doi: 10.1126/science.1181263. [DOI] [PubMed] [Google Scholar]
- 10.Pease L.F., III, Sorci M., Belfort G. Probing the nucleus model for oligomer formation during insulin amyloid fibrillogenesis. Biophys. J. 2010;99:3979–3985. doi: 10.1016/j.bpj.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmit J.D., Ghosh K., Dill K. What drives amyloid molecules to assemble into oligomers and fibrils? Biophys. J. 2011;100:450–458. doi: 10.1016/j.bpj.2010.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ladiwala A.R.A., Lin J.C., Tessier P.M. Resveratrol selectively remodels soluble oligomers and fibrils of amyloid Abeta into off-pathway conformers. J. Biol. Chem. 2010;285:24228–24237. doi: 10.1074/jbc.M110.133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stine W.B., Jr., Dahlgren K.N., LaDu M.J. In vitro characterization of conditions for amyloid-beta peptide oligomerization and fibrillogenesis. J. Biol. Chem. 2003;278:11612–11622. doi: 10.1074/jbc.M210207200. [DOI] [PubMed] [Google Scholar]
- 14.Sandberg A., Luheshi L.M., Härd T. Stabilization of neurotoxic Alzheimer amyloid-beta oligomers by protein engineering. Proc. Natl. Acad. Sci. USA. 2010;107:15595–15600. doi: 10.1073/pnas.1001740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kayed R., Head E., Glabe C.G. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 16.Campioni S., Mannini B., Chiti F. A causative link between the structure of aberrant protein oligomers and their toxicity. Nat. Chem. Biol. 2010;6:140–147. doi: 10.1038/nchembio.283. [DOI] [PubMed] [Google Scholar]
- 17.Bolognesi B., Kumita J.R., Yerbury J.J. ANS binding reveals common features of cytotoxic amyloid species. ACS Chem. Biol. 2010;5:735–740. doi: 10.1021/cb1001203. [DOI] [PubMed] [Google Scholar]
- 18.Tiwari A., Liba A., Hayward L.J. Metal deficiency increases aberrant hydrophobicity of mutant superoxide dismutases that cause amyotrophic lateral sclerosis. J. Biol. Chem. 2009;284:27746–27758. doi: 10.1074/jbc.M109.043729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romero D., Aguilar C., Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. USA. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith J.F., Knowles T.P.J., Welland M.E. Characterization of the nanoscale properties of individual amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2006;103:15806–15811. doi: 10.1073/pnas.0604035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carulla N., Caddy G.L., Dobson C.M. Molecular recycling within amyloid fibrils. Nature. 2005;436:554–558. doi: 10.1038/nature03986. [DOI] [PubMed] [Google Scholar]
- 22.Chatani E., Lee Y.-H., Goto Y. Ultrasonication-dependent production and breakdown lead to minimum-sized amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2009;106:11119–11124. doi: 10.1073/pnas.0901422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vernaglia B.A., Huang J., Clark E.D. Guanidine hydrochloride can induce amyloid fibril formation from hen egg-white lysozyme. Biomacromolecules. 2004;5:1362–1370. doi: 10.1021/bm0498979. [DOI] [PubMed] [Google Scholar]
- 24.Hirota-Nakaoka N., Hasegawa K., Goto Y. Dissolution of beta2-microglobulin amyloid fibrils by dimethylsulfoxide. J. Biochem. 2003;134:159–164. doi: 10.1093/jb/mvg124. [DOI] [PubMed] [Google Scholar]
- 25.Byrne N., Angell C.A. Formation and dissolution of hen egg white lysozyme amyloid fibrils in protic ionic liquids. Chem. Commun. (Camb.) 2009;9:1046–1048. doi: 10.1039/b817590j. [DOI] [PubMed] [Google Scholar]
- 26.Malisauskas M., Weise C., Morozova-Roche L. Lability landscape and protease resistance of human insulin amyloid: a new insight into its molecular properties. J. Mol. Biol. 2010;396:60–74. doi: 10.1016/j.jmb.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 27.Knowles T.P.J., Waudby C.A., Dobson C.M. An analytical solution to the kinetics of breakable filament assembly. Science. 2009;326:1533–1537. doi: 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- 28.Dische F.E., Wernstedt C., Watkins P.J. Insulin as an amyloid-fibril protein at sites of repeated insulin injections in a diabetic patient. Diabetologia. 1988;31:158–161. doi: 10.1007/BF00276849. [DOI] [PubMed] [Google Scholar]
- 29.Ivanova M.I., Sievers S.A., Eisenberg D. Molecular basis for insulin fibril assembly. Proc. Natl. Acad. Sci. USA. 2009;106:18990–18995. doi: 10.1073/pnas.0910080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heldt C.L., Sorci M., Belfort G. Detection and reduction of microaggregates in insulin preparations. Biotechnol. Bioeng. 2011;108:237–241. doi: 10.1002/bit.22902. [DOI] [PubMed] [Google Scholar]
- 31.Sorci M., Grassucci R.A., Belfort G. Time-dependent insulin oligomer reaction pathway prior to fibril formation: cooling and seeding. Proteins. 2009;77:62–73. doi: 10.1002/prot.22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heldt C.L., Zhang S., Belfort G. Asymmetric amyloid fibril elongation: a new perspective on a symmetric world. Proteins. 2011;79:92–98. doi: 10.1002/prot.22861. [DOI] [PubMed] [Google Scholar]
- 33.Whitmore L., Wallace B.A. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 34.Compton L.A., Johnson W.C.J., Jr. Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal. Biochem. 1986;155:155–167. doi: 10.1016/0003-2697(86)90241-1. [DOI] [PubMed] [Google Scholar]
- 35.Janes R.W. Reference datasets circular dichroism and synchrotron radiation circular dichroism spectroscopy of proteins. In: Wallace B.A., Janes R.W., editors. Modern Techniques in Circular Dichroism and Synchrotron Radiation Circular Dichroism Spectroscopy. IOS Press; Amsterdam, The Netherlands: 2008. pp. 183–201. [Google Scholar]
- 36.Lednev I.K., Ermolenkov V.V., Xu M. Deep-UV Raman spectrometer tunable between 193 and 205 nm for structural characterization of proteins. Anal. Bioanal. Chem. 2005;381:431–437. doi: 10.1007/s00216-004-2991-5. [DOI] [PubMed] [Google Scholar]
- 37.Heldt C.L., Hernandez R., Carbonell R.G. A colorimetric assay for viral agents that produce cytopathic effects. J. Virol. Methods. 2006;135:56–65. doi: 10.1016/j.jviromet.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 38.Guinier A., Fournet G. Wiley; New York: 1955. Small-Angle Scattering of X-Rays. [Google Scholar]
- 39.Haas J., Vöhringer-Martinez E., Grubmüller H. Primary steps of pH-dependent insulin aggregation kinetics are governed by conformational flexibility. ChemBioChem. 2009;10:1816–1822. doi: 10.1002/cbic.200900266. [DOI] [PubMed] [Google Scholar]
- 40.Sethuraman A., Han M., Belfort G. Effect of surface wettability on the adhesion of proteins. Langmuir. 2004;20:7779–7788. doi: 10.1021/la049454q. [DOI] [PubMed] [Google Scholar]
- 41.Pocker Y., Biswas S.B. Conformational dynamics of insulin in solution. Circular dichroic studies. Biochemistry. 1980;19:5043–5049. doi: 10.1021/bi00563a017. [DOI] [PubMed] [Google Scholar]
- 42.Tiyaboonchai W., Woiszwillo J., Middaugh C.R. Insulin containing polyethylenimine-dextran sulfate nanoparticles. Int. J. Pharm. 2003;255:139–151. doi: 10.1016/s0378-5173(03)00055-3. [DOI] [PubMed] [Google Scholar]
- 43.Hua Q.-X., Jia W., Weiss M.A. A protein caught in a kinetic trap: structures and stabilities of insulin disulfide isomers. Biochemistry. 2002;41:14700–14715. doi: 10.1021/bi0202981. [DOI] [PubMed] [Google Scholar]
- 44.Lednev I.K. Vibrational spectroscopy: biological applications of ultraviolet Raman spectroscopy. In: Uversky V.N., Permyakov E.A., editors. Protein Structures, Methods in Protein Structures and Stability Analysis. Nova Science Publishers; New York: 2007. pp. 1–26. [Google Scholar]
- 45.Shashilov V.A., Sikirzhytski V., Lednev I.K. Quantitative methods for structural characterization of proteins based on deep UV resonance Raman spectroscopy. Methods. 2010;52:23–37. doi: 10.1016/j.ymeth.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 46.Podestà A., Tiana G., Manno M. Early events in insulin fibrillization studied by time-lapse atomic force microscopy. Biophys. J. 2006;90:589–597. doi: 10.1529/biophysj.105.068833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bouchard M., Zurdo J., Robinson C.V. Formation of insulin amyloid fibrils followed by FTIR simultaneously with CD and electron microscopy. Protein Sci. 2000;9:1960–1967. doi: 10.1110/ps.9.10.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krebs M.R.H., Bromley E.H.C., Donald A.M. The mechanism of amyloid spherulite formation by bovine insulin. Biophys. J. 2005;88:2013–2021. doi: 10.1529/biophysj.104.051896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novitskaya V., Bocharova O.V., Baskakov I.V. Amyloid fibrils of mammalian prion protein are highly toxic to cultured cells and primary neurons. J. Biol. Chem. 2006;281:13828–13836. doi: 10.1074/jbc.M511174200. [DOI] [PubMed] [Google Scholar]
- 50.Vilasi S., Iannuzzi C., Sirangelo I. Effect of trehalose on W7FW14F apomyoglobin and insulin fibrillization: new insight into inhibition activity. Biochemistry. 2008;47:1789–1796. doi: 10.1021/bi701530w. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y.B., Schubert D. Steroid hormones block amyloid fibril-induced 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) formazan exocytosis: relationship to neurotoxicity. J. Neurochem. 1998;71:2322–2329. doi: 10.1046/j.1471-4159.1998.71062322.x. [DOI] [PubMed] [Google Scholar]
- 52.Maji S.K., Perrin M.H., Riek R. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Datki Z., Juhász A., Penke B. Method for measuring neurotoxicity of aggregating polypeptides with the MTT assay on differentiated neuroblastoma cells. Brain Res. Bull. 2003;62:223–229. doi: 10.1016/j.brainresbull.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 54.Lee S., Fernandez E.J., Good T.A. Role of aggregation conditions in structure, stability, and toxicity of intermediates in the Abeta fibril formation pathway. Protein Sci. 2007;16:723–732. doi: 10.1110/ps.062514807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu M., Ermolenkov V.V., Lednev I.K. Hen egg white lysozyme fibrillation: a deep-UV resonance Raman spectroscopic study. J Biophotonics. 2008;1:215–229. doi: 10.1002/jbio.200710013. [DOI] [PubMed] [Google Scholar]
- 56.Sikirzhytski V., Topilina N.I., Lednev I.K. Genetic engineering combined with deep UV resonance Raman spectroscopy for structural characterization of amyloid-like fibrils. J. Am. Chem. Soc. 2008;130:5852–5853. doi: 10.1021/ja8006275. [DOI] [PubMed] [Google Scholar]
- 57.Martins I.C., Kuperstein I., Rousseau F. Lipids revert inert Abeta amyloid fibrils to neurotoxic protofibrils that affect learning in mice. EMBO J. 2008;27:224–233. doi: 10.1038/sj.emboj.7601953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tjernberg A., Markova N., Hallén D. DMSO-related effects in protein characterization. J. Biomol. Screen. 2006;11:131–137. doi: 10.1177/1087057105284218. [DOI] [PubMed] [Google Scholar]
- 59.Monera O.D., Kay C.M., Hodges R.S. Protein denaturation with guanidine hydrochloride or urea provides a different estimate of stability depending on the contributions of electrostatic interactions. Protein Sci. 1994;3:1984–1991. doi: 10.1002/pro.5560031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Topilina N.I., Sikirzhytsky V., Welch J.T. Charge distribution and amyloid fibril formation: insights from genetically engineered model systems. Biomacromolecules. 2010;11:1721–1726. doi: 10.1021/bm901467a. [DOI] [PubMed] [Google Scholar]
- 61.Yu L., Edalji R., Olejniczak E.T. Structural characterization of a soluble amyloid beta-peptide oligomer. Biochemistry. 2009;48:1870–1877. doi: 10.1021/bi802046n. [DOI] [PubMed] [Google Scholar]
- 62.Paravastu A.K., Qahwash I., Tycko R. Seeded growth of beta-amyloid fibrils from Alzheimer's brain-derived fibrils produces a distinct fibril structure. Proc. Natl. Acad. Sci. USA. 2009;106:7443–7448. doi: 10.1073/pnas.0812033106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Glabe C.G. Structural classification of toxic amyloid oligomers. J. Biol. Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


