Abstract
The recently discovered aging-dependent large accumulation of point mutations in the human fibroblast mtDNA control region raised the question of their occurrence in postmitotic tissues. In the present work, analysis of biopsied or autopsied human skeletal muscle revealed the absence or only minimal presence of those mutations. By contrast, surprisingly, most of 26 individuals 53 to 92 years old, without a known history of neuromuscular disease, exhibited at mtDNA replication control sites in muscle an accumulation of two new point mutations, i.e., A189G and T408A, which were absent or marginally present in 19 individuals younger than 34 years. These two mutations were not found in fibroblasts from 22 subjects 64 to 101 years of age (T408A), or were present only in three subjects in very low amounts (A189G). Furthermore, in several older individuals exhibiting an accumulation in muscle of one or both of these mutations, they were nearly absent in other tissues, whereas the most frequent fibroblast-specific mutation (T414G) was present in skin, but not in muscle. Among eight additional individuals exhibiting partial denervation of their biopsied muscle, four subjects >80 years old had accumulated the two muscle-specific point mutations, which were, conversely, present at only very low levels in four subjects ≤40 years old. The striking tissue specificity of the muscle mtDNA mutations detected here and their mapping at critical sites for mtDNA replication strongly point to the involvement of a specific mutagenic machinery and to the functional relevance of these mutations.
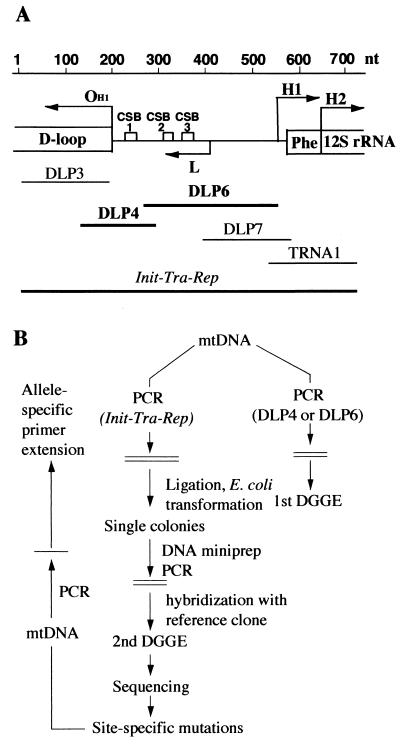
Recently, the discovery of an aging-dependent large accumulation of point mutations in the control region for mtDNA replication of human skin fibroblasts has been reported (1). Particularly striking was the demonstration, in a generally high proportion of molecules (up to 50%), of a T to G transversion at position 414 in the original Cambridge sequence (2), within the promoter for the synthesis of the RNA primer of mtDNA heavy (H)-strand synthesis (3) and for light (L)-strand transcription (4). This mutation was present in more than 50% of the individuals above 65 years of age and absent in younger individuals. In the present work, to investigate the occurrence of these aging-dependent mutations in other cell types, in particular, in postmitotic cells, a screening was carried out for the detection in human muscle of aging-related specific point mutations in the DLP4 and DLP6 segments of the main mtDNA control region, which were the segments previously found to carry the fibroblast mtDNA mutations (1). These two mtDNA segments correspond to one of the hypervariable portions of the main control region (5), and were chosen, for the purpose of analysis, as containing each a uniform melting domain (Y.M., unpublished data). They carry critical sequences for mtDNA replication (Fig. 1A). In particular, DLP4 contains the primary origin of H-strand mtDNA synthesis (OH1) (3), whereas DLP6 contains the promoter and start site for H-strand replication RNA primer synthesis (6) and the two evolutionarily conserved sequence blocks CSB2 and CSB3 (7).
Figure 1.
(A) Shown are the portion of the main control region of human mtDNA containing the initiation sites for rRNA-encoding DNA (rDNA; H1) and whole H-strand transcription (H2) and for L-strand transcription and synthesis of the primer of H-strand synthesis (L; ref. 6) and the primary origin for H-strand DNA synthesis (OH1; ref. 3), as well as the map positions of the CSB1, CSB2, and CSB3 (conserved sequence blocks 1, 2, and 3; ref. 7), of the DLP4 and DLP6 segments chosen for DGGE analysis in this work, of the overlapping DLP3, DLP7 (1), and TRNA1 segments (8), and of the large fragment, encompassing all of the above segments (Init-Tra-Rep), used for cloning. (B) Scheme of the approaches followed to carry out a preliminary screening of the mtDNA samples for the presence of mutations in the DLP4 and/or DLP6 segment by first-round DGGE; then, to identify and quantify the mutations by cloning of the whole Init-Tra-Rep fragment; followed by second-round DGGE and sequencing (1); and, finally, to carry out a large-scale screening of mtDNA samples for the identified mutations by allele-specific termination of primer extension (1). Phe, tRNAPhe gene.
In the present study, a large-scale analysis of mtDNA from biopsied or autopsied muscle samples showed the absence or minimal presence of the T414G fibroblast mtDNA mutation. By contrast, surprisingly, the aging-dependent accumulation of two new mutations, i.e., A189G and T408A, which were virtually absent in fibroblasts and, in a few individuals tested, also in other tissues, was detected in the muscle of most of 30 individuals 53 to 92 years old, which included four with partial muscle denervation. These two new mtDNA mutations, which occurred at the same critical sites for replication previously shown to be targeted by mutations in fibroblasts, were absent or marginally present in 23 individuals younger than 40 years.
Materials and Methods
Source of Tissue Samples.
One large group of tissue samples (group A) was obtained by biopsy or at autopsy from genetically unrelated individuals without any history of neuromuscular diseases. In particular, muscular biopsy samples (quadriceps) were obtained from 40 healthy volunteers who were free of any acute or chronic illness, and had normal blood cell counts and chemistry panels. Henceforth, these and other individuals used as sources of material are designated by their age in years (y), followed in some cases by a number to distinguish multiple individuals of the same age. The 40 volunteers mentioned above included 31 males [19 y, African-American (AA); 20 y-1, Caucasian (C); 20 y-2, C; 20 y-3, C; 21 y-1, C; 21 y-2, AA; 22 y-1, Indian; 22 y-2, Hispanic (H); 22 y-3, C; 24 y-1, C; 24 y-2, C; 24 y-3, C; 25 y-1, C; 25 y-2, C; 26 y, C; 28 y, C; 30 y-1, Native American; 32 y, H; 33 y, C; 34 y, C; 60 y-1, C; 60 y-2, C;, 61 y, C; 62 y, C; 65 y-1, C; 65 y-2, C; 65 y-3, C; 70 y-1, H; 70 y-2, C; 73 y, C; 74 y, C; and 75 y, C], and nine females, all Caucasian (76 y; 77 y-1; 77 y-2; 78 y; 81 y; 82 y; 83 y; 84 y-2; and 92 y). These biopsies were obtained during the course of studies reviewed and approved by the Institutional Review Boards of Charles Drew University of Medicine and Science and Harbor-University of California at Los Angeles Research and Education Institute, Los Angeles, CA, and of Washington University School of Medicine, St. Louis, MO. The autopsy samples of muscle and other tissues came from five individuals who also had no known clinical history of neuromuscular diseases [53 y, female (F), C, deltoid muscle (m); 79 y, F, H, intercostal m., heart, liver, spleen; 84 y-1, male (M), C, intercostal m., psoas m., liver, lymph node, spleen; 88 y, M, C, deltoid m.; and 90 y, F, H, deltoid m., skin]. An additional group (group B) of muscle samples (quadriceps) came from diagnostic biopsies, performed, with informed consent, on eight patients with clinical polyneuropathy, all of whom had mild to prominent denervation, with partial reinnervation (30 y-2, M, Chinese; 39 y-1, F, C; 39 y-2, F, Japanese; 40 y, F, C; 80 y, F, C; 81 y-1, M, C; 81 y-2, M, C; and 81 y-3, F, C). The fibroblast mtDNA samples used in the present work were previously described (1), except those from three fibroblast cultures obtained from the NIGMS Human Genetic Mutant Cell Repository (Camden, NJ) [29 y (GMO2674A) and 96 y (GM00731A)] and from the National Institute on Aging Cell Repository (Camden, NJ) [75 y-2 (AG13348)].
DNA Extraction and mtDNA Purification.
Nucleic acid isolation from frozen biopsied muscle samples was carried out by homogenization in the TRI Reagent (Molecular Research Center, Cincinnati) with a Brinkmann homogenizer (model PT10/35), and extraction from the homogenate according to the manufacturer's recommendations. Isolation of total nucleic acids from frozen autopsied muscle samples was performed by homogenization with a pestle and mortar in liquid N2, followed by proteinase K/SDS treatment and phenol/chloroform extraction.
Highly purified mtDNA was isolated from total nucleic acids by nearly complete digestion of nuclear DNA and nucleocytosolic RNA with the restriction enzymes BglII and DraIII, which do not cut mtDNA, with ribonuclease A, and with exonuclease III, as previously described (1).
Denaturant Gradient Gel Electrophoresis (DGGE).
First-round DGGE analysis of the PCR products of the DLP4 and DLP6 segments of highly purified mtDNA (Fig. 1A) was carried out, as previously described (1, 8), for the identification of the mtDNA fragments carrying mutations.
Cloning, Second-Round DGGE, and Sequencing.
The mtDNA samples selected by the first-round DGGE analysis were used to clone the large Init-Tra-Rep fragment, which contains all of the initiation sites for transcription and replication of mtDNA, and, in particular, encompasses the overlapping DLP3, DLP4, DLP6 (1), and TRNA1 (8) segments (Fig. 1A). This cloning, second-round DGGE of the PCR products of the DLP4 and DLP6 segments of the Init-Tra-Rep plasmid clones, and sequencing of selected secondary PCR products (Fig. 1B) were performed as previously detailed (1).
Allele-Specific Termination of Primer Extension.
Quantification of the specific point mutations was carried out by allele-specific termination of primer extension, with minor modifications of the original protocol (9). The following combinations of template, oligodeoxynucleotides, and deoxynucleoside and dideoxynucleoside triphosphates were used for each specific point mutation: (i)[T414G], template, DLP6; primer, 5′GGTGACTGTTAAAAGTGC (positions 434–417 in Cambridge sequence); dATP, dGTP, dTTP, ddCTP; (ii) [A189G], template, DLP4; primer, 5′-GCATTAATTAATTAACACACTTTAGTAAG (positions 222–194); dATP, dGTP, dTTP, ddCTP; and (iii) [T408A], template, DLP6; primer, 5′-CCAGCCTAACCAGATTTCAAATTTTATC (positions 377–404); dCTP, dGTP, dTTP, ddATP.
The DLP4 or DLP6 segments were amplified by PCR with the Expand High Fidelity PCR System (Boehringer Mannheim), and the amplified fragments were separated from free nucleotides. The template DNA and the corresponding gel-purified 5′-32P-labeled primer were mixed at a 1:1 molar ratio in a solution containing Sequenase buffer, 6.7 mM DTT, 100 μM dNTPs, and 100 μM ddNTP (dideoxynucleoside 5′-triphosphate). The mixtures were overlaid by mineral oil, heated at 100°C for 5 min, slowly cooled down to 65°C (at rate of −1°C/min), kept at 65°C for 10 min, and then chilled to 0°C. After addition of 1 μl of 1:8 dilution of Sequenase version 2.0 (United States Biochemical), the samples were incubated at 45°C for 5 min. The products were sequentially denatured at 100°C for 5–20 min in the presence of 18% formamide, loaded and separated on a 50-cm-long 20% polyacrylamide-6 M urea gel. The intensity of the bands was quantified by using a PhosphorImager and the imagequant program.
Results
For a preliminary analysis of the DLP4 and DLP6 mtDNA segments, muscle samples were obtained from biopsy material of three healthy individuals 70 to 74 years of age and from autopsy material of four individuals 53 to 88 years of age. All these individuals and others analyzed subsequently, who lacked any known clinical history of neuromuscular disease, are referred to as belonging to group A (see Materials and Methods). DGGE analysis revealed mutations in the DLP4 and DLP6 segments of all mtDNA samples tested. To identify and quantify these mutations, and at the same time to determine the colinearity of the mutations occurring in the DLP4 and DLP6 segments of each mtDNA sample, the large fragment Init-Tra-Rep, which contains all of the initiation sites for transcription and replication of mtDNA, and, in particular, encompasses DLP4 and DLP6 (Fig. 1A), was PCR-amplified from each selected mtDNA sample and cloned in Escherichia coli (1). After separate PCR amplification of the DLP4 and DLP6 segments from 48 Init-Tra-Rep plasmid clones derived from each mtDNA sample, a second round of DGGE analysis and subsequent sequencing of the secondary PCR products were carried out (ref. 1; Fig. 1B).
As shown in Table 1 (which is published as supplemental data on the PNAS web site, www.pnas.org), none of the specific point mutations found in the DLP4 segment of fibroblast mtDNA (1) was detected in the corresponding cloned mtDNA segment from the eight muscle samples analyzed. As mentioned above, these samples included biopsy tissue from individuals 70 y-1, 73 y, and 74 y, and autopsy tissue from individuals 53 y, 79 y, 84 y-1, and 88 y. The most striking observation was of an accumulation of an A to G transition at position 189 (2), not previously found in fibroblasts (1). It occurred in 15% to 42% of the mtDNA molecules of all muscle samples, except that from 79 y. In the latter individual, the intercostal muscle sample exhibited an A189C transversion in all its cloned DLP4 segments, presumably an inherited polymorphism, as previously described (10). At position 185, a G to A transition was found in a large proportion of mtDNA molecules (31%) in 74 y, whereas a G187A transition in the same individual and an A insertion at position 191 in 73 y were found, in both cases, in single clones. These findings pointed to the sequence 185–191 being a likely hot spot for mutations in muscle mtDNA.
The T414G transversion and the other specific point mutations previously detected in DLP6 of fibroblast mtDNA (1) were also not found in the cloned DLP6 segments from all of the eight muscle samples analyzed (Table 1). However, most surprising was finding a new T to A transversion at position 408, present in 2% to 11% of the mtDNA in all samples, except that from 74 y. There was no correlation between the frequency of the T408A mutation and that of the A189G mutation in the same mtDNA samples (Table 1). In the five cloned muscle mtDNA samples, derived from autopsy tissue of individuals 53 y, 84 y-1 (intercostal and psoas muscles), and 88 y, and from biopsy tissue of individual 70 y-1, which carried both the A189G and the T408A mutations (Table 1), only two Init-Tra-Rep mtDNA clones [from 84 y-1 (i) and 84 y-1 (p)], of a total of 8 clones exhibiting the T408A mutation, were found to carry at the same time the A189G transition. Besides the T408A transversion, a T407C transition (2%), a C411G transversion (2%) and a C411A transversion (2%) were found in some muscle samples (Table 1). These results pointed to the 407–411 sequence as being another hot spot for mutations in muscle mtDNA. Also observed in the sample from 79 y were a few variations in length of the homopolymeric (polyC) tract HT D310 (11), a few random point mutations or single nucleotide insertions, and a 27-nt duplication (Table 1).
In other experiments, the DLP4 and DLP6 segments of quadriceps muscle mtDNA from ten biopsied individuals 19 to 34 years old of Group A were screened by first-DGGE for heteroplasmic point mutations. The ten mtDNA samples that exhibited the highest amount of abnormal bands in the PCR-amplified DLP4 or DLP6 segments were further analyzed by the cloning-sequencing approach described above. This analysis revealed the presence of the A189G mutation in 6–8% of mtDNA from three individuals (21 y-1, 33 y, and 34 y), and of an A189C transversion in 83% of mtDNA from a 28-year-old individual, while failing to detect the presence of the T408A transversion in three individuals (21 y-1, 28 y, and 34 y).
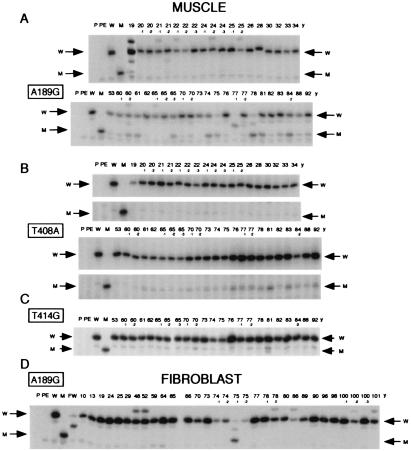
The discovery, by the DGGE-cloning-sequencing analysis described above, of an aging-dependent accumulation of what appeared to be specific nucleotide substitutions in muscle mtDNA at positions 189 and 408 opened the way to using the method of allele-specific termination of primer extension (1, 9) for rapid screening of a large number of muscle samples for these mutations (Fig. 1B). Fig. 2A shows such an analysis of the A189G mutation in highly purified mtDNA from biopsy muscle samples of 40 genetically unrelated male or female individuals between 19 and 92 years old, and from autopsy muscle samples of individuals 53 y and 88 y. All these individuals lacked any known history of neuromuscular diseases. It appears that, whereas the muscle mtDNA samples from most of the 23 individuals older than 53 years exhibited a clear signal of varying intensity, and often strong relative to the wild-type signal, corresponding to the A189G transition, only three of the mtDNA samples from the 19 individuals younger than 34 years exhibited a very faint signal (19 y, 21 y-2, and 24 y-3).
Figure 2.
Autoradiograms showing the electrophoretic patterns obtained in the analysis of different mutations by allele-specific termination of primer extension. (A) Detection of the A189G mutation in DLP4 segments PCR-amplified from muscle mtDNA of individuals without any history of neuromuscular disease (group A, see Materials and Methods), comparing 19 individuals aged 19 to 34 years and 23 individuals aged 53 to 92 years old. (B) Detection of the T408A mutation in DLP6 segments amplified from muscle mtDNA of the individuals indicated in A. A section of the gel has been cut out for space considerations. The extended primer, which terminated at the site of the mutation, corresponds to the lower band of each doublet (indicated by M arrow), whereas the upper band is a spurious product visible also in the lane for the wild-type DLP6 (W). (C) Detection of the T414G mutation in DLP6 fragments amplified from muscle mtDNA of the individuals indicated in A. (D) Detection of the A189G mutation in DLP4 segments amplified from fibroblast mtDNA of 32 individuals from 20-week fetal (FW) to 101 years old. In A and D, some samples exhibited consistently in repeated runs a relatively minor band migrating slower than the extended primer terminated on the wild-type template (W), which, most likely, resulted from a conformational change in the template associated with a polymorphism(s) in DLP4; this hypothesis is supported by the sequencing data for individuals 22 y-1 and 25 y-2 (data not shown), for 79 y (Table 1), and for 48 y (1), and by the presence of the same band in multiple tissues from 79 y and 90 y (see below, Fig. 3). In A, C, and D, the weak abnormal bands migrating faster than the mutant or wild-type band, but which were present also in the PE lane and/or W lane (see below), were due to spurious products. In the lanes for 19 y, 21 y, 60 y-2, 75 y, 77 y-1, 77 y-2, and 78 y of A and in the lanes for FW and 75 y-1 of D, a band [migrating approximately mid-way between the extension product corresponding to the wild-type sequence (terminating at position 187) and that corresponding to the mutant sequence (terminating at position 189)] represents the extended primer prematurely terminated on the wild-type template, due presumably to an A to G transition at position 188. The number above each lane represents the age of the individual analyzed. P, primer; PE, primer + Sequenase; W and M, primer extension products obtained on plasmid DNA carrying a cloned DLP4 or DLP6 fragment with wild-type and, respectively, mutant sequence.
Fig. 2B shows the primer extension analysis of the T408A transversion in the same muscle mtDNA samples tested above. It appears that almost all of the patterns from the 23 individuals 53 to 92 years old exhibited a clear band of varying intensity corresponding to the T408A mutation. By contrast, the mtDNA samples from 19 individuals younger than 34 years did not exhibit the same mutation, or carried only traces of it. In agreement with the results of the DGGE-cloning-sequencing experiments discussed above, primer extension analysis of the fibroblast-specific T414G transversion showed that the 23 individuals aged 53 to 92 years did not carry this mutation in muscle mtDNA, or carried it in minimal amounts (Fig. 2C).
Previous work (1) had failed to detect the presence of the A189G mutation in fibroblasts from four old individuals and ten individuals ≤48 years of age (1). In full confirmation of that result, the analysis of fibroblast mtDNA samples from 32 differently aged individuals, of whom 22 were 64 to 101 years of age (all listed in ref. 1, except 29 y, 75 y-2, and 96 y), by allele-specific termination of primer extension revealed the absence of the A189G transition in nearly all samples (Fig. 2D). One individual (75 y-1) exhibited the mutation in 84% of mtDNA, possibly representing an inherited polymorphism (Fig. 2D). The absence of the T408A mutation in mtDNA from fibroblasts of 29 individuals had also been previously reported (1).
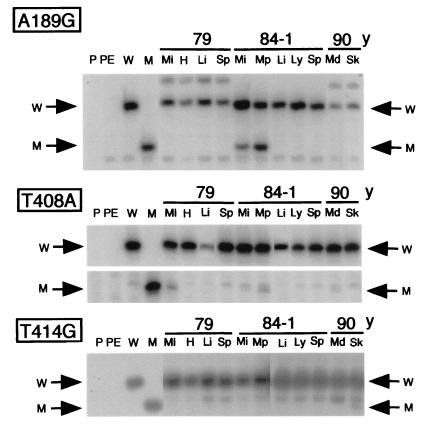
The striking tissue specificity of the aging-dependent muscle mtDNA mutations revealed by the data described above was confirmed by experiments in which different tissues from the same individual were analyzed. As shown in Fig. 3, the A189G and/or T408A mutations were present in muscle from 79 y, 84 y-1, and 90 y, but not in other tissues, whereas the T414G mutation was present only in skin.
Figure 3.
Primer extension data for the detection of the A189G, T408A, and T414G mutations in DLP4 and DLP6 segments amplified from mtDNA of different tissues of 79 y, 84 y-1, and 90 y. Mi, intercostal muscle; Mp, psoas muscle; Md, deltoid muscle; H, heart; Li, liver; Sp, spleen; Ly, lymph node; and Sk, skin. Symbols W and M are explained in Fig. 2 legend.
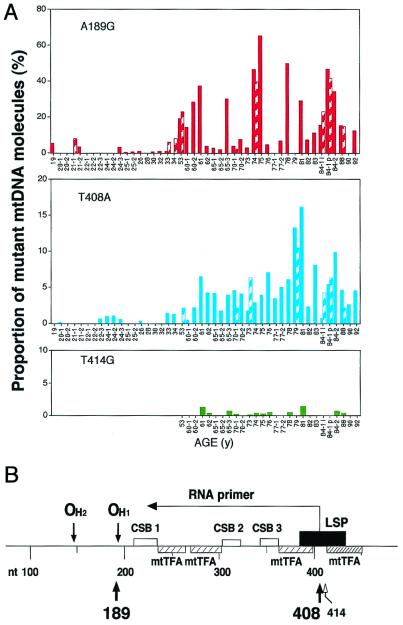
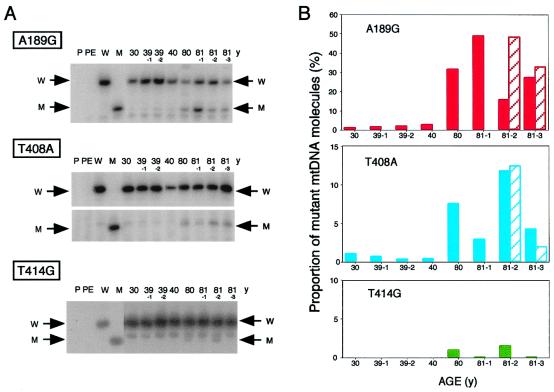
Fig. 4A shows the quantification of the specific point mutations detected in group A individuals. It appears that 14 of the 26 individuals aged 53 to 92 years (54%) carried the A189G mutation in a proportion between 11 and 64% of their muscle mtDNA molecules, and 4 more in a proportion between 5 and 10%. By contrast, the majority of the 19 younger individuals did not carry this mutation or carried it in minimal proportion of the mtDNA molecules (<1%), with only 6 individuals exhibiting it 3 to 8% of their mtDNA. The great majority of the individuals older than 53 years (73%) carried the T408A transversion in 3 to 16% of their muscle mtDNA. By contrast, this mutation was present in only 8 of the 19 young individuals, and, in these, only in <1.4% of the mtDNA molecules. From the data presented in Fig. 4, it appears that the frequencies of the A189G and T408A mutations, as determined by the primer extension method, are in substantial agreement with those obtained by the DGGE-cloning-sequencing approach (1). Fig. 4A also shows the quantification of the fibroblast-specific T414G transversion in muscle mtDNA from the 26 individuals 53 to 92 years-old. It appears that less than half of these muscle samples carried the mutation in marginal levels, with only 2 (from 61 y and 81 y) exhibiting it in >1% of the molecules (1.35 and 1.44%, respectively). In a recently reported work, by using a very sensitive method (12), the T414G mutation has been detected in some muscle mtDNA samples from old individuals, but not in brain samples; however, no quantification of the mutation in the muscle was presented.
Figure 4.
(A) Diagram summarizing the age distribution and frequency of the A189G, T408A, and T414G mutations in mtDNA from skeletal muscles of individuals of a wide range of ages of group A (see Materials and Methods). The number below each tick on the abscissae axes indicates the age of the individuals analyzed (in years). Solid bars represent the primer extension data, the striped bars, the data from the DGGE-cloning-sequencing analysis. (B) Scheme of a portion of the mtDNA main control region showing the positions of the two muscle-specific mtDNA mutations detected in the present work (thick arrows) and of the fibroblast-specific mtDNA T414G mutation (thin arrow). The positions of binding of the mitochondrial transcription factor A (the densely hatched rectangle indicates a position of high affinity binding), and the site of the promoter for L-strand transcription (LSP) are shown. Other symbols are explained in Fig. 1A legend.
In the present work, eight additional samples were tested (group B; see Materials and Methods). These samples were obtained from four older individuals (one 80 years old and three 81 years old) and four younger ones (30 to 40 years old), which exhibited symptoms of various degrees of muscle weakness, and revealed, in histochemical studies, a slight to prominent denervation, with partial reinnervation. As illustrated in Fig. 5 A and B, the muscle samples from all four older individuals showed, by primer extension analysis, an accumulation of the A189G transition in a substantial to large proportion of their mtDNA (17 to 49%), whereas this mutation was present at a very low frequency (1.5 to 3.2%) in the muscle mtDNA samples from all four younger individuals. Similarly, the samples from the four older subjects exhibited the T408A transversion (Fig. 5 A and B) in proportion ranging from 3 to 12% of the mtDNA molecules, whereas the samples from all four younger subjects showed very low levels of this mutation (0.4–1.1%). The A189G transition and the T408A transversion in 81 y-2 and 81 y-3 were also quantified by the cloning-sequencing approach, with similar results. The proportion of mtDNA molecules carrying the A189G mutation in the biopsied quadriceps muscle from the four individuals 80 years or older with partial denervation (averaging 31.2 ± 5.8%) was significantly higher than that found in the quadriceps muscle biopsied from five old normal individuals (81 y, 82 y, 83 y, 84 y-2, and 92 y), averaging 18.8 ± 4.8%. The partially denervated biopsy muscle samples from three of the four older individuals exhibited also very low levels of the T414G mutation, corresponding to 0.5 to 2.3% of mtDNA molecules.
Figure 5.
(A) Primer extension data for the detection of the A189G, T408A, and T414G mutations in mtDNA from biopsied quadriceps muscle of eight individuals exhibiting partial muscle denervation (group B in Materials and Methods). (B) Diagram summarizing the frequency of the A189G, T408A, and T414G mutations in the eight individuals. Bars are as in Fig. 4.
Discussion
The highly discriminative procedure used in the present work for the purification of mtDNA (1) and the absence of any PCR products, when total DNA from mtDNA-less ρ0206.143B cells was amplified with primers specific for the DLP4 or DLP6 segments (1) or for the Init-Tra-Rep (data not shown), would exclude any role of nuclear mtDNA pseudogenes in the results described here.
The age distribution and tissue specificity of the specific point mutations detected in the muscle mtDNA control region strongly suggest that these mutations were not inherited. The A189G transition has been previously described as polymorphisms (12). However, in the present work, none of the 54 muscle samples analyzed carried this mutation as a polymorphism. The failure to find this mutation in 29 fibroblast mtDNA samples also strongly argues against an inherited phenomenon. The presence of the A189G mutation in heteroplasmic form has been recently reported in muscle, but not in blood, brain, and heart, from 4 old individuals (13). However, no data for young individuals were presented in that paper. The T408A transversion has not been previously reported. No differences were observed in the present work in the prevalence of the two types of muscle-specific mtDNA mutations, A189G and T408A, nor in the proportion of mutated mtDNA, between men and women.
Whatever may be the mechanism underlying the first appearance of the muscle mtDNA mutations described here, e.g., oxygen radical-induced mtDNA damage, γ-DNA polymerase errors, or a phenomenon akin to the SOS response (1), an amplification of the mutations must occur. This amplification is presumably due to a replicative advantage of the mtDNA molecules carrying them, as previously shown for pathogenetic point mutations (14) and for deletions (15, 16). However, to account for the high proportion of mutated mtDNA found in muscle fibers, especially in the case of the A189G transition, one would have to assume multiple independent initiating events within mitochondria of different fibers. The observation in the present work that the muscles exhibiting partial denervation underwent a similar or, in the case of A189G, a more pronounced aging-dependent accumulation of the two muscle-specific mtDNA mutations than the muscles from healthy old individuals indicates that the aging-related mtDNA structural changes occur also in this pathological context, with the denervation or its underlying processes possibly playing a synergistic role in the genesis and/or amplification of the A189G mutation. More extensive data are, however, required to validate this possibility.
The A189G transition occurs very close to the primary site of H-strand DNA synthesis initiation in human cells, at position 191 (OH1; ref. 3; Fig. 4B). It is interesting that, in fibroblast mtDNA, the segments around both the primary and the secondary origin of DNA synthesis (at position 147; ref. 3) were previously found to be sites of an aging-dependent large accumulation of mutations (1).
The T408A transversion occurs in the middle of the promoter for mtDNA H-strand primer synthesis and for L-strand transcription, just one nucleotide downstream of the transcription start site (nucleotide 407), and at a position immediately adjacent to a segment with high affinity for the mitochondrial transcription factor A (ref. 3; Fig. 4B). It is quite significant that this position is very close to the site of the main fibroblast-specific T414G mutation, thus defining a clear mutational hot-spot segment of mtDNA. The observation in the present study that the T408A transversion occurred consistently in the DLP6 mtDNA segment of individuals 53 to 92 years of age in a much lower proportion of molecules (3 to 16%, in 73% of the individuals), as compared with the A189G mutation in the DLP4 segment (5 to 64%, in 69% of the individuals; Fig. 3), raises the possibility that the T408A mutation occurs in a minor subpopulation of muscle fibers, possibly not carrying the A189G mutation. This possibility is strongly supported by the observation of the absence of any correlation between the frequency of the T408A mutation and that of the A189G mutation in the same mtDNA samples (Figs. 4A and 5B, and Table 1). Despite this evidence, single-fiber PCR amplification of mtDNA or in situ hybridization experiments with sequence-specific oligonucleotide or PNA probes will be necessary to provide direct evidence for the occurrence of the two mutations in different muscle fibers, and, possibly, in different fiber types.
The striking tissue specificity of the aging-dependent accumulation of point mutations detected in the previous study (1) and in the present work in the main control region of mtDNA and their localization in critical sites for H-strand mtDNA synthesis strongly suggest the involvement of cell type-specific proteins or cofactors. These proteins or cofactors may affect the susceptibility of these mtDNA sequences to oxidative damage or to replicative errors, or provide replicative advantage to the mutant molecules. It is significant, in this context, that the muscle-specific and fibroblast-specific aging-related mtDNA mutations occur at or near the site of DNA attachment to the inner mitochondrial membrane (17), which is a likely site of nucleo-mitochondrial interactions. In the cited work, after Triton X-100 lysis of HeLa cell mitochondria, the great majority (≈95%) of mtDNA molecules could be isolated as carrying a protein structure near the origin of replication (17). It is plausible that this protein structure includes proteins involved in the initiation of mtDNA replication and/or in mtDNA copy number control and playing a crucial role in the nucleo-mitochondrial cross-talk relevant to the above phenomena. It will be necessary to investigate the occurrence of any such proteins and their possible role in the first appearance of the tissue-specific mutations detected in fibroblasts and muscle and/or in their aging-dependent amplification.
Supplementary Material
Acknowledgments
We thank Drs. A. Chomyn and T. Suzuki for valuable discussions, and B. Keeley and L. Bellavia for expert technical assistance. This work was supported by National Institute on Aging [National Institutes of Health (NIH)] Grant AG12117 (to G.A.); and by NIH Grant 1R01AG14369, Research Centers in Minority Institutions (RCMI) Clinical Research Initiative (P20RR11145), and RCMI Grants G12RR03026 and U54RR14616 (to S.B.).
Abbreviations
- H-strand
heavy strand
- L-strand
light strand
- DGGE
denaturant gradient gel electrophoresis
- Init-Tra-Rep
initiation sites for transcription and replication
References
- 1.Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Science. 1999;286:774–779. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- 2.Anderson S, Bankier A T, Barrell B G, de Bruijn M H, Coulson A R, Drouin J, Eperon I C, Nierlich D P, Roe B A, Sanger F, et al. Nature (London) 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 3.Ghivizzani S C, Madsen C S, Nelen M R, Ammioni C V, Hauswirth W W. Mol Cell Biol. 1994;14:7717–7730. doi: 10.1128/mcb.14.12.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montoya J, Christianson T, Levens D, Rabinowitz M, Attardi G. Proc Natl Acad Sci USA. 1982;79:7195–7199. doi: 10.1073/pnas.79.23.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenberg B D, Newbold J E, Sugino A. Gene. 1983;21:33–49. doi: 10.1016/0378-1119(83)90145-2. [DOI] [PubMed] [Google Scholar]
- 6.Attardi G. Int Rev Cytol. 1985;93:93–145. doi: 10.1016/s0074-7696(08)61373-x. [DOI] [PubMed] [Google Scholar]
- 7.Walberg M W, Clayton D A. Nucleic Acids Res. 1981;9:5411–5421. doi: 10.1093/nar/9.20.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michikawa Y, Hofhaus G, Lerman L S, Attardi G. Nucleic Acids Res. 1997;25:2455–2463. doi: 10.1093/nar/25.12.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofhaus G, Attardi G. Mol Cell Biol. 1995;15:964–974. doi: 10.1128/mcb.15.2.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kogelnik A M, Lott M T, Brown M D, Navathe S B, Wallace D C. Nucleic Acids Res. 1998;26:112–115. doi: 10.1093/nar/26.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauswirth W W, Clayton D A. Nucleic Acids Res. 1985;13:8093–8104. doi: 10.1093/nar/13.22.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murdock D G, Christacos N C, Wallace D C. Nucleic Acids Res. 2000;28:4350–4355. doi: 10.1093/nar/28.21.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calloway C D, Reynolds R L, Herrin G L, Jr, Anderson W W. Am J Hum Genet. 2000;66:1384–1397. doi: 10.1086/302844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoneda M, Chomyn A, Martinuzzi A, Hurko O, Attardi G. Proc Natl Acad Sci USA. 1992;89:11164–11168. doi: 10.1073/pnas.89.23.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsson N-G, Holme E, Kristiansson B, Oldfors A, Tulinius M. Pediatr Res. 1999;28:131–135. doi: 10.1203/00006450-199008000-00011. [DOI] [PubMed] [Google Scholar]
- 16.Khrapko K, Bodyak N, Thilly W G, van Orsouw N J, Zhang X, Coller H A, Perls T T, Upton M, Vijg J, Wei J Y. Nucleic Acids Res. 1999;27:2434–2441. doi: 10.1093/nar/27.11.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albring M, Griffith J, Attardi G. Proc Natl Acad Sci USA. 1977;74:1348–1352. doi: 10.1073/pnas.74.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.