Abstract
The transcription factor Batf controls TH17 differentiation by regulating the expression of both RORγt and RORγt target genes such as Il17. Here, we report the mechanism by which Batf controls in vivo class switch recombination (CSR). In T cells, Batf directly controls expression of the transcription factors Bcl-6 and c-Maf, both of which are needed for development of T follicular helper (TFH) cells. Restoring TFH activity to Batf−/− T cells in vivo requires co-expression of both Bcl-6 and c-Maf. In B cells, Batf directly controls the expression of both activation-induced cytidine deaminase (AID) and of IH-CH germline transcripts. Thus, Batf functions at multiple hierarchical levels across two cell types to globally regulate in vivo switched antibody responses.
Introduction
Batf and Batf3, two AP1 family transcription factors, provide distinct lineage-specific functions within the immune system1,2. Batf3 is required specifically for development of the CD8α+ lymphoid-resident dendritic cell (DC) subset responsible for cross presentation in vivo1 and of the related CD103+ peripheral DC subset3. In contrast, Batf is required for the differentiation of TH17 cells2. Batf3 expression was highly restricted to conventional dendritic cells and fully differentiated TH1 cells, but not other myeloid or lymphoid lineages. Importantly Batf3−/− T cells show no apparent defect in the development of any known T helper subset. While Batf is expressed in TH17 cells, TH1 and TH2 cells, and in activated B cells, the development of TH1 and TH2 cells, mature B cells, and all dendritic cell subsets is normal in Batf −/− mice2.
Mice lacking Batf fail to induce the TH17 transcription factor RORγt, and fail to express TH17-specific cytokines such as IL-17A2. Batf not only controls TH17 development through regulating RORγt expression, but also directly controls TH17-specific gene targets, since reconstitution of Batf −/− T cells with RORγt fails to completely restore IL-17 expression. Consistent with this observation, Batf directly binds to regulatory regions surrounding the IL-17 gene locus. The mechanism of gene regulation by Batf appears to arise from the formation of a heterodimer with Jun proteins that exerts transcriptionally unique, non-redundant actions on genes involved in the TH17 development.
Immunization of Batf −/− mice with MOG peptide fails to induce EAE in contrast to wild-type mice2, consistent with a requirement for TH17 development in EAE4. This defect is largely due to a T cell-intrinsic property of Batf −/− T cells, since transfer of wild-type T cells into Batf −/− mice restores their ability to manifest severe EAE after MOG immunization. However, the onset of disease in such mice is slightly delayed relative to wild-type mice, suggesting additional defects in Batf −/− mice beyond the defect in TH17 differentiation.
In the present study, we examined Batf −/− mice for development and activity of lymphocyte populations beyond TH17 cells. A recent study of independently generated Batf−/− mice reported loss of TFH cells, reduced antibody production for switched isotypes, and reduced expression of activation induced cytidine deaminase (AID) in B cells5. However, that study did not examined the molecular basis of the loss of TFH function in Batf−/− mice, nor identify the full range of B-cell specific defects involved in class switching. Here, we have identified several additional actions of Batf that influence both TFH function and class switching in B cells. We show that Batf is required for the expression of two major transcription factors already known to regulate TFH development, Bcl-66–8 and c-Maf9. Importantly, co-expression of both Bcl-6 and c-Maf are required to restore any TFH activity to Batf−/− T cells. In addition, we find that ectopic expression of AID in Batf−/− B cells does not restore class switching, and that Batf is also required for expression of IH-CH germline transcripts, which are a known prerequisite for isotype switching10,11. These results show that Batf functions in vivo as a global regulator of class switching through its dual requirements in TFH cells and B cells, and by operating at multiple transcriptional levels within each of these cell types.
Results
Cell-intrinsic TFH defects in Batf−/− mice
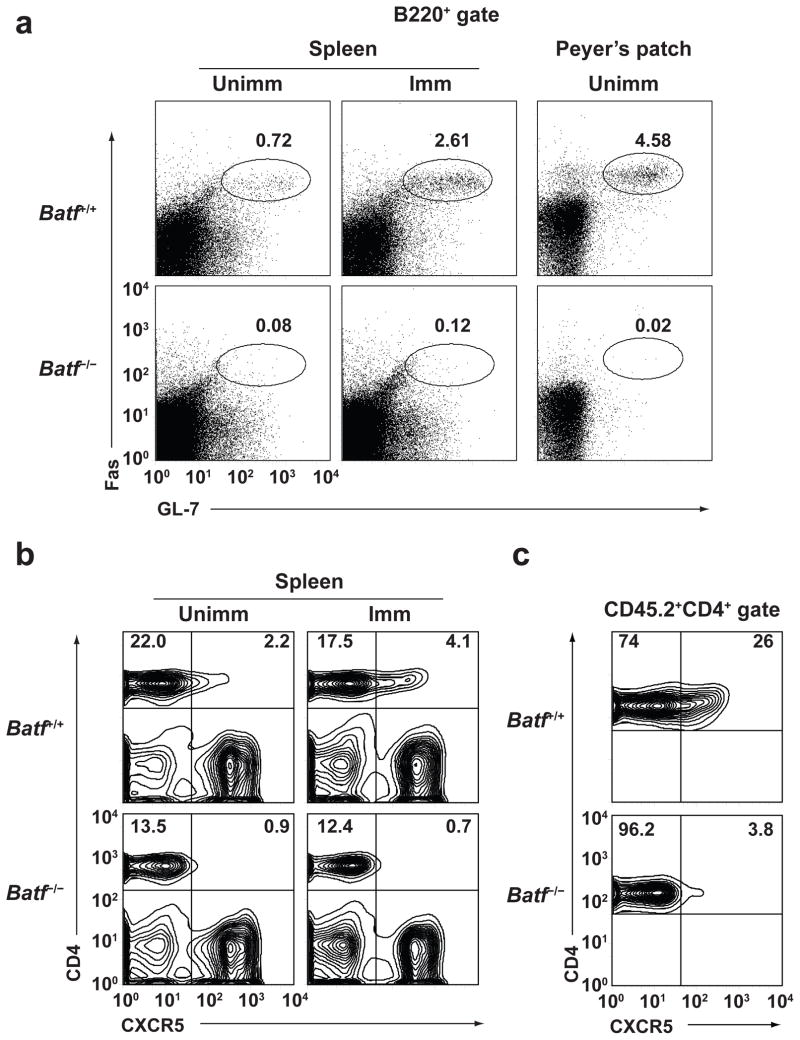
Unimmunized Batf−/− mice have slightly increased serum IgM concentrations, but greatly reduced amounts of all other isotypes (Supplementary Fig. 1). Batf −/− mice showed normal antigen-specific IgM production to T-independent TNP-Ficoll immunization and T-dependent NP-CGG immunization, but virtually absent production of antigen-specific IgG3 and IgG1 antibody, respectively (Supplementary Fig. 2) and failed to develop PNA-positive germinal centers in response to immunization with sheep red blood cells (SRBC) (Supplementary Fig. 3a). B cells in unimmunized and immunized Batf−/− mice failed to express Fas or GL7 characteristic of germinal center (GC) B cells in Spleen or Peyer’s patches (Fig. 1a, Supplementary Fig. 3b–c), while Peyer’s patch T cells lacked CXCR5 expression (Fig. 1b, Supplementary Fig. 3d) consistent with a defect in T follicular helper (TFH) cells, a CD4+ T cell subset specialized in providing B cell help12,13.
Figure 1.
Defects in germinal center B cells and TFH cells in Batf−/− mice. (a) Germinal center B cells in spleen or Peyer’s patches from unimmunized or SRBC-immunized mice were stained with anti-B220, anti-GL7, and anti-Fas. Shown are two-color histograms gated on B220+ cells. Numbers indicate the percentage Fas+GL7+ cells in the indicated gate. Data are representative of three experiments. (b) Splenocytes were stained on day 7 for CD4 and CXCR5 from mice unimmunized mice or mice immunized with SRBC. Data are representative of three experiments. (c) Flow cytometry of CD4 and CXCR5 expression in spleens of SJL mice that received transfers of either CD4+CD62L+ CD45.2+ cells from Batf+/+ and Batf−/− mice and immunized with SRBC 7 days before staining. The data are gated on CD45.2+ cells. Data are representative of two experiments.
To determine whether the loss of switched antibody in Batf−/− mice results from a defect in B cells or T cells, we assessed antibody responses after mixed adoptive transfer of wild-type or Batf−/− T cells and B cells into Rag2−/− recipients (Supplementary Fig. 4). Co-transfer of wild-type B cells and T cells restored the development of Fas+ GL7+ GC B cells in Rag2-recipients, while transfer of Batf−/− T cells and Batf−/− B cells did not. Batf−/− B cells co-transferred with wild-type T cells restored a Fas+ GL7+ phenotype to Batf−/− B cells, while wild-type B cells co-transferred with Batf−/− T cells failed to acquire a Fas+ GL7+ phenotype after immunization (Supplementary Fig. 4a). Antigen-specific IgM antibody was induced under all combinations of T and B cell co-transfer (Supplementary Fig. 4b), but both wild-type or Batf−/− B cells that were co-transferred with Batf−/− T cells failed to generate antigen-specific IgG antibody responses (Supplementary Fig. 4c), suggesting that the reduced production of switched antibodies in Batf−/− mice can result from a T-cell intrinsic defect. However, a significant (P<0.05) reduction in antigen-specific IgG produced also occurred in mice that received Batf−/− B cells co-transferred with wild-type T cells (Supplementary Fig. 4c), suggesting there is a B-cell intrinsic defect in Batf−/− mice for switched antibody responses as well.
We found that Batf−/− mice have greatly reduced CXCR5+ CD4+ T cells, not only without immunization as recently reported5, but also after immunization (Fig. 1b, Supplementary Fig. 5a). To test whether this defect was T-cell intrinsic, we transferred wild-type or Batf−/− T cells into normal SJL background recipients and measured their induction of CXCR5 after immunization (Fig. 1c and Supplementary Fig. 5c). Wild-type T cells, but not Batf−/− T cells, induced high expression of CXCR5 protein, suggesting that the defect in TFH cells in Batf−/− T mice is T-cell intrinsic.
Batf-dependent Bcl-6 and c-Maf expression by TFH cells
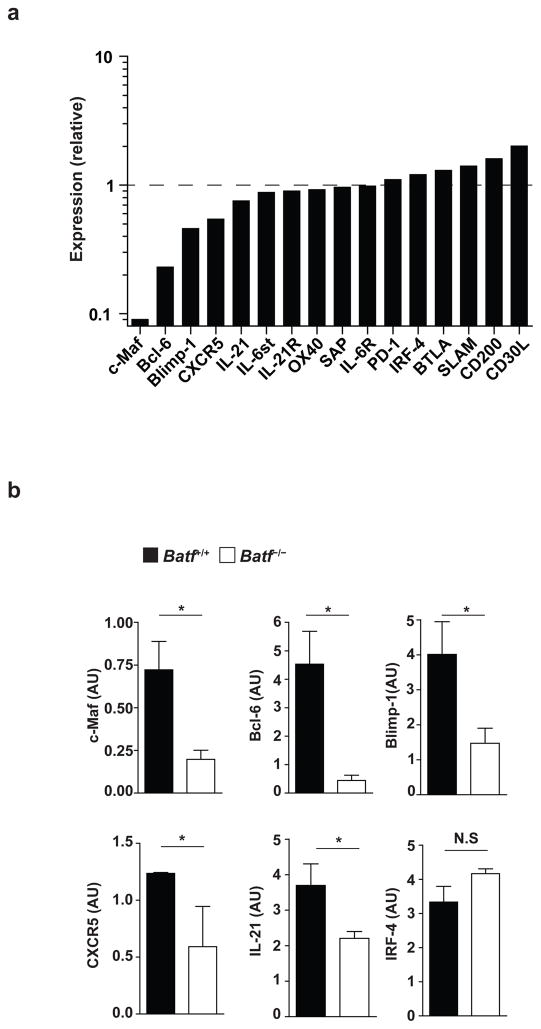
We surveyed global gene expression in wild-type and in Batf−/− CD4+ T cells on day 1 or day 3 after in vitro activation in conditions that promote differentiation of TFH-like cells14 (Supplementary Fig. 6). The transcription factors c-Maf and Bcl-6, both of which are required for TFH differentiation6–9, showed a striking reduction in Batf−/− CD4+ T cells relative to wild-type cells (Fig. 2a), each being reduced approximately 5- to 10-fold using both microarray analysis and quantitative real-time PCR (qPCR) (Fig. 2b, Supplementary Table 1). CXCR5, IL-21, and Blimp1, which is downregulated in TFH and acts as a Bcl-6 antagonist7, are only slightly reduced in Batf−/− T cells. Many genes that are highly expressed in or required for TFH development or activity, such as SAP15, IRF416, PD-117, or BTLA18, are not decreased or altered in Batf−/− T cells compared to wild-type T cells. Thus, Batf appears to control the expression of the two major TFH-specific transcription factors c-Maf and Bcl-6. Expression of IRF4, also required for TFH development16, was not altered in Batf−/− T cells, indicating that Batf controls a specific subset of the transcription factors involved in TFH development (Fig. 2a, b).
Figure 2.
Batf is required for c-Maf and Bcl-6 expression by in vitro activated T cells. (a) Relative expression levels of the indicated genes involved in follicular helper T cell development or function were determined for Batf−/− T cells by microarray analysis of Batf−/− and Batf+/+ CD4+ T cells stimulated for 3 days with anti-CD3 and anti-CD28 in the presence of IL-6, anti-IL-4, anti-IFN-γ, and anti-TGF-β. Relative values are presented from one microarray experiment with the expression in Batf+/+ CD4+ T cells defined as 1. (b) Quantitative RT-PCR analysis of c-Maf, Bcl-6, Blimp-1, CXCR5, IL-21, and IRF4 mRNA expression in Batf+/+ or Batf−/− T cells stimulated as in a. Results are normalized to HPRT mRNA expression and are presented as arbitrary unit (AU). *P<0.05 (unpaired student t-test). Data are combined from three independent experiments (mean and s.e.m.).
Batf−/− T cells were previously reported to show a defect in TH2 development based on reduced IL-4 mRNA measured by qPCR of T cells from C57BL/6 background mice5. However, we find that BALB/c background Batf−/− T cells activated in TH2-inducing conditions expressed normal levels of GATA-3 mRNA and produced wild-type amounts of IL-4 protein (Supplementary Fig. 7). These cells also exhibited normal TH1 development and the expected defect in RORγt expression and TH17 development 2.
Direct regulation of Bcl-6 and c-Maf by Batf
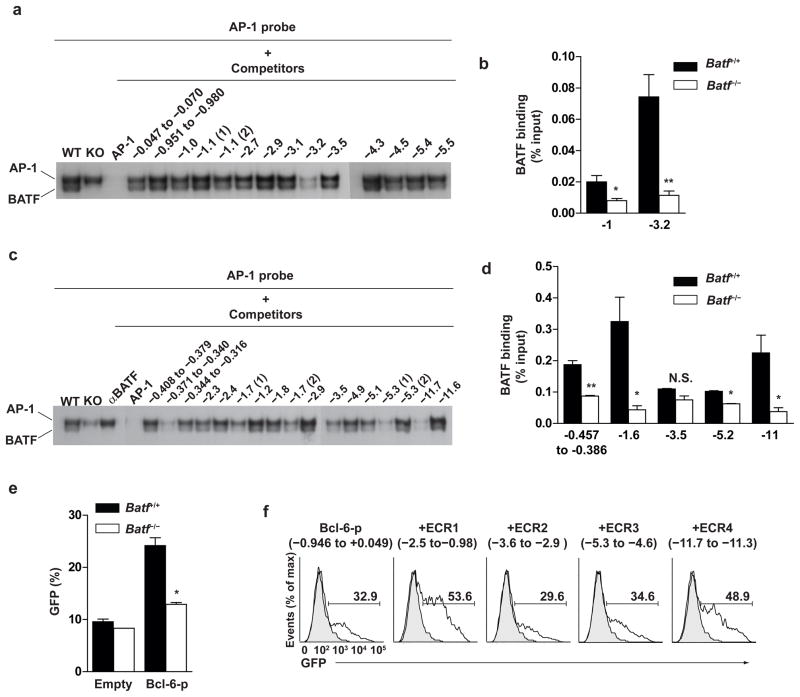
To ask whether Batf directly or indirectly controls Bcl-6 and c-Maf in T cells, we attempted to identify conserved non-coding regions in these loci that might harbor functional regulatory elements (Supplementary Fig. 8a). Within three highly conserved regions upstream of the c-Maf promoter, (−0.98 to −2.5 kb; −2.9 to −3.6 kb; and −4.6 to −5.3kb), we tested all potential TGAC/G Batf binding sites2 (Supplementary Table 2) in electrophoretic mobility shift assays (EMSA) for competition with authentic Batf (Fig. 3a). One region located at −3.2 kb efficiently competed for Batf binding. Using a chromatin immunoprecipitation (ChIP) assay, we found that this region also bound to Batf in vivo in wild-type primary T cells, but not in Batf−/− T cells, activated in the presence of IL-6 (Fig. 3b).
Figure 3.
Batf directly binds to conserved elements in the c-Maf and Bcl-6 loci in vivo. (a) Purified Batf+/+ or Batf−/− CD4+ T cells were cultured with anti-CD3 and anti-CD28 with IL-6 for 24 h and treated with PMA/ionomycin for last 4 hours. Nuclear extracts were analyzed by EMSA using a consensus AP-1 probe2. Annealed double-stranded oligonucleotides of 29 to 32 bp in length from the indicated regions of c-Maf locus were used as competitors as described2. Data are representative of two experiments. (b) CD4+ T cells were stimulated as in a and ChIP analysis of the indicated regions was performed using anti-BATF antibody2. *P<0.05 and **P<0.005 (unpaired student t-test). Data are from two independent experiments (mean and s.e.m.). (c) EMSA was performed as in a using annealed double-stranded oligonucleotides from the indicated regions of Bcl-6 locus as competitors. Data are representative of two experiments. (d) CD4+ T cells were stimulated as in a and ChIP analysis of the indicated regions of the Bcl-6 locus was performed. *P<0.05 and **P<0.005 (unpaired student t-test). Data are from two independent experiments (mean and s.e.m.). (e) CD4+ T cells were cultured with anti-CD3 and anti-CD28 in the presence of IL-6, anti-IL-4, anti-IFN-γ, and anti-TGF-β and infected with hCD4-pA GFP-RV or hCD4-pA GFP-Bcl-6-promoter-RV. hCD4 and GFP expression was examined on day 3 after stimulation. Shown are the percentages of GFP+ cells within the hCD4+ cell gate. *P<0.05(unpaired student t-test). Data are from two independent experiments (mean and s.e.m.). (f) Phoenix E cells were transfected with the Bcl-6 reporter plasmid hCD4-pA GFP-Bcl-6-promoter-RV, or Bcl-6 reporter plasmid containing the indicated 5′ conserved regions of Bcl-6 locus (ECR1, ECR2, ECR3, ECR4). hCD4 and GFP expression was examined 2 days after transfection. Shown is a single color histogram of GFP expression for hCD4+ cells. Data are representative of three experiments.
Within five highly conserved non-coding regions upstream of the Bcl-6 promoter (Supplementary Fig. 8b), we similarly tested all 17 potential Batf binding sites using EMSA. We identified five of these 17 sites that efficiently competed for Batf binding (Fig. 3c). We tested these five regions using ChIP assays, and found that three, located within the proximal promoter, at −1.6 kb, and at −11 kb, also bound to Batf in vivo (Fig. 3d). The proximal Bcl-6 promoter region, containing an in vivo Batf binding site, was also functionally responsive to Batf in primary T cells, since its activity was reduced in Batf−/− T cells relative to wild-type T cells (Fig. 3e, and Supplementary Fig. 9). Two upstream regions, located between −0.98 to −2.5 kb (ECR1) and −11.3 to −11.7kb (ECR4) and which bound Batf in vivo, also exhibited functional enhancer activity in cooperation with the proximal Bcl-6 promoter in primary T cells (Fig. 3f). In contrast, the two regions that did not bind Batf in vivo by CHIP assay also did not exhibit enhancer activity (Fig. 3f).
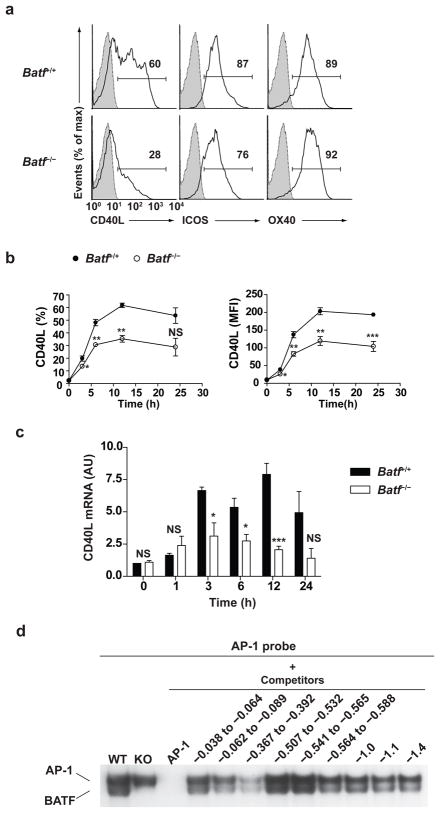
CD40 ligand (CD40L), ICOS and OX40 are receptors that are expressed by activated T cells and are involved in T-dependent activation of GC B cells19. We examined whether expression of these receptors was altered in Batf−/− T cells (Fig. 4). In T cells activated by anti-CD3 and anti-CD28, CD40L expression was selectively reduced in Batf−/− T cells relative to wild-type T cells, whereas ICOS and OX40 expression were normal (Fig. 4a). Both the frequency CD40L+ T cells (Fig. 4b, left panel) and level of CD40L surface expression (Fig. 4b, right panel) were reduced by approximately 40% in Batf−/− T cells relative to wild-type T cells. This reduction was due to decreased transcription, since CD40L mRNA was similarly reduced in Batf−/− T cells relative to controls (Fig. 4c), suggesting a possible direct involvement of Batf in its control. Within 3 conserved non-coding regions in the CD40L locus (Supplementary Fig. 8c), only 1 of 9 potential Batf binding sites, located at −367 to −392 of the proximal promoter, strongly competed for Batf bind by EMSA assay (Fig. 4d).
Figure 4.
Batf is required for optimal CD40 ligand expression. (a) Purified naïve Batf+/+ or Batf−/− CD4+ T cells were cultured with anti-CD3 and anti-CD28 for 12 h (CD40L analysis) or 24 h (ICOS and OX40 analysis) and analyzed by FACS. Shown are single color histograms for the indicated protein gated on CD4+ cells. Shaded histogram represents isotype control staining. Data are representative of three experiments. (b) CD40L expression by cells as described in (a) was analyzed by FACS at 3, 6, 12, and 24 hour after stimulation. Shown is the percentage of CD40L+ cells (left) and Mean fluorescence intensity (MFI) (right). *P<0.05, **P<0.01, and ***P<0.005 (unpaired student t-test). NS, not significant. Data are from three independent experiments (mean and s.e.m.). (c) CD40L expression by cells as described in (a) was analyzed by real-time PCR at 1, 3, 6, 12, and 24 hour after stimulation. *P<0.05 and ***P<0.005 (unpaired student t-test). NS, not significant. Data are from two independent experiments (mean and s.e.m.). (d) EMSA was performed as described in Figure 3a using the indicated double-stranded oligonucleotides from the CD40L locus as competitors. Data are representative of two experiments.
Bcl-6 and c-Maf restore Batf−/− TFH activity
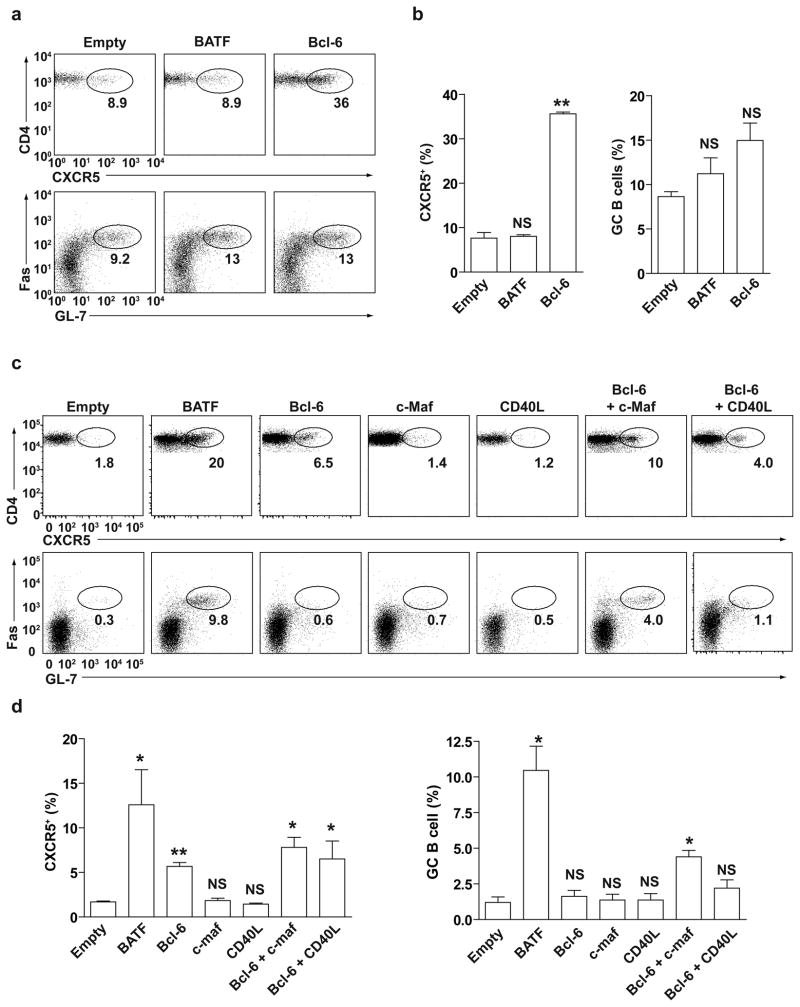
We next asked if the requirement for Batf in TFH activity was solely due to controlling Bcl-6 and c-Maf expression. We expressed various combinations of factors into wild-type or Batf−/− T cells and tested for restoration of CXCR5 expression and TFH activity in vivo after adoptive transfer into Rag2−/− mice (Fig. 5). When expressed in wild-type T cells, Bcl-6, but not Batf, increased the percentage of T cells that express CXCR5 (Fig. 5a, b), and these T cells only slightly augmented GC B cell development (Fig. 5b) as previously described6–8.
Figure 5.
Co-expression of Bcl-6 and c-Maf partially restores TFH activity in Batf−/− T cells. CD4+ T cells purified from Batf+/+ mice (a, b) or Batf−/− mice (c, d) were activated with anti-CD3 and anti-CD28 and infected with the indicated retroviruses. Infected T cells were mixed with BALB/c B cells and transferred into Rag2−/− mice subsequently immunized with SRBC. CXCR5 expression by GFP+CD4+ T cells and Fas and GL7 expression by B was analyzed by FACS at 7 days after immunization. (a) FACS analysis for CD4 and CXCR5 (upper panels) expression by Batf+/+ T-cells infected with the empty retrovirus GFP-RV (Empty), BATF-expressing (BATF) or Bcl-6-expressing (Bcl-6) retrovirus. Data shown is gated on GFP+ cells. FACS analysis for Fas and GL7 (lower panels) gated on co-transferred B220+ cells. Data are representative of two experiments. Transferred T cells were between 60% to 80% were GFP+. (b) Frequency of CXCR5+ Batf+/+ CD4+ T cells (left) or Fas+GL7+ B220+ cells (right) 7 days after immunization. **P<0.005, versus empty-RV control (unpaired student t-test). NS, not significant. Data are from two independent experiments (mean and s.e.m.). (c) FACS analysis of CD4 and CXCR5 (upper panels) expression by Batf−/− T cells infected with the indicated retroviruses; GFP-RV (Empty); BATF-GFP; Bcl-6-GFP; c-Maf-GFP; CD40L-GFP; Bcl-6-hCD4 and c-Maf-GFP; or Bcl-6-hCD4 and CD40L-GFP RV. Data shown is gated on GFP+ cells (for single infections) or GFP+hCD4+ cells (for double infections). FACS analysis of Fas and GL7 expression (lower panels) is gated on B220+ cells. Data are representative of two experiments. In double retroviral infections, roughly 50% of transferred T cells were hCD4+GFP+. (d) Frequency of CXCR5+ Batf+/+ CD4+ T cells (left) or Fas+GL7+ B220+ cells (right) 7 days after immunization. *P<0.05 and **P<0.005, versus empty-RV control (unpaired student t-test). NS, not significant. Data are from two independent experiments (mean and s.e.m.).
We next determined the activity of factors in restoring TFH function when expressed in Batf−/− T cells. Batf-expressing retrovirus restored CXCR5 surface expression in Batf−/− T cells to levels similar or greater than in wild-type T cells (Fig. 5c, upper panels, Fig. 5d) and strongly promoted the development of GC B cells as assayed by Fas and GL7 expression (Fig. 5c, lower panels, and Fig. 5d). In contrast, the Bcl-6-expressing retrovirus did not increase surface CXCR5 expression in Batf−/− T cells to extent induced by Batf, and failed to promote GC B cell development (Fig. 5c, d). Likewise, expression of c-Maf by retrovirus failed to induce either CXCR5 by Batf−/− T cells or to promote GC B cell development (Fig. 5c, d). However, co-expression of Bcl-6 together with c-Maf in Batf−/− T cells induced CXCR5 expression to amounts greater than that by Bcl-6 alone, and promoted a significant (P<0.05) increase in GC B cell development (Fig. 5c, d).
Since expression of Bcl-6 alone into Batf−/− T cells restored significant (P<0.005) amounts of CXCR5 expression but did not promote GC B cell development, we wondered if c-Maf might contribute to the functional activity of CXCR5-expressing T cells by independently inducing expression of CD40L by Batf−/− T cells. To test this, we co-expressed CD40L with Bcl-6 in Batf−/− T cells and examined their in vivo TFH activity (Fig. 5c, d). Expression of CD40L alone failed to augment CXCR5 expression by T cells or GC B cell development, and when co-expressed with Bcl-6, failed to augment the effects of Bcl-6 alone. Thus, the action of c-Maf in TFH activity does not appear to lie simply in the regulation of CD40L expression, and likely involves control of additional targets.
Batf directly regulates AID expression
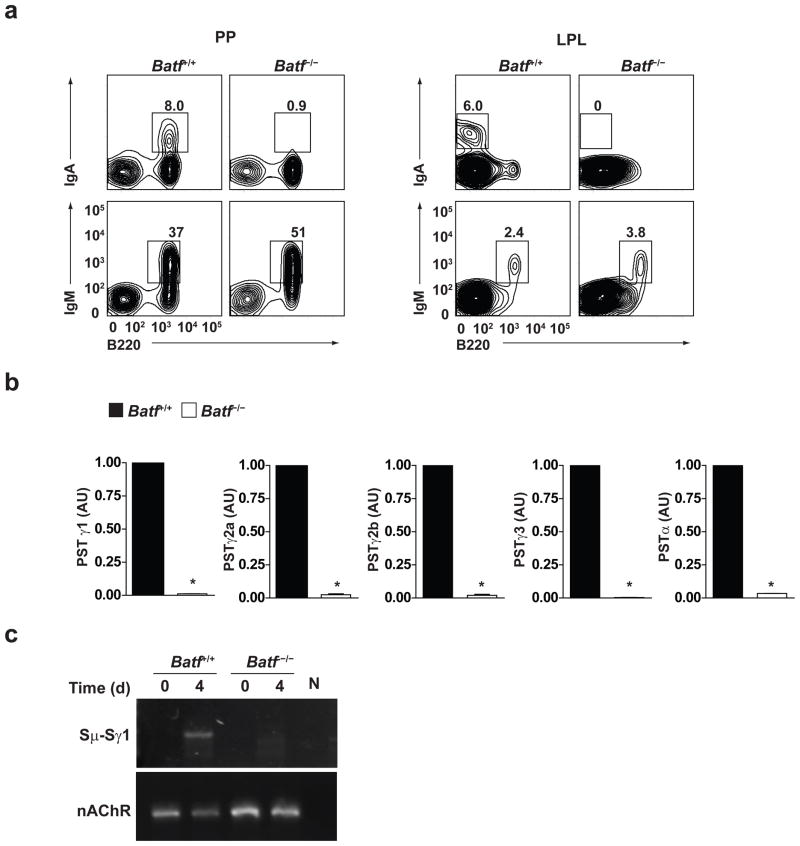
Because the defect in switched antibody responses in Batf−/− B cells was B cell-intrinsic (Supplementary Fig. 4c), we examined the secretion and surface expression of antibody using an in vitro class switching system (Supplementary Fig. 10a, b). We found dramatic reductions for each of the switched isotypes, IgG1, IgG2a, IgG2b, IgG3, and IgA. This defect occurs in vivo, shown by absent surface IgA expression of B cells from Peyer’s patches and lamina propria (Fig. 6a), but is not caused by altered B cell proliferation, a requirement for class switching20–22 or plasmacyte differentiation, since Batf−/− B cells proliferate normally and express the plasma cell marker CD138 (syndecan)23,24 (Supplementary Fig. 10c, 11).
Figure 6.
Impaired class switching in Batf−/− B cells. (a) FACS analysis for IgA, IgM and B220 expression by lymphocytes from Peyer’s patches (PP) or Lamina propria (LPL) of Batf+/+ or Batf−/− mice was performed. Data are representative of two experiments. (b) Quantitative RT-PCR analysis of post-switched Iμ-CH transcripts (PST) in Batf+/+ or Batf−/− B-cells stimulated for 4 days as in Supplementary figure 10a, b. Expression is normalized to hprt expression and is presented relative to the expression in Batf+/+ B cells, set at 1. **P<0.0001 (unpaired student t-test). Data are from three independent experiments. (c) Digestion-circularization PCR assay of genomic DNA isolated from Batf+/+ or Batf−/− B-cells unstimulated (d0) or stimulated with LPS plus IL-4 for 4 days (d4) for measurement of Sμ-Sγ1 recombination. Data are representative of two experiments.
Defective isotype switching is directly evident at the level of DNA rearrangements (Fig. 6b). B cells were activated under conditions that induce switching to the various isotypes and analyzed for expression of Iμ-CH post-switched transcripts (PST) for each of these isotypes25. There was a dramatic reduction in the level of post switched transcripts for each of the isotypes examined in Batf−/− B cells compared to wild-type B cells (Fig. 6b). This appears to results from a failure of switching at the level of DNA recombination. For example, for B cells induced to undergo IgG1 switching, wild-type B cells had DNA rearrangements between the Sμ and Sγ1 regions, but such DNA rearrangements were not detected in Batf−/− B cells (Fig. 6c).
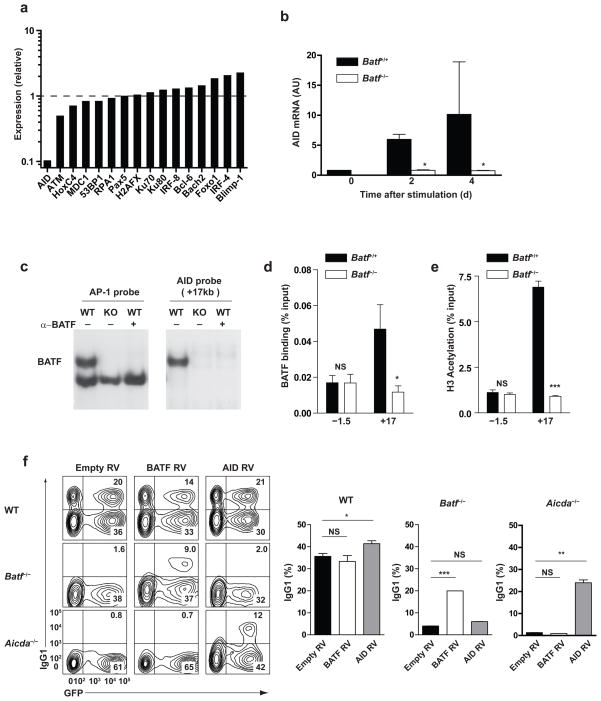
Although AID expression was reported to be reduced in Batf−/− B cells5, the basis for this reduction was not determined. Further, the ability of AID to restore switching defects in Batf−/− B cells was not demonstrated, so that defects beyond reduced AID expression could contribute to defective class switching in Batf−/− B cells. Therefore, to identify mechanisms contributing to defective CSR, we compared global gene expression of wild-type and Batf−/− B cells activated in vitro with LPS (Fig. 7a, Supplementary Fig. 12). Aicda mRNA was reduced by approximately 10-fold in Batf−/− B cells relative to wild-type B cells by microarray and q-PRC (Fig. 7a, b). Among 15 additional proteins known to be involved in class switching, AID was the only one to be significantly reduced in Batf−/− B cells (Fig. 7a). And in contrast to T cells, Batf−/− B cells did not show any known transcription factors that were reduced significantly relative to wild-type cells (Supplementary Fig. 12). For example, expression of Bach2 and IRF4, which are both required for isotype switching26, 27, were not reduced in Batf−/− B cells, suggesting that Batf might directly control AID expression rather than controlling expression of subordinate transcription factors as it does in TFH cells.
Figure 7.
Batf directly regulates AID mRNA expression. (a) Relative expression levels of genes involved in class switch recombination determined by microarray analysis of Batf+/+ or Batf−/− B cells activated for 2 days with LPS. Expression is shown as a ratio of expression in Batf−/− B cells compared to Batf+/+ B cells. Data are from one microarray experiment. (b) Quantitative RT-PCR analysis of AID mRNA expression in Batf+/+ or Batf−/− B-cells stimulated for 2 or 4 days with LPS. Results are normalized to hprt expression and are presented as arbitrary unit (AU). *P<0.05 (unpaired student t-test). Data are from three independent experiments. (c) EMSA analysis was performed using B-cell nuclear extract from Batf+/+ B cells (WT) or Batf−/− B cells (KO) stimulated with LPS plus IL-4 for 24 hours and either a consensus AP-1 probe or +17 kb (2) AID probe (Supplementary Table 2) in the presence (+) or absence (−) of anti-BATF antibody. (d) B cells were stimulated as in c and ChIP performed with DNA precipitated using anti-BATF antibody and amplified using primers specific to −1.5 kb or +17 kb regions of AID locus. *P<0.05 (unpaired student t-test). NS, not significant. Data are from two independent experiments (mean and s.e.m.). (e) ChIP was performed as in d but DNA was precipitated using anti-acetylated histone H3. ***P<0.005 (unpaired student t-test). NS, not significant. Data are from two independent experiments (mean and s.e.m.). (f) FACS analyses (left) was performed on wild-type, Batf−/− or AID−/− B cells on the C57/BL6 background, activated with LPS and IL-4 and infected with the indicated retrovirus, and analyzed after 3 days. Data are representative data of two experiments. Frequency (right) of IgG1 positive B220+ B cells as determined in (f) from three biological replicates (mean and s.e.m.). *P<0.05, **P<0.01, and ***P<0.005, versus empty-RV control value (unpaired student t-test). NS, not significant.
To test this, we identified potential TGAC/G Batf binding sites2 located within 5 conserved non-coding regions near the AID locus (Supplementary Fig. 13a, Supplementary Table 2) and tested the ability of these sites to bind compete for Batf binding in EMSA using a consensus AP-1 probe. T cell nuclear extracts contain a Batf-JunB complex that migrates faster than the Fos-Jun AP-1 heterodimer2, and which shifts with anti-Batf antibody and is missing in Batf−/− T cell extracts (Supplementary Fig. 14a). B cell extracts contain this same Batf complex, which is shifted by anti-Batf antibody and missing in Batf−/− B cells extracts (Supplementary Fig. 14a), and which is also a heterodimer with JunB because it is shifted by antibody against JunB, but not to Fos, c-Jun, JunD, ATF1, or ATF3 (Supplementary Fig. 14b). Interestingly, the Fos-Jun AP-1 complex present in T cell extracts is not present in B cells, but is replaced by a faster migrating complex that contains only JunB among the proteins assayed, perhaps representing a JunB homodimer. The absence of the Fos-Jun complex in activated B cells may result from loss of Fos expression in activated B cells (Supplementary figure 14c) as Fos transcription is essentially extinguished by 1 day after activation, while Batf transcription has dramatically increased relative to unstimulated B cells.
Of 13 potential Batf binding elements within the AID locus, one competed very efficiently for Batf binding by EMSA (Supplementary Fig. 15). This site is located 17kb downstream of the Aicda gene within a region previously identified as being required for AID expression in vivo28. Importantly, when used directly as an EMSA probe, this site bound to a single complex that was shifted by anti-Batf antibody and was missing in Batf−/− B cells extracts, and did not bind the faster migrating JunB homodimer formed with the consensus AP-1 probe (Fig. 7c). Using ChIP analysis, the chromatin region containing this Batf binding site also bound to Batf in vivo in activated wild-type B cells, but not Batf−/− B cells (Fig. 7d). Further, this same region shows highly acetylated histone H3K14, a mark of active chromatin, only in wild-type B cells, but not in Batf−/− B cells, activated by LPS and IL-4 (Fig. 7e). These results together suggest that Batf may directly regulate AID expression via interactions at the +17kb regulatory region28.
To test if the only role of Batf in CRS is to promote AID expression in activated B cells, we re-expressed AID by retrovirus into Batf −/− B cells and measured isotype switching after in vitro activation with LPS plus IL-4 (Fig. 7f). As controls, Batf−/− B cells and AID−/− B cells were also transduced with either empty retrovirus, Batf-expressing retrovirus, or an AID-expressing retrovirus and activated under the same conditions. Empty retrovirus failed to restore switching by either Batf−/− or AID−/− B cells, as expected. The Batf-expressing retrovirus restored IgG1 isotype switching in Batf−/− B cells, but not in AID-deficient B cells, also as expected (Fig. 7f). The AID-expressing retrovirus restored IgG1 isotype switching in AID-deficient B cells, as expected, but importantly failed to restore IgG1 isotype switching in Batf−/− B cells (Fig. 7f). This result indicates that Batf is required at an additional point in isotype switching beyond its role in AID expression.
IH-CH germline transcription requires Batf
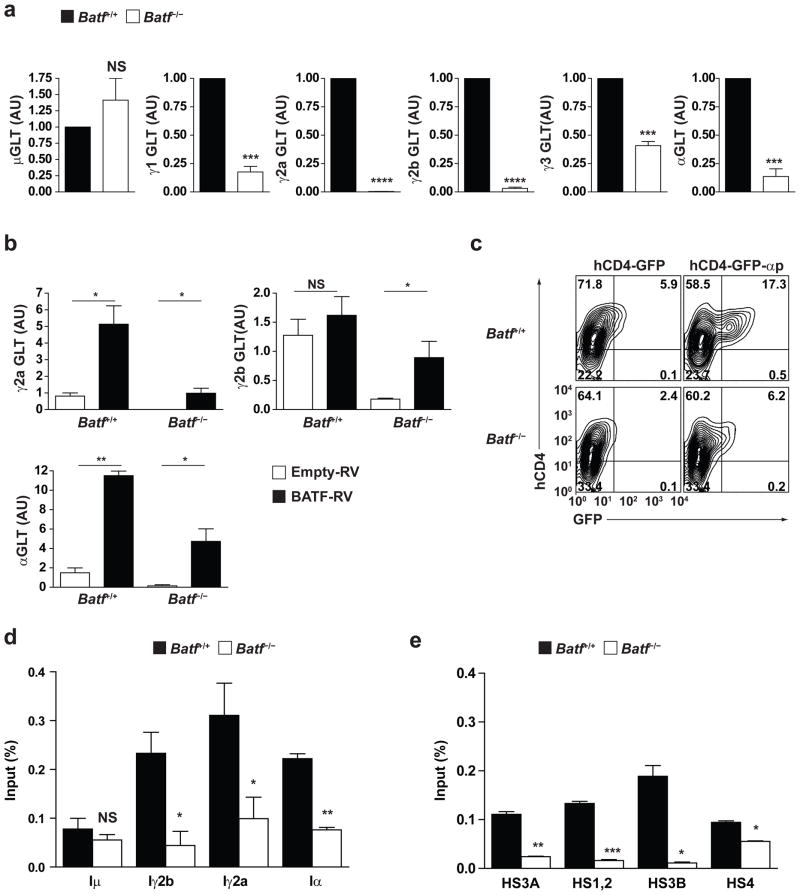
Class switch recombination (CSR) requires germline transcription from I promoters upstream of each switch region to allow AID access to switch region DNA29,30,31,32. Thus, we examined germline IH-CH transcripts (GLT) initiated from various I promoters in wild-type and Batf−/− B cells activated under conditions that induce switching to various isotypes (Fig. 8a). Germline transcripts initiated from the Iμ promoter (μGLT) were detected in both wild-type and Batf−/− B cells at similar levels, which is expected because Batf−/− B cells express IgM normally. In contrast, GLT levels of all other isotypes were substantially reduced in Batf−/− B cells relative to wild-type B cells (Fig. 8a). γ1GLT was reduced by 80%, while γ2aGLT and γ2bGLT were virtually undetectable. This result suggests that Batf is also required for normal germline transcription of all antibody isotypes.
Figure 8.
Batf is required for germline transcription. (a) Quantitative RT-PCR analysis was carried out for the indicated germline IH-CH transcripts (GLT) in Batf+/+ (black) or Batf−/− (white) B cells stimulated for 2 days as in Fig. 6d. Expression is normalized to hprt expression and is presented as relative to the expression in Batf+/+ B cells defined as 1. ***P<0.005 and ****P<0.0001 (unpaired student t-test). NS, not significant. Data are from three independent experiments. (b) Batf+/+ or Batf−/− B cells were activated with LPS, infected with empty-GFP RV (white) or BATF-GFP retrovirus (black), infected GFP+ cells purified by sorting and cultured for 2 days with LPS plus IFN-γ (γ2a GLT), or LPS plus TGF-β (γ2b GLT and αGLT) as indicated, and then quantitative RT-PCR analysis for the indicated germline IH-CH transcripts (GLT) was carried out. Expression is normalized to hprt expression. *P<0.05 and **P<0.01 (unpaired student t-test). NS, not significant. Data are from two independent experiments. (c) Batf+/+ or Batf−/− B cells were stimulated with αCD40 plus IL-4 and TGF-β and infected with either the empty reporter virus (hCD4-GFP) or the Iα promoter-reporter virus (hCD4-GFP-αp). FACS analysis for hCD4 and GFP expression on day 4 after stimulation is shown. Numbers represent the percentages of cells in indicated gate. (d, e) ChIP analysis of BATF binding to the indicated I promoters (d) or indicated regions of the 3′ IgH enhancer (e) in Batf+/+ (black) or Batf−/− (white) B cells cultured for 2 days with LPS. *P<0.05 and **P<0.01 (unpaired student t-test). NS, not significant. Data are from two independent experiments.
To determine if Batf over-expression could augment GLT, we transduced empty or Batf-expressing retroviruses into wild-type B cells activated with LPS alone or with IFN-γ or TGF-β and assayed for γ2aGLT, αGLT and γ2bGLT (Fig. 8b). Batf increased the level of GLT from each I promoter region compared to empty retrovirus, with the greatest increases seen for γ2aGLT and αGLT. Batf caused an increase by approximately 6-fold in γ2aGLT in B cells activated with LPS plus IFN-γ. When transduced into Batf−/− B cells, Batf-expressing retrovirus restored GLT to levels comparable to wild-type B cells (Fig. 8b).
To test whether Batf can directly regulate germline transcription from I promoters, we examined the activity of the Iα promoter using a reporter construct stably transduced into primary wild-type and Batf−/− B cells (Fig. 8c). The Iα reporter was active in wild-type B cells stimulated with anti-CD40 antibody in the presence of TGF-β and IL-4, but this activity was substantially reduced in Batf−/− B cells activated under the same conditions. This result suggests that the Iα promoter is Batf-dependent. Using ChIP analysis, we detected in vivo Batf binding to Iγ2b, Iγ2a, and Iα, promoter regions in wild-type but not Batf−/− B cells (Fig. 8d). Germline transcription of each CH gene is controlled by elements within the I promoters as well as within the 3′ IgH enhancer29,30. In addition, Batf was detected to bind in vivo to the HS3A, HS1,2 and HS3B regions of the 3′ IgH enhancer in wild-type but not Batf−/− B cells (Fig. 8e). These results suggest that Batf may interact with regulatory elements within I promoters and 3′ enhancer of IgH locus to control germline transcription.
Discussion
We previously reported that Batf−/− mice have a defect in TH17 development based on its direct regulation of both RORγt and of RORγt target genes such as IL-17A2. An independent report subsequently showed Batf−/− mice have decreased antibody production based on defects in T cells and B cells5. Here, we determined the molecular mechanisms that underlie the requirement for Batf in CSR based on actions in both T cells and B cells. And as we previously showed that Batf regulates both the TH17 transcription factor RORγt33 as well as directly acting on RORγt targets2, we now point out that this dual level of action of Batf is also evident in its control of TFH differentiation in T cells and of CSR in B cells.
In T cells, Batf appears to directly control the expression of two of the three major transcription factors needed for TFH development, Bcl-6 and c-Maf. Batf−/− T cells have dramatically reduced express two of the transcription factors recognized as important for TFH differentiation, the TFH master factor Bcl-6, and the bZIP transcription factor c-Maf13. Co-expression of Bcl-6 or c-Maf into Batf−/− T cells partially restored TFH activity in vivo, but it is likely that additional direct Batf targets remain to be identified that will explain the residual TFH deficit in these Batf−/− T cells. In B cells, Batf is required for class switch recombination not only because it directly controls expression of AID, but also because it is required for normal IH-CH germline transcription through all target isotype switch regions via direct interactions with I region promoters and elements of the 3′ IgH enhancer. While decreased AID expression was previously used to explain the decrease in isotype switching in Batf−/− B cells5, we discovered an additional defect that prevents CSR even when AID expression is restored. These finding substantially broaden our understanding of how Batf regulates antibody production.
In TFH cells, Batf is required to express two critical TFH-specific transcription factors, Bcl-6 and c-Maf. Bcl-6 is required for normal TFH differentiation and function, since Bcl6−/− T cells fail to express CXCR5 or to support germinal center formation6–8. Over-expression of Bcl-6 causes an upregulation of TFH markers in vitro and in vivo including CXCR5, ICOS, and PD-1 and induced TFH differentiation in vivo6–8. Until now, Bcl-6 was thought to be both necessary and sufficient for development of TFH cells. We now show that Bcl-6 is insufficient for TFH development in the absence of Batf. A recent report has also documented an important role for c-Maf in TFH differentiation9. Ablation of c-Maf reduced CXCR5+CD4+ T cell numbers and IL-21 expression9, while over-expression of c-Maf enhances IL-21 expression34, which may promote TFH development or expansion. Thus, by controlling both Bcl-6 and c-Maf expression, Batf acts at the top of a transcriptional hierarchy controlling TFH differentiation.
In B cells, Batf also acts at two hierarchical levels to control CSR. Batf is required for expression of AID as reported previously5, which we now show is likely due to strong binding to the required enhancer at located at +17kb. In addition, we show that Batf is required for transcription through the switch regions, which seems to involve direct interactions with both I region promoters and elements of the 3′ enhancer of IgH locus. Germline transcription is required for CSR to allow AID access to DNA to induce targeted DNA breaks within the switch regions. Re-expression of AID by retrovirus into Batf−/− B cells did not restore CSR, likely because of insufficient levels of GLT from I region promoters. Batf appears to physically associate with most of the I region promoters and with several regions of the 3′ enhancer, which are themselves required for GLT and CSR35.
It is interesting that the small AP-1 family members Batf and Batf3 exert such specific and lineage-restricted actions1,2,5 because they had previously been thought to act simply as negative regulators of AP-1 activity36–38. Batf3 expression is largely restricted to dendritic cells and Batf3−/− mice exhibit a defect in the development of a subset of dendritic cells. Batf acts more broadly, being required for TH17 and TFH cells, which notably both require IL-6 for their development18,39, and many genes regulated by Batf in T cells are dependent on IL-6 for their induction 2. The challenge now is to understand how Batf and Batf3 exert their specific transcriptional effects distinctly from other AP-1 family members such as Fos and Atf3 which are unable to substitute for these genes in their cell-type specific actions.
Supplementary Material
References
- 1.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schraml BU, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edelson BT, et al. Peripheral CD103(+) dendritic cells form a unified subset developmentally related to CD8 alpha(+) conventional dendritic cells. Journal of Experimental Medicine. 2010;207:823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Langrish CL, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betz BC, et al. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. Journal of Experimental Medicine. 2010;207:933–942. doi: 10.1084/jem.20091548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nurieva RI, et al. Bcl6 Mediates the Development of T Follicular Helper Cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston RJ, et al. Bcl6 and Blimp-1 Are Reciprocal and Antagonistic Regulators of T Follicular Helper Cell Differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu D, et al. The Transcriptional Repressor Bcl-6 Directs T Follicular Helper Cell Lineage Commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Bauquet AT, et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bottaro A, et al. S-Region Transcription Per-Se Promotes Basal Ige Class Switch Recombination But Additional Factors Regulate the Efficiency of the Process. EMBO Journal. 1994;13:665–674. doi: 10.1002/j.1460-2075.1994.tb06305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, et al. A Selective Defect in Igg2B Switching As A Result of Targeted Mutation of the I-Gamma-2B Promoter and Exon. EMBO Journal. 1993;12:3529–3537. doi: 10.1002/j.1460-2075.1993.tb06027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King C. New insights into the differentiation and function of T follicular helper cells. Nature Reviews Immunology. 2009;9:757–766. doi: 10.1038/nri2644. [DOI] [PubMed] [Google Scholar]
- 13.Linterman MA, Vinuesa CG. Signals that influence T follicular helper cell differentiation and function. Seminars in Immunopathology. 2010;32:183–196. doi: 10.1007/s00281-009-0194-z. [DOI] [PubMed] [Google Scholar]
- 14.Nurieva RI, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi H, et al. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 2008;455:764–7U2. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon H, et al. Analysis of Interleukin-21-Induced Prdm1 Gene Regulation Reveals Functional Cooperation of STAT3 and IRF4 Transcription Factors. Immunity. 2009;31:941–952. doi: 10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haynes NM, et al. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. The Journal of Immunology. 2007;179:5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- 18.Nurieva RI, et al. Generation of T Follicular Helper Cells Is Mediated by Interleukin-21 but Independent of T Helper 1, 2, or 17 Cell Lineages. Immunity. 2008 doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King C, Tangye SG, Mackay CR. T follicular helper (T-FH) cells in normal and dysregulated immune responses. Annual Review of Immunology. 2008;26:741–766. doi: 10.1146/annurev.immunol.26.021607.090344. [DOI] [PubMed] [Google Scholar]
- 20.Hasbold J, et al. Cell division number regulates IgG1 and IgE switching of B cells following stimulation by CD40 ligand and IL-4. European Journal of Immunology. 1998;28:1040–1051. doi: 10.1002/(SICI)1521-4141(199803)28:03<1040::AID-IMMU1040>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 21.Deenick EK, Hasbold J, Hodgkin PD. Switching to IgG3, IgG2b, and IgA is division linked and independent, revealing a stochastic framework for describing differentiation. Journal of Immunology. 1999;163:4707–4714. [PubMed] [Google Scholar]
- 22.Rush JS, et al. Expression of activation-induced cytidine deaminase is regulated by cell division, providing a mechanistic basis for division-linked class switch recombination. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13242–13247. doi: 10.1073/pnas.0502779102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanderson RD, Lalor P, Bernfield M. B lymphocytes express and lose syndecan at specific stages of differentiation. Cell Regul. 1989;1:27–35. doi: 10.1091/mbc.1.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blink EJ, et al. Early appearance of germinal center-derived memory B cells and plasma cells in blood after primary immunization. J Exp Med. 2005;201:545–554. doi: 10.1084/jem.20042060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu CC, Paul WE, Max EE. Quantitation of Immunoglobulin Mu-Gamma-1 Heavy-Chain Switch Region Recombination by A Digestion Circularization Polymerase Chain-Reaction Method. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6978–6982. doi: 10.1073/pnas.89.15.6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muto A, et al. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429:566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- 27.Klein U, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 28.Crouch EE, et al. Regulation of AID expression in the immune response. Journal of Experimental Medicine. 2007;204:1145–1156. doi: 10.1084/jem.20061952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudley DD, et al. Mechanism and control of V(D)J recombination versus class switch recombination: Similarities and differences. Advances in Immunology. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- 30.Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Annual Review of Immunology. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhuri J, et al. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 32.Ramiro AR, et al. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nature Immunology. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- 33.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 34.Hiramatsu Y, et al. c-Maf activates the promoter and enhancer of the IL-21 gene, and TGF-beta inhibits c-Maf-induced IL-21 production in CD4(+) T cells. Journal of Leukocyte Biology. 2010;87:703–712. doi: 10.1189/jlb.0909639. [DOI] [PubMed] [Google Scholar]
- 35.Vincent-Fabert C, et al. Genomic deletion of the whole IgH 3′ regulatory region (hs3a, hs1,2, hs3b, hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood. 2010 doi: 10.1182/blood-2010-01-264689. [DOI] [PubMed] [Google Scholar]
- 36.Iacobelli M, Wachsman W, McGuire KL. Repression of IL-2 promoter activity by the novel basic leucine zipper p21SNFT protein. The Journal of Immunology. 2000;165:860–868. doi: 10.4049/jimmunol.165.2.860. [DOI] [PubMed] [Google Scholar]
- 37.Echlin DR, et al. B-ATF functions as a negative regulator of AP-1 mediated transcription and blocks cellular transformation by Ras and Fos. Oncogene. 2000;19:1752–1763. doi: 10.1038/sj.onc.1203491. [DOI] [PubMed] [Google Scholar]
- 38.Williams KL, et al. Characterization of murine BATF: a negative regulator of activator protein-1 activity in the thymus. Eur J Immunol. 2001;31:1620–1627. doi: 10.1002/1521-4141(200105)31:5<1620::aid-immu1620>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 39.Korn T, et al. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448:484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranganath S, et al. GATA-3-dependent enhancer activity in IL-4 gene regulation. Journal of Immunology. 1998;161:3822–3826. [PubMed] [Google Scholar]
- 41.Ranganath S, Murphy KM. Structure and specificity of GATA proteins in Th2 development. Molecular & Cellular Biology. 2001;21:2716–2725. doi: 10.1128/MCB.21.8.2716-2725.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sedy JR, et al. B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nat Immunol. 2005;6:90–98. doi: 10.1038/ni1144. [DOI] [PubMed] [Google Scholar]
- 43.Zhu H, et al. Unexpected characteristics of the IFN-gamma reporters in nontransformed T cells. Journal of Immunology. 2001;167:855–865. doi: 10.4049/jimmunol.167.2.855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.