SUMMARY
Treatment of tuberculosis, a complex granulomatous disease, requires long-term multidrug therapy to overcome tolerance, an epigenetic drug resistance that is widely attributed to nonreplicating bacterial subpopulations. Here, we deploy Mycobacterium marinum-infected zebrafish larvae for in vivo characterization of antitubercular drug activity and tolerance. We describe the existence of multi-drug tolerant organisms that arise within days of infection, are enriched in the replicating intracellular population, and are amplified and disseminated by the tuberculous granuloma. Bacterial efflux pumps that are required for intracellular growth mediate this macrophage-induced tolerance. This newly discovered tolerant population also develops when Mycobacterium tuberculosis infects cultured macrophages, suggesting that it contributes to the burden of drug tolerance in human tuberculosis. Efflux pump inhibitors like verapamil reduce this tolerance. Thus, the addition of this currently approved drug, or more specific inhibitors, to standard antitubercular therapy may shorten the duration of curative treatment.
INTRODUCTION
Despite more than 50 years of effective antitubercular drugs, TB eradication remains elusive (Sacchettini et al., 2008) due to the complexity of curative treatment regimens (Connolly et al., 2007; Donald and McIlleron, 2009). Long-term therapy is required to prevent relapses with genetically drug-sensitive bacilli that become transiently resistant in the host, a phenomenon called tolerance. The best-case regimen of six months was made possible by drug combinations that presumably reduce the tolerant population (Donald and McIlleron, 2009). This so-called “short course therapy” represented a major advance as prior regimens lasted 12–18 months. However, adherence to six months of multidrug treatment is difficult, leading to relapses that perpetuate the epidemic and fuel the development of genetic resistance (Sacchettini et al., 2008). Thus, an urgent goal of antitubercular drug discovery is to overcome tolerance (Connolly et al., 2007; Sacchettini et al., 2008). Several new drugs are in development (Sacchettini et al., 2008), yet most lack this key treatment-shortening property. This failure highlights a poor understanding of TB tolerance mechanisms (Barry et al., 2009; Connolly et al., 2007; Sacchettini et al., 2008; Warner and Mizrahi, 2006).
Drug tolerance in TB is best contextualized to the in vivo lifestyle of mycobacteria. Mtb resides within complex immunological structures called granulomas, either within macrophages or extracellularly, in the necrotic core (caseum) (Figure S1) (Barry et al., 2009; Connolly et al., 2007). Multiple granuloma types, reflecting different levels of local disease activity, may coexist in a given patient (Figure S1) (Barry et al., 2009; Canetti, 1955; Connolly et al., 2007; Rich, 1946). A longstanding model is that tolerant bacteria are sequestered in a subset of granulomas wherein their replication and metabolism are uniformly slowed (Figure S1) (Barry et al., 2009; Connolly et al., 2007). Newer models posit that quiescent, drug tolerant bacteria are present in all lesion types and may be induced by deterministic mechanisms responsive to stress (e.g. hypoxia) and/or the stochastic formation of persister cells, some of which may be nonculturable under standard laboratory conditions (Figure S1) (Barry et al., 2009; Connolly et al., 2007; Garton et al., 2008; Mukamolova et al., 2010; Sacchettini et al., 2008; Warner and Mizrahi, 2006). However, all tolerance models and consequently drug discovery efforts are centered on the assumption that slowed growth and/or metabolic quiescence is the primary mediator of tolerance.
In this study we demonstrate that growing mycobacteria develop multi-drug tolerance soon after they infect macrophages. This understanding was gained by a two-pronged approach: first we used zebrafish larvae infected with Mycobacterium marinum (Mm) to spatially and temporally characterize the responses of individual animals to frontline antitubercular drugs. Mm infections feature drug tolerance and require long-term treatment in both humans and fish (Aubry et al., 2002; Decostere et al., 2004). Using the larval model, we discovered that tolerant bacteria arise within individual macrophages soon after infection and are then expanded and disseminated by tuberculous granulomas. Guided by these findings, we used cultured macrophage infection models combined with bacterial efflux pump mutants and pharmacological inhibitors to identify both a mechanism and a therapy for macrophage-induced tolerance in TB.
RESULTS
A zebrafish larval model for in vivo characterization of antitubercular drug activity
To assess antitubercular drug activity in the model, we infected larvae intravenously with GFP-expressing Mm and maintained them in the presence of antitubercular drugs. Concentrations that represented multiples of the in vitro minimum inhibitory concentration (MIC) for Mm were used, and the drug-supplemented water was changed daily (Table S1, Figure S2A–E and Experimental Procedures). We established minimal isoniazid (INH), rifampicin (RIF), ethambutol (EMB) and moxifloxacin (MOX) concentrations that were nontoxic and normalized host survival when administered within one day post infection (dpi), which we term the minimum effective concentration (MEC) (Figures 1A, S2B–E and Tables S1 and S2). Bacterial burdens of infected larvae can be assessed visually by fluorescence microscopy, or quantitatively by larval lysis and enumeration of bacterial colony forming units (CFU). To facilitate rapid, serial quantification of bacterial burden, we developed software that enumerates fluorescent pixels in images of infected larvae, or fluorescent pixel count (FPC), and showed it to be an accurate predictor of bacterial burden (Figure S3 and Experimental Procedures). Increased survival was associated with lower bacterial burdens at four days post treatment (dpt) as judged by fluorescence microscopy, and confirmed quantitatively by FPC and CFU analyses (Figures 1A–1F, S3 and Table S2). Findings consistent with human TB data were also obtained with streptomycin (STM, Figure S2C–E), which has resurged as a cornerstone of treatment for extremely drug resistant (XDR) TB (Donald and McIlleron, 2009). In summary, clinically relevant antitubercular drugs were efficacious in the zebrafish infection model with the expected exception of pyrazinamide to which Mm is innately resistant (Figure S2F–G).
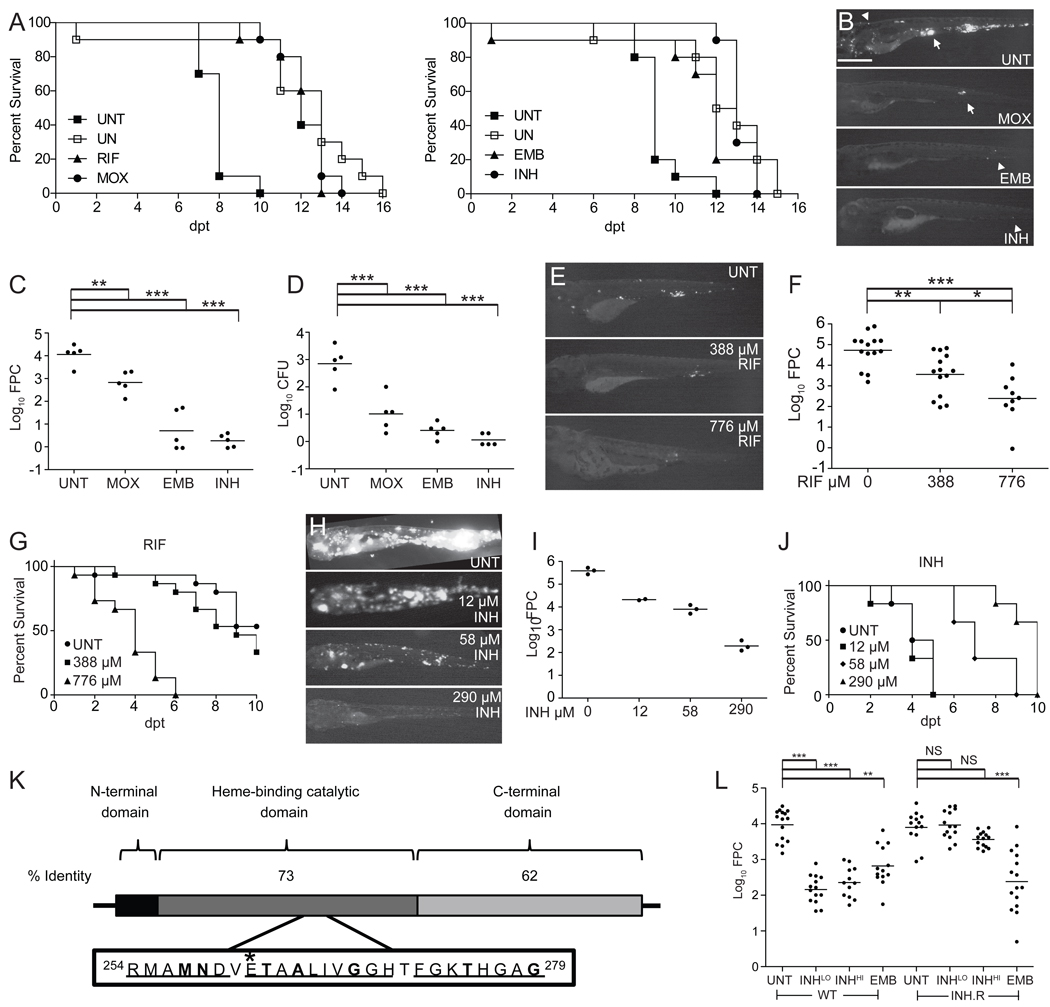
Figure 1. The Mm-larval zebrafish infection model replicates the specificity and activity of clinically relevant antitubercular drugs.
(A–D) Larvae were soaked in the MEC of RIF (388 µM), MOX (62.3 µM), EMB (1442 µM) or INH (290 µM) (Table S1). (A) Survival of uninfected (UN) larvae vs. those infected with 800 Mm and immediately treated with RIF, MOX, EMB, INH or left untreated (UNT). Left panel in the presence of 1% DMSO (see Figure S2A). Survival of treated infected larvae was significantly different from UNT larvae for all drugs (Table S2). Results are representative of at least two independent experiments.
(B–D) Larvae were infected with 155 Mm and left untreated (UNT) or treated with MOX, EMB, or INH for 4 days, prior to assessment of bacterial burdens by fluorescence microscopy (B, representative larvae are shown), FPC (C) or CFU enumeration of the lysed larvae immediately after imaging (D). Arrow, granuloma; arrowhead, single infected macrophage. Scale bar 500 µm. For C and D, individual larvae (points) and means (bars) are shown. Significance testing by one way ANOVA with Dunnett’s post test.
(E and F) Larvae infected with 46 Mm were soaked for 3 days in 388 or 776 µM RIF or left untreated. Representative fluorescence images (E) and bacterial burdens (F) of survivors are shown. Significance testing by one-way ANOVA with Tukey’s post-test.
(G) Survival of uninfected larvae upon treatment with 0, 388 or 776 µM RIF added 2 dpf. P=0.0010 for 0 vs. 776; 0 vs. 388 µM, NS by Log-rank test. N=15 per group.
(H–J) Larvae infected with 1800 Mm were left untreated or immediately soaked in 12, 58 or 290 µM INH. Representative fluorescence images (H) and bacterial burdens (I) of survivors at 4 dpt are shown. Mean ba cterial burdens (bars) compared by one-way ANOVA with Tukey’s post-test resulted in P < 0.001 for all comparisons, with the exception of 12 vs. 58 µM INH, which was not significant (P > 0.05). (J) Survival curve, N= 6 larvae per group. P=0.0018, Log-rank test for trend comparing all curves and P=0.0010 for comparison of untreated vs. 58 µM or untreated vs. 290 µM.
(K) Functional domains of KatG. Mtb KatG is 740aa and Mm KatG is 743aa. Boxed inset of Mtb catalytic domain shows regions of identity with Mm (underlined) and point mutations (in bold) that confer INH resistance in Mtb (Sandgren et al., 2009). *Position of the single amino acid substitution (E265V) in the INH-resistant Mm strain corresponds to the E261 position of Mtb KatG.
(L) Bacterial burdens of 3 dpt larvae infected with 300 WT or INH-resistant (INH.R) Mm and soaked in 290 (LO) or 2320 µM (HI) INH, 1442 µM EMB, or left untreated (UNT) beginning 1 dpi. Median log10FPC (bars) compared using Kruskal-Wallis test with Dunn's post-test. For all panels, *, P<0.05; **, P<0.01; ***, P<0.001, NS, not significant.
To further probe the model’s relevance to human TB, we used the MECs as starting points from which to examine drug potency and dose-dependent activity. In human pulmonary TB studies, the early bactericidal activity (EBA) of antitubercular drugs, defined as their ability to reduce sputum bacterial counts in pulmonary TB patients over the first several days, has been useful to guide drug dosing (Donald and Diacon, 2008; Jindani et al., 2003). Each drug has distinctive EBA characteristics. For RIF, doses above the currently used therapeutic dose increase the EBA, raising the question of whether higher doses would have greater efficacy (Donald and Diacon, 2008). In the larvae, treatment with the RIF MEC caused a 1.2 log10 reduction in bacterial burden, while treatment with twice the MEC caused an additional 1.2 log10 reduction despite increased toxicity (Figure 1E–G). We similarly observed dose-dependent activity for MOX: stepwise increases in concentration produced greater diminutions in bacterial burdens (Figure S2H). In contrast to RIF, INH EBA does not increase when the conventional dose is doubled. We too observed no further reduction in bacterial counts at double the INH MEC (data not shown). Also, stepwise two-fold reductions from the conventional dose continue to show activity in human TB, with a graded EBA decrease down to 1/16th the therapeutic dose (Donald and Diacon, 2008). Similarly, we found a graded concentration-dependent bactericidal activity down to 1/24th the MEC (Figure 1H and 1I) and a therapeutic benefit at 1/5th the MEC (Figure 1J).
Finally, since drug-resistant TB is an increasing problem (Sacchettini et al., 2008), we asked if the model could differentiate Mm strains with altered drug susceptibility. INH resistance is usually the first to occur in human TB and often the first step in the progression to XDR TB (Donald and McIlleron, 2009). INH is a pro-drug activated by the bacterial catalase KatG, and the vast majority of INH-resistant clinical Mtb isolates have katG mutations (Sandgren et al., 2009). We identified a spontaneous Mm INH-resistant katG mutant with an in vitro INH MIC of 464 µM, 8-fold higher than for wild-type (Figure 1K; Supplemental Experimental Procedures). The relative resistance of the Mm katG mutant was detectable in vivo: in larvae infected with wild-type Mm, maximal bacterial killing was observed at 290 µM whereas an eight-fold higher concentration was ineffective against the katG mutant (Figure 1L). INH resistance was specific: EMB, which has a distinct target (Belanger et al., 1996), showed similar efficacy against both strains (Figure 1L). In summary, the zebrafish larval model reliably replicates the activity of antitubercular compounds and their dosing characteristics in humans.
Drug tolerance occurs prior to granuloma formation
Our observation that residual cultivatable bacteria persisted four dpt, despite initiating treatment within a day of infection when the bacteria were in individual macrophages (Figures 1B–D) suggested the early presence of drug-tolerant bacteria, prior to granuloma formation. We pursued this observation in detail for INH because its tolerance in human TB has been studied extensively (Donald and Diacon, 2008; Donald and McIlleron, 2009; Jindani et al., 2003). We confirmed that the residual bacteria were tolerant, rather than genetically resistant to INH: the INH susceptibility of individual bacilli recovered from six treated larvae (10.3 ±1.2 CFU per larva) was unchanged from the parental strain.
INH tolerance is seen in human EBA studies where monotherapy produces biphasic killing. The majority of the bacteria are killed with two days of treatment, after which the rate of killing drops leaving a population of drug tolerant bacteria 14 dpt (Figure 2A)(Donald and Diacon, 2008; Jindani et al., 2003). We conducted an EBA-like study in the larvae, starting treatment after granulomas had formed, at three dpi. Serial quantitative tracking of infection by fluorescence microscopy revealed that INH killing kinetics mirror those observed in human EBA studies (Figure 2A–C).
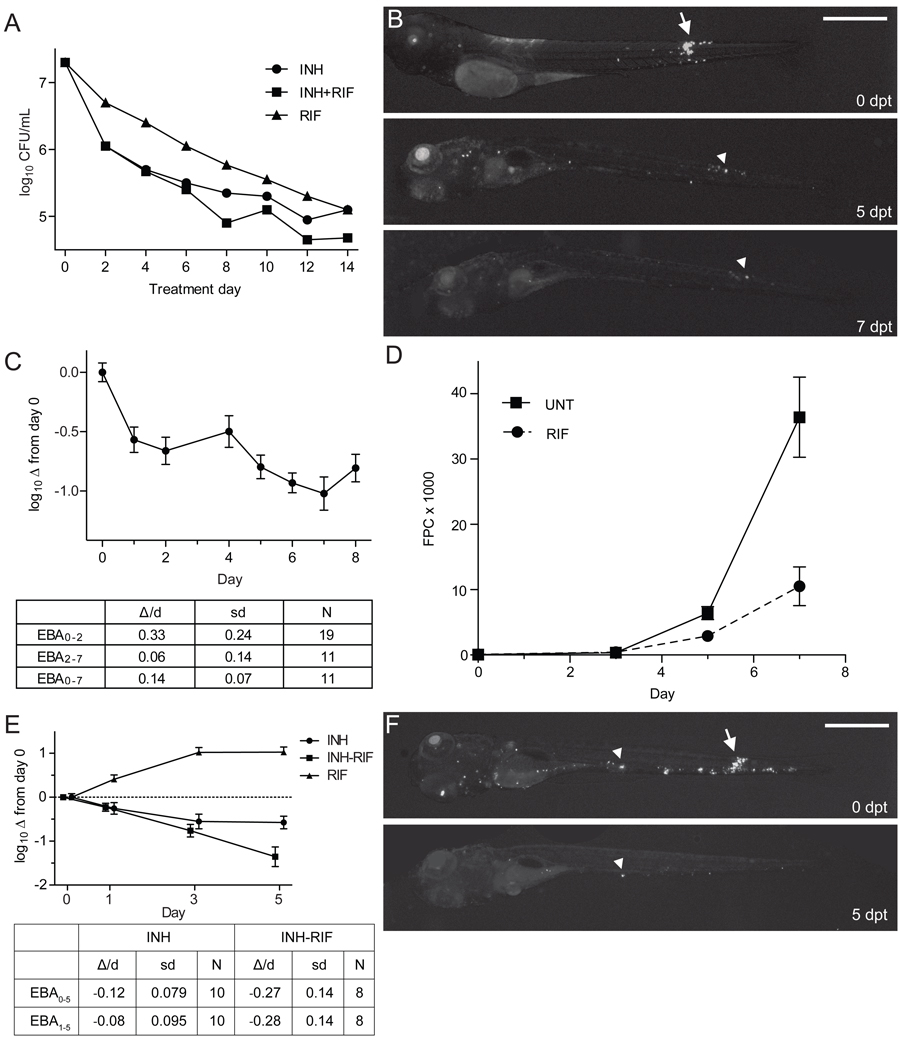
Figure 2. INH treatment of Mm-infected larvae results in a biphasic EBA with persistence of tolerant organisms.
(A) Authors’ rendition of human clinical EBA data (see Figure 1 of (Jindani et al., 2003)) showing the rate of clearance of Mtb from sputum in patients with previously untreated, smear-positive pulmonary tuberculosis, upon treatment with INH and/or RIF.
(B and C) Twenty larvae were infected with 300 Mm and treated with 290 µM INH beginning 3 dpi. Each larva was imaged daily for 8 dpt, and bacterial burdens quantified by FPC. (B) Representative larva imaged at the beginning of treatment (0 dpt), and again at five and seven dpt. Arrow, granuloma, arrowhead, macrophage containing persistent bacteria. (C) EBA curve for INH-treated larvae, showing the mean log10 FPC change from day 0. Error bars represent SEM. EBA was calculated as described in the Experimental Procedures.
(D–F) Ten larvae per group were infected with 300 Mm and were serially imaged for enumeration of bacterial burden by FPC. (D) Larvae were treated with 388 µM RIF or left untreated, beginning 1 dpi. Mean FPC and SEM are shown. (E) Larvae were treated with 290 µM INH, 388 µM RIF, or a combination of both drugs beginning at 3 dpi. Data analyzed as in (C). (F) Representative INH-RIF treated larvae annotated as in (B).
By tracking individual animals, we determined the location of the persistent bacteria (Figure 2B). Several infected macrophages had very little diminution in fluorescence, suggesting they contained tolerant bacteria, and their varying positions on consecutive days suggested their continued movement during treatment. There were also residual infected macrophages (Figure 2B, arrowhead) from granulomas that had dissolved with treatment (Figure 2B, arrow). Thus, tolerant bacteria were present both in individual macrophages and in granuloma macrophages.
In human TB, the addition of RIF to INH-containing regimens has shortened time to sterilization and thus treatment length. This is reflected in EBA studies where RIF is not rapidly bactericidal on its own and the additional activity of the INH-RIF combination is seen only in the slower phase of killing, when INH-tolerant bacteria are implicated (Figure 2A)(Donald and Diacon, 2008; Jindani et al., 2003). Therefore, RIF’s treatment-shortening effect is attributed to its capacity to reduce INH tolerant bacteria (Donald and McIlleron, 2009). Reminiscent of the human data, RIF in the larvae was not rapidly bactericidal on its own (Figure 2D), yet increased INH efficacy in the slow phase of its EBA curve (Figure 2E). Residual bacteria persisted after combination therapy, albeit fewer than with INH monotherapy (Figure 2F). In sum, the model demonstrates drug tolerance akin to that seen in human TB and the well-known synergistic effect of RIF in reducing tolerance. Furthermore, it reveals tolerant bacteria within individual macrophages at the earliest stages of infection.
Drug tolerant bacteria are expanded and disseminated by granuloma formation
All models of TB tolerance invoke metabolically quiescent bacteria (Barry et al., 2009; Connolly et al., 2007; Sacchettini et al., 2008; Warner and Mizrahi, 2006). Yet during human TB treatment individual lesions can expand, or new ones appear at distant sites, despite overall clinical improvement and reduction in lesion size and number (Akira et al., 2000; Barry et al., 2009; Bobrowitz, 1980; Canetti, 1955). Importantly, bacteria from new lesions are drug sensitive and the patients ultimately cured with no change in therapy (reviewed in (Akira et al., 2000)). These observations are consistent with an uneven distribution of drug tolerant bacteria that expand locally or after dissemination. Indeed even, before the advent of effective TB therapy, it was appreciated that within a given patient, some lesions resolve whilst others worsen, suggesting distinct local host responses in the course of natural infection (Canetti, 1955; Rich, 1946).
We wondered if the drug tolerant bacteria we had observed within scattered macrophages are the ones that are then amplified and disseminated by mechanisms of granuloma formation (Davis and Ramakrishnan, 2009). However, we first had to determine if our model recapitulated the differential progression of natural infection. We found that in the first week of infection, some granulomas progressed while others resolved (Figure 3A). Lesions in the head region (containing the organs) were more likely to progress whereas those in the tail region (primarily muscle) were more likely to resolve (Figure 3B and 3C). The effect was even more striking at later developmental stages (days 7 to 11) when organogenesis is more advanced (Figure 3C). By one month, granulomas were exclusively within organs (Figure 3D and 3E), reminiscent of the observation that human TB seldom involves muscle (Rich, 1946).
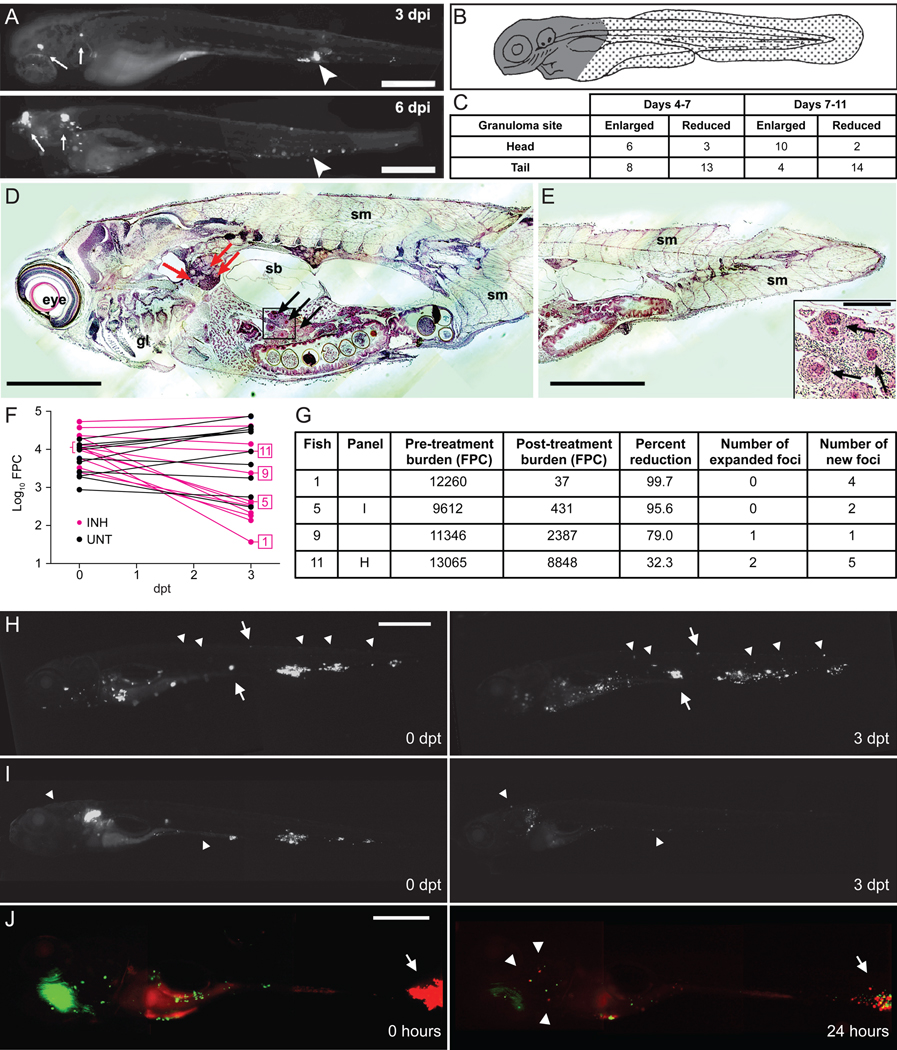
Figure 3. Mm infections are dynamic during antibiotic treatment.
(A–D) Tracking of untreated Mm infections in individual larvae. (A) Fluorescence images of a representative larva at 3 and 6 dpi. Arrows, enlarging granulomas, arrowheads, shrinking granulomas. Scale bar, 400 µm. (B) Cartoon indicating "head" region (gray) primarily containing organs, and "tail" region (stippled) comprised mostly of muscle. (C) Enumeration of expanding and contracting granulomas over time. Differential region-specific outcomes of granulomas were statistically significant for changes occurring between days 7 and 11 (P=0.0022), but not for changes occurring between days 4 and 7 (P=0.2360, Fisher’s exact test).
(D and E) Hematoxylin and eosin staining showing Mm in caseating granulomas in a 33 day-old fish that was infected at 1 dpf. Red and black arrows indicate granulomas in pronephros and liver, respectively. gl, gill; sb, swim bladder; sm, somite. Scale bar 300 µm. Higher ma gnification of granulomas in boxed inset of (D) is shown on bottom right of (E). Scale bar 50 µm.
(F–I) Larvae infected with varying Mm inocula for 4 days were then treated with 290 µM INH or left untreated for an additional 3 days. Larvae were imaged at 4 and 7 dpi (0 and 3 dpt). (F) Pre- and post-treatment log10 FPC values for individual larvae are plotted with data points from the same individual connected. (G) Raw FPC values before and after treatment, percent change, and the presence of expanding and new foci are reported for representative fish indicated in (F). (H and I) Fluorescence images of fish 11 (H) and fish 5 (I) as reported in (F and G), shown before and after treatment. Arrows, enlarging granulomas, and arrowheads, new foci. Scale bars 500 µm.
(J) A single larva was infected with 500 Mm constitutively expressing the Kaede photoactivatible GFP for 4 days. Composite red and green fluorescence images immediately after photoactivation of a granuloma (left panel) and 24 hours later (right panel). Arrows, photoactivated granuloma, arrowheads, single macrophages containing red fluorescent bacteria. Scale bar 250 µm.
Next, to study the effect of drug treatment on individual lesions, we infected larvae with a range of inocula to achieve varying infection burdens (Figure 3F) and then began INH treatment for half the larvae. We quantified and spatially localized infection in each animal at the start of treatment and three days later by fluorescence microscopy. As expected, untreated larvae demonstrated an increase in bacterial burden (+0.2176 log10 FPC, P=0.0008, Student's t-test) whereas treatment reduced the bacterial burden (−1.001 log10 FPC).
Spatial monitoring of the untreated larvae confirmed the expected differential progression of lesions. Ten of the 12 treated larvae had a reduction in bacterial burdens ranging from 32.3% to 99.7% (Figure 3F and 3G). We found local “nonresponsiveness” amongst the responding larvae, analogous to that observed in humans (Figure 3G–3I). For example, fish 11 (Figure 3G and 3H) had an overall 32.3% reduction in bacterial burden, yet two individual infected macrophages expanded into two infected cells and a granuloma and multiple new foci appeared. New foci developed even in animals with an overall >90% reduction in bacterial burdens (e.g. Fish 5, Figure 3G and 3I).
Finally, we determined whether new infection foci resulted from the egress of macrophages (containing tolerant bacteria) from existing granulomas, a phenomenon we have documented during untreated infection (Davis and Ramakrishnan, 2009). We infected larvae with bacteria expressing the fluorescent reporter Kaede that changes from green to red upon photoactivation (Davis and Ramakrishnan, 2009). We selectively photoactivated the largest granuloma and immediately began INH treatment (Figure 3J, left panel). We then assessed if macrophages harboring the photoactivated bacteria left granulomas after initiating treatment. Within 24 hours, three infected macrophages had left the granuloma and dispersed far from the initial site (Figure 3J, right panel). Importantly, the original granuloma had shrunk substantially showing its overall response to therapy, consistent with the human data (Bobrowitz, 1980). The bacteria within the departed macrophages were both red and green fluorescent, suggesting that they were synthesizing new Kaede protein and thus metabolically active, again consistent with the human data where the new foci had viable bacteria that were then able to expand.
In summary, as is the case in human TB, individual lesions in Mm-infected zebrafish larvae respond variably to treatment and dissemination occurs during effective therapy. The detailed temporal monitoring possible in this model suggests a mechanism for these long-standing observations in humans: drug treatment favors growth of tolerant bacteria within individual macrophages, which can expand into granulomas by recruiting new macrophages and/or migrate to disseminate infection.
Macrophage Residence Induces Drug Tolerance in Mm and Mtb
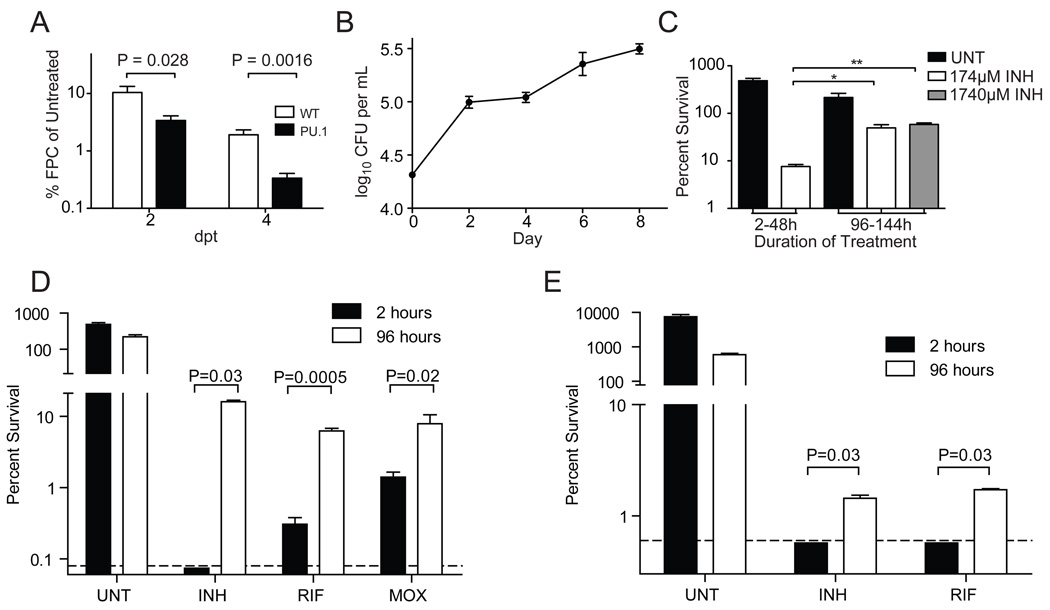
The drug tolerant bacteria observed within larval macrophages can be explained by three non-mutually exclusive mechanisms: 1) pre-existing drug tolerant persisters, 2) induction of tolerance upon exposure to the macrophage environment, and 3) induction of tolerance upon exposure to drug. Subtherapeutic concentrations of multiple drugs produce tolerance in actively growing Mtb cultures (de Steenwinkel et al., 2010; Morris et al., 2005; Viveiros et al., 2002) and we found this to also be the case for Mm (Figure S4A). To probe the mechanism(s) of tolerance in the context of host infection, we examined the development of INH tolerance in larvae depleted of macrophages by anti-sense knock down of the PU.1 myeloid transcription factor; in the absence of macrophages the Mm grow extracellularly (Clay et al., 2007). Wild-type and macrophage-depleted (PU.1) larvae were infected at one dpf and INH treatment was begun the following day. By two dpt, when INH tolerance is first apparent (Figure 2C), wild-type larvae had 10.4% ± 2.9% of the bacteria in the untreated controls, whereas PU.1-depleted larvae had 3.4% ± 0.7%, a 3.1-fold reduction (Figure 4A). The improved relative efficacy of INH in the PU.1-deficient larvae was even greater at 4 dpt with a 5.7-fold relative reduction (1.9% ± 0.4% residual bacteria in wild-type vs. 0.3% ± 0.07%) (Figure 4A). These data suggested that the tolerant bacterial population was enriched by macrophage residence.
Figure 4. Antibiotic tolerance is induced by macrophage residence.
A) Wild-type and PU.1-morphant larvae were infected with 300 Mm. One dpi, larvae were treated with 290 µM INH, or left untreated. Larvae were imaged at two and four dpt and bacterial burdens determined by FPC. For each timepoint, FPC of each treated larva was normalized to the mean FPC of the untreated control group. N=20 wild-type or 12 PU.1 morphant larvae per group. P values determined using Student’s t-test.
(B and C) J774A.1 macrophages we re infected with Mm and were left untreated, or were treated with INH prior to lysis and enumeration of CFU. (B) Growth of Mm in the untreated control wells. (C) Survival of intracellular Mm upon exposure to 174 or 1740 µM INH during the time periods indicated (two-48 hours, or 96–144 hours) prior to macrophage lysis and enumeration of CFU. Percent survival was compared using one-way ANOVA with Dunnett’s post-test.
(D) Mm were used to infect THP-1 macrophages for 2 or 96 hours prior to being released by macrophage lysis. CFU were enumerated at the time of release, and again following 48 hours exposure to 174 µM INH, 1.21 µM RIF, 7.48 µM MOX, or left untreated. For the purpose of display, values below the limit of detection (0.08%, dashed line) were arbitrarily set to 0.074%. P values were determined using Student’s t-test (RIF and MOX), or the Mann-Whitney rank test (INH).
(E) Mtb strain H37Rv was used to infect J774A.1 macrophages, and were grown and treated as described for (D), except that the concentration of INH was 4.4 µM, reflecting the greater inherent susceptibility of this organism to INH. For the purpose of display, values below the limit of detection (0.6%, dashed line) were arbitrarily set to 0.57%. P values were determined using the Mann-Whitney rank test. In all panels, error bars represent SEM.
To determine the mechanism of this macrophage-dependent tolerance, we turned to a cell culture infection model using J774 mouse or THP1 human macrophage cell lines (Volkman et al., 2004). Cells infected with Mm for either two or 96 hours were treated with 174 µM INH (three-fold the in vitro MIC) for an additional 48 hours and then lysed to obtain intracellular bacterial counts. After two hours of macrophage infection, 7.6% ± 0.8% Mm survived the subsequent 48 hours of INH treatment whereas after a 96-hour infection period, 49.5% ± 8.1% survived treatment, representing a 6.5-fold increase in persisting bacteria (Figure 4B–C). There was no additional killing with 10-fold increased INH (1740 µM) (Figure 4C). The INH MIC for the bacteria recovered at 96 hours was unchanged from the parental strain, confirming that they had not acquired genetic resistance.
Our findings suggested that intramacrophage residence induces drug tolerance. Alternatively, drug activity could be limited intracellularly due to the residence of a bacterial subpopulation in a protected niche or to drug modification by the host cell. To differentiate between these possibilities, bacteria were grown in macrophages for two or 96 hours, released from the infected macrophages and then incubated in bacterial growth medium with and without drugs for an additional 48 hours prior to plating. Even with direct exposure to 174 µM INH, the proportion of tolerant bacteria was > 200-fold higher at 96 hours post infection than after two hours (Figure 4D and Table S3). Macrophage-conditioned Mm also developed tolerance to RIF with a 20.6-fold increase in survival and to MOX with a 4.8-fold increase in survival (Figure 4D and Table S3). Dilution of the macrophage lysates prior to exposure to antibiotics did not affect the proportion of tolerant organisms (Figure S4B) excluding the possibility that tolerance was simply due to increased bacterial density at 96-hours (Figure 4B). Finally, we found that Mtb also develops macrophage-induced drug tolerance: 96 hour vs. two hour infection yielded > 2.3-fold increase in INH survival and > 2.8-fold increase in RIF survival (Figure 4E and Table S3). Thus, macrophage residence rapidly induces Mm and Mtb to become tolerant to multiple drug classes.
Drug Tolerance is Associated with a Replicating Intracellular Population
Current models invoke host-induced bacteriostasis as a mechanism for drug tolerance during infection. Indeed, both Mtb and Mm exhibit drug tolerance under conditions of slowed growth in vitro (Figure S4C)(Paramasivan et al., 2005). Moreover, the net growth of Mm in macrophages appeared to decline slightly between 48 and 96 hours (Figure 4B), raising the possibility that tolerance might result from macrophage-induced bacteriostasis. However, both our zebrafish studies and prior human clinical data showed that bacteria expand and disseminate in vivo in the face of drug treatment. Thus, even if tolerance is initially associated with nongrowing persisters, they must somehow then retain the tolerance phenotype upon resuming growth.
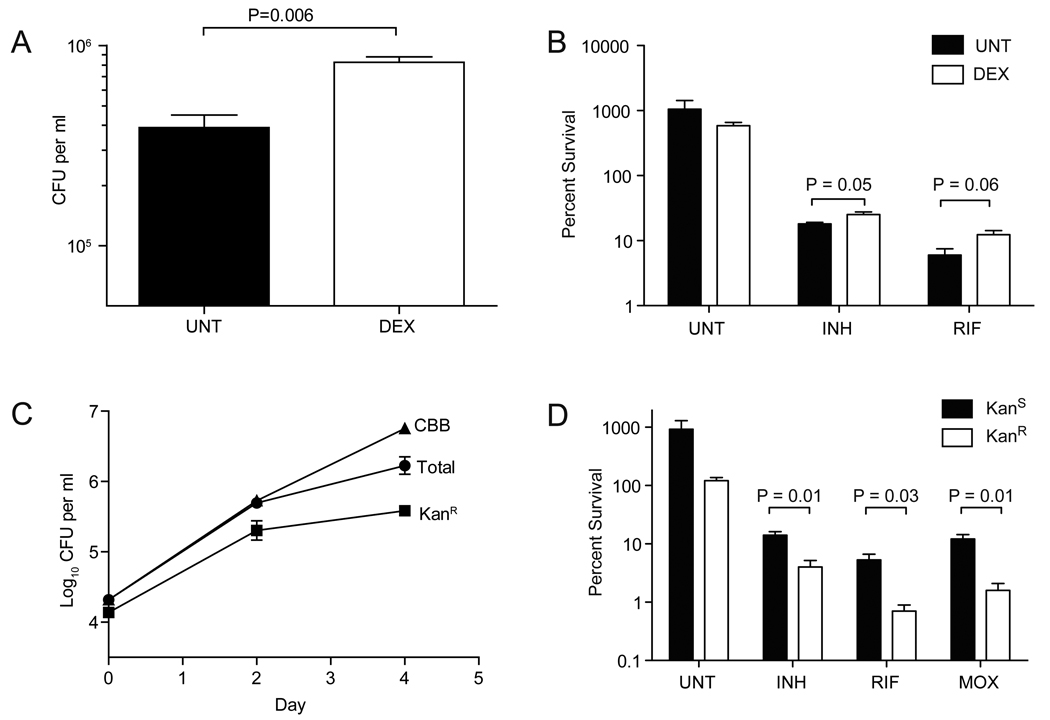
To probe the replicative state of the tolerant bacteria, we manipulated intracellular growth by treating infected macrophages with dexamethasone (DEX), a broad-spectrum anti-inflammatory agent that increases net intracellular bacteria (Rook et al., 1987). DEX treatment resulted in a 2.1-fold increase in intracellular Mm at 96 hours that was accompanied by a 1.4-fold and 2.1-fold increase in the proportion of bacteria tolerant to INH and RIF, respectively (Figure 5A–B and Table S4). These experiments revealed that tolerance is not diminished by increased bacterial growth, and suggested rather that it is enhanced in growing intracellular bacteria.
Figure 5. Growing bacteria are enriched for antibiotic tolerance.
(A and B) J774A.1 macrophages were infected with Mm and were treated with 100 nM dexamethasone at t=0. Macrophages were lysed at 96 hpi to release bacteria. (A) Total CFU at time of release. (B) Percent survival of released bacteria upon 48 hours exposure to 174 µM INH, 1.21 µM RIF, or left untreated.
(C) J774A.1 macrophages were infected with Mm/pBP10 and total and KanR CFU enumerated at 48 and 96 hpi. The cumulative bacterial burden (CBB) was calculated as described in the Supplemental Experimental Procedures. (D) Mm/pBP10 grown intracellularly for 96 hours (described in (C)) were released by macrophage lysis and then treated for an additional 48 hours with 174 µM INH, 1.21 µM RIF, 7.48 µM MOX, or left untreated, prior to enumeration of total and KanR CFU. KanS CFU were calculated as the total CFU minus the mean KanR CFU. In all panels, error bars represent SEM, and P values were determined using Student’s t-test.
We then directly compared the numbers of drug tolerant bacteria in non-growing and growing intracellular populations. We transformed Mm with the unstable plasmid pBP10 that is lost at a constant rate from dividing, but not non-dividing mycobacteria (Figure S5) (Gill et al., 2009). Using Mm/pBP10, we showed the rate of plasmid loss per generation to be unchanged under different growth conditions (Figure S5 and Supplemental Experimental Procedures). We next used Mm/pB10 to examine bacterial growth in macrophages. The generation time of the intracellular population was not decreased over the 96-hour assay period (Figure 5C and Figure S5). Furthermore, the apparent slowing of growth between 48 and 96 hours (Figure 4B) was in fact due to a large increase in bacterial death, which overshadowed a more modest increase in growth rate in this time period (Figure S5E).
We then compared the proportion of drug tolerant bacteria in the population that had retained the plasmid (KanR ; i.e. bacteria that had not yet replicated and daughter cells that had retained the plasmid upon replication) to that in the population that had lost the plasmid (KanS ; i.e. completed at least one round of replication). Drug tolerant bacteria were enriched in the KanS population: 3.5-fold for INH, 7.6-fold for RIF, and 7.6-fold for MOX (Figure 5D and Table S4). Together, these results suggested that drug tolerance was not associated with macrophage-induced bacteriostasis but rather that it was enhanced in the growing intracellular population.
Macrophage-induced bacterial efflux pumps mediate drug tolerance
One explanation for the multiclass drug tolerance we observed is a reduction in steady state intrabacterial drug concentration, which could be achieved by decreased drug entry and/or increased efflux. The induction of efflux pumps causes single or multidrug resistance in many bacteria, including Mtb (De Rossi et al., 2006; Li and Nikaido, 2009; Louw et al., 2009), so we investigated their potential role in macrophage-induced tolerance. Mtb and Mm encode numerous efflux pumps that can mediate resistance to antitubercular drugs when overexpressed (Li and Nikaido, 2009). Efflux pump activity has been invoked to explain multiple aspects of drug resistance in mycobacteria: 1) intrinsic resistance, 2) acquired multi-drug resistance, and 3) tolerance induced by antimicrobial exposure (De Rossi et al., 2006; Gupta et al., 2010; Louw et al., 2009; Morris et al., 2005; Viveiros et al., 2002). Moreover, several mycobacterial efflux pumps and their regulators are induced during macrophage infection (Fontan et al., 2008; Morris et al., 2005; Nguyen and Thompson, 2006; Ramon-Garcia et al., 2009; Rohde et al., 2007; Schnappinger et al., 2003; Zahner et al., 2010). Finally, efflux pump inhibitors enhance the activity of certain drugs on drug-resistant Mtb; these include the currently approved pump inhibitors verapamil (VER), reserpine (RES) and thioridazine (Li and Nikaido, 2009).
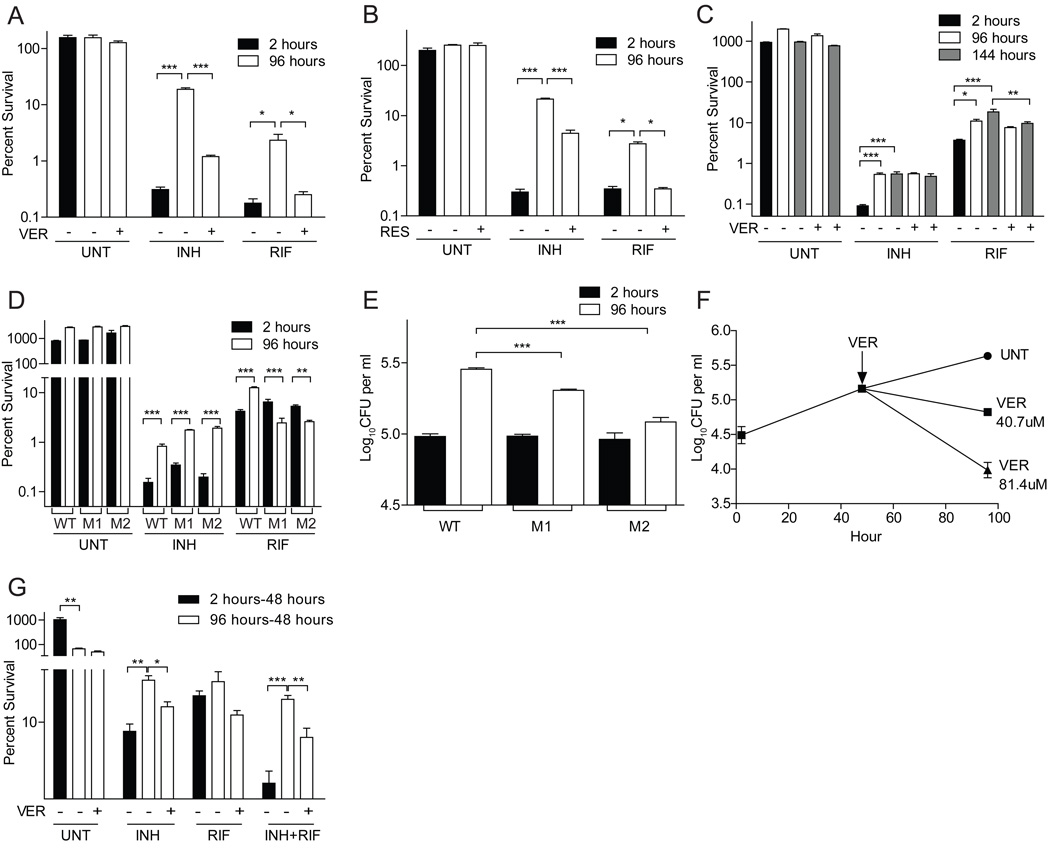
To determine if bacterial efflux pumps mediated macrophage-induced drug tolerance, we added subinhibitory VER and RES in conjunction with antibiotics to macrophage-released Mm and found that tolerance was reduced (Figure 6A–B and Table S5). VER produced a 15.6-fold reduction in INH survival and a 9.2-fold reduction in RIF survival; RES produced a 4.8-fold reduction in INH survival and a 7.9-fold reduction in RIF survival. These effects were specific to macrophage-induced tolerance: VER did not reduce stationary phase induced tolerance to INH or RIF (Figure S6A).
Figure 6. Bacterial efflux pumps confer tolerance within macrophages.
(A and B) THP-1 macrophages were infected with Mm and lysed at two or 96 hpi. The released bacteria were treated for an additional 48 hours with 174 µM INH, 1.21 µM RIF or left untreated, in the presence or absence of 81.4 µM verapamil (A) or 65.7 µM reserpine (B), prior to enumeration of CFU.
(C) THP-1 macrophages were infected with Mtb strain CDC1551 and lysed at two, 96 or 144 hpi. The released bacteria were then treated with antibiotics for 48 hours as described in panel (A), except that the concentration of INH was 4.4 µM, as described for Figure 4E.
(D and E) THP-1 macrophages were infected with Mtb strains JHU1258c-715 (“M1”), JHU1258c-833 (“M2”) and the isogenic wild-type control, CDC 1551, for two or 96 hours prior to lysis and enumeration of CFU. (D) Released bacteria were treated as described in panel (C).
(F) THP1 cells were infected with Mm for 48 hours, prior to addition of 0 (UNT), 40.7 or 81.4 µM VER for an additional 48 hours. P<0.001 using one-way ANOVA, with Dunnett’s post-test comparing each treatment group to the untreated control after 48 hours of VER treatment.
(G) THP1 cells infected with Mm for two hours or 96 hours were incubated for an additional 48 hours with 174 µM INH, 1.21 µM RIF or both, and in the presence or absence of 40.7µM VER. Cells were lysed and CFU were enumerated at the end of the 48 hour treatment. Percent survival was calculated relative to the mean intracellular counts present at the start of antibiotic exposure. For all panels, error bars represent SEM. Significance testing was performed using one-way ANOVA with Dunnett’s (A,B, D and E) or Bonferroni (C and D) post-tests.
In Mtb, VER reduced tolerance to RIF but not INH (Figure 6C and Table S5). RIF survival was reduced by 1.4-fold and 1.9-fold, respectively, in bacteria grown intracellularly for 96 and 144 hours prior to challenge. This result suggests that Mtb possesses VER-resistant pumps distinct from those used by Mm. Consistent with this idea, INH exposure induces transcription of more efflux pumps than does RIF in MDR-TB (Gupta et al., 2010). Moreover, drug-induced INH tolerance in Mtb is sensitive to RES but not to VER (Colangeli et al., 2005; Viveiros et al., 2002).
Our data suggested that distinct efflux pumps mediate INH and RIF tolerance in Mtb. The Mtb Rv1258c efflux pump is transcriptionally induced upon: 1) macrophage infection (Morris et al., 2005; Schnappinger et al., 2003) and 2) exposure to subinhibitory concentrations of RIF, but not INH, in an Mtb isolate resistant to both drugs (Siddiqi et al., 2004). Hypothesizing that Rv1258c mediates macrophage-induced RIF tolerance, we tested two Mtb strains with distinct transposon insertions in Rv1258c. After 96 hours of macrophage residence, both mutants retained INH, but lost RIF, tolerance (Figure 6D and Table S5). Indeed, they became hypersusceptible to RIF: while the wild-type had a 3.0-fold increase in tolerance, the mutants had 2.5- and 2.0-fold reductions in tolerance. Moreover, the mutants were compromised for intra-macrophage growth (Figure 6E), suggesting that this efflux pump is required for both intracellular growth and RIF tolerance.
To confirm that the loss of tolerance in the Rv1258c mutants was not simply a function of intracellular growth attenuation, we tested two additional mutants with macrophage growth defects: Mm mutants lacking the RD1/ESX-1 secretion system or the cell surface/secreted Erp protein retained macrophage-inducible antibiotic tolerance (Figure S6B–C). The Erp mutant developed RIF tolerance despite being RIF-hypersusceptible at baseline (Cosma et al., 2006b; Gao et al., 2003). This would suggest that efflux pump induction due to intracellular residence allowed the bacteria to extrude sufficient RIF to become more resistant than at baseline, despite growth attenuation. In contrast, the Rv1258c-deficient bacteria become hypersusceptible to RIF because their intracellular damage is coupled to an inability to expel RIF.
Finally, we showed that the Mtb Rv1258c mutants retained stationary phase-induced RIF tolerance, again showing specificity of this efflux pump in mediating macrophage-induced tolerance (Figure S6D). In sum, these results suggest that bacterial efflux pumps are induced upon macrophage infection to promote bacterial growth but also mediate drug tolerance. This mechanistic understanding explains the enrichment of tolerant bacteria in the growing intracellular population.
VER reduces intracellular Mm growth and tolerance
If VER reduces tolerance by inhibiting bacterial efflux pumps induced upon intracellular residence, it should reduce tolerance when administered directly to infected macrophages. It should also inhibit intracellular growth as suggested by the Rv1258c mutants. Indeed, efflux-pump and potassium-transport inhibitors have been found to promote intracellular killing of multi-drug resistant Mtb (Amaral et al., 2007). We similarly found that incubation of Mm-infected macrophages with VER reduced intracellular bacterial growth at concentrations that did not impact bacteria in axenic culture (Figure 6F and S6A). Incubation of infected macrophages with VER also inhibited tolerance (Figure 6G). 96 hour Mm-infected macrophages were treated with INH alone or INH and VER for an additional 48 hours before lysis and bacterial enumeration; the addition of VER reduced tolerance by 2.0 fold (Figure 6G and Table S5). Finally, when added to a synergistic combination of INH and RIF, VER further reduced tolerance by 2.2-fold (Figure 6G and Table S5), suggesting the potential of VER to increase the efficacy of existing treatment regimens. Taken together, the finding that mutations in Rv1258c and treatment with the efflux pump inhibitor VER lead to the same two phenotypes (growth attenuation and loss of tolerance) indicates that efflux pumps are required for both processes. Moreover, the observation that VER treatment phenocopies the Rv1258c mutation argues that it is the loss of Rv1258c function in the mutants, rather than polar effects on downstream genes, that is responsible for the observed phenotypes.
Macrophage-induced tolerance persists after bacteria are rendered extracellular
Both the zebrafish larval model and the human EBA studies reveal the existence of tolerant bacteria. However, in contrast to the intracellular bacteria present in the larvae, the tolerant bacteria assessed in the human studies are thought to be predominantly extracellular, residing within the necrotic cores of open cavitary lesions (Figure S1). Necrosis results from death of the granuloma’s macrophage core, and is sustained by continued influx and lysis of infected and uninfected macrophages (Cosma et al., 2004; Dannenberg, 2003). So for macrophage-induced tolerance to play a significant role in human cavitary TB, it must persist after the bacteria are rendered extracellular by macrophage lysis. To test this model, we lysed Mm out of macrophages and monitored the proportion of tolerant organisms over time. Mm retained tolerance for at least 120 hours after macrophage release despite continued replication in the lysate (Figure S7A–B). Whether tolerance is retained due to slow turn over of the efflux pump or persistence of the original macrophage stimulus in the lysates, remains to be determined. Regardless of the mechanism, these findings argue that extracellular bacteria in cavitary TB may be rendered tolerant by prior growth in macrophages.
DISCUSSION
Drug tolerance presents a significant challenge to the eradication of TB. Here, we exploit complementary infection models to demonstrate that macrophages and granulomas both play a role in the induction and expansion of drug tolerant bacteria.
The zebrafish larval model has enabled reassessment of fundamental dogmas about the role of macrophages and granulomas in TB pathogenesis (Davis and Ramakrishnan, 2009; Tobin and Ramakrishnan, 2008; Tobin et al., 2010; Volkman et al., 2010). These studies found that rather than serving as strictly host protective structures, granulomas are co-opted by mycobacteria for their expansion and dissemination. We now show that tolerant bacteria arising within individual macrophages similarly exploit granulomas for their amplification and dissemination during the course of drug treatment. This finding helps to explain decades-old observations that human TB lesions can expand, and new ones appear, in the face of overall clinical and radiological response (Akira et al., 2000; Bobrowitz, 1980; Canetti, 1955).
The core finding of this study are that drug tolerant bacteria originate in macrophages dependent on the activity of bacterial efflux pumps (Figure 7). Within a few days of macrophage infection, a bacterial subpopulation arises that manifests tolerance to multiple drugs independent of drug exposure. The association between intramacrophage growth and drug tolerance suggests that a common mechanism promotes both. This is borne out by our finding that Rv1258c is required for both macrophage growth and intracellular RIF tolerance in Mtb. The activation of bacterial efflux pumps in response to membrane and oxidative stress and antimicrobial peptides in vitro (Morris et al., 2005; Nguyen and Thompson, 2006; Ramon-Garcia et al., 2009; Zahner et al., 2010) suggests that their induction during intracellular residence may protect the bacteria from these same conditions in vivo. For example, efflux pumps mediate intrinsic resistance of Neisseria meningitidis to human antimicrobial peptides (Tzeng et al., 2005). The finding that DEX-treated macrophages induce tolerance may shed some light on the intracellular stimuli involved. DEX-treatment suppresses macrophage oxidative bursts while preserving antimicrobial peptide expression (Duits et al., 2001; Ehrchen et al., 2007). Antimicrobial peptides, which are induced in Mtb-infected human macrophages and are required for macrophage antimycobacterial activity (Liu et al., 2007; Rivas-Santiago et al., 2008), may mediate the induction of bacterial efflux pumps and thereby tolerance.
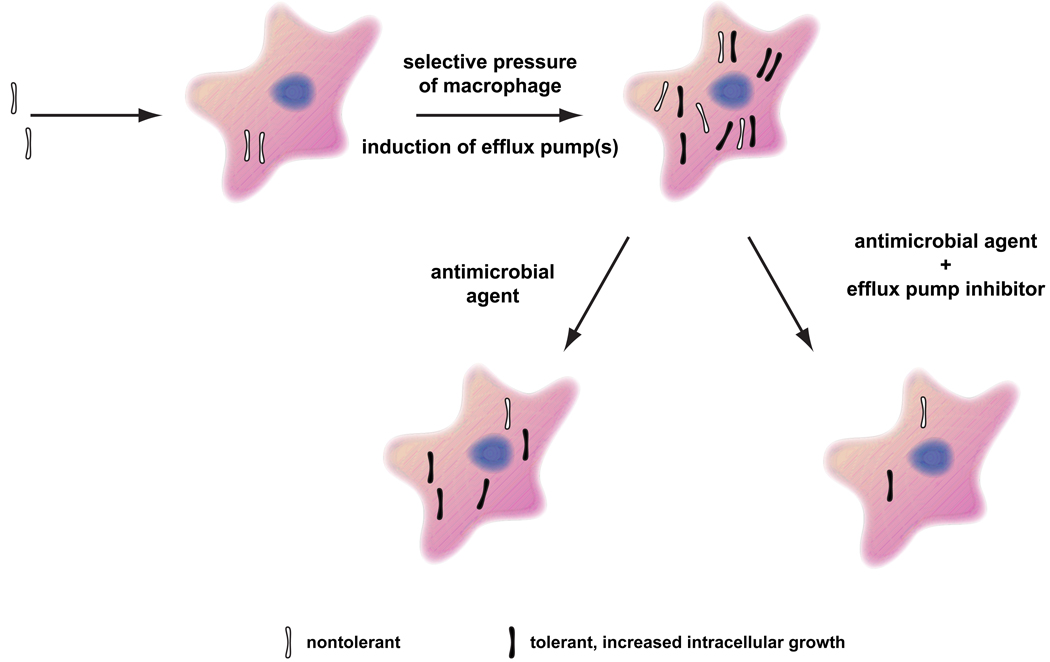
Figure 7. Model for the mechanism of antibiotic tolerance in TB and its treatment.
Nontolerant bacteria are phagocytosed by macrophages soon after infection wherein they induce efflux pumps to counter macrophage defenses. These efflux pumps render bacteria tolerant to multiple antitubercular drugs. The tolerant bacteria are associated with the growing population because of their enhanced ability to counter macrophage defenses. Antitubercular drug treatment spares tolerant bacteria and the addition of efflux pump inhibitors reduces their numbers.
Our in vivo results show that tolerant bacteria arise within a subset of infected macrophages during treatment. This may be due to macrophage heterogeneity, again underscoring the complexity of the macrophage-Mycobacterium interface. Individual macrophages may cause distinct and/or differential induction of bacterial efflux pumps, and thus, variable tolerance within the bacterial population. Additionally, antibiotics have long been thought to act in co-operation with cellular host defenses, an idea further supported by the increasing recognition of the complex, multidimensional mechanisms of antibiotic activity (Kohanski et al., 2010).
How do our findings relate to human clinical studies? Among the readily cultivable bacilli in the sputum of humans with active cavitary TB, two distinct populations have been described: drug-susceptible and drug-tolerant (Jindani et al., 2003). The metabolic status of the tolerant population has been the source of much debate. Spurred by our discovery of early tolerance in zebrafish larvae, we revisited human EBA and radiological studies, and realized that growing, tolerant populations must mediate the progression and expansion of tubercular lesions that occur in the face of treatment (Bobrowitz, 1980; Jindani et al., 2003). The association between tolerance and intracellular growth in the zebrafish larval and macrophage models provides a mechanism for the earlier human studies. Further strengthening this link is our finding that tolerance persists for days after the bacteria are released from macrophages suggesting that the bacteria present in the necrotic core of macrophage-lined cavities may utilize similar tolerance mechanisms.
Slowly growing bacteria that are not cultivable under standard conditions have also been observed in human sputum (Garton et al., 2008; Mukamolova et al., 2010). These too appear to be drug tolerant, but their relative impact on treatment duration and clinical relapse is unclear (Mukamolova et al., 2010). Importantly, tolerance models centered on nonreplicating bacteria do not account for the recent finding that diarylquinolones, which are equally bactericidal for exponentially-growing and dormant bacteria in culture (Koul et al., 2008), only shorten the time to cure from six months to four in the mouse model of TB, which features slowed bacterial growth (Ibrahim et al., 2009). While this shortening may be extremely important clinically, it does not support the notion that tolerance, and thus the long duration required for therapy, is mediated solely by dormant bacterial populations. One possibility is that the growing, tolerant bacteria revealed in this work are responsible for the substantial residual tolerance observed after diarylquinolone-containing therapies.
In summary, we have identified a population of drug tolerant bacteria that are likely to be substantially present in TB patients as they are induced by macrophages, which host dynamic mycobacterial populations throughout infection. Current drug discovery efforts, with their emphasis on quiescent tolerant bacteria, fail to take into account the role of this population in tolerance. Our identification of pharmacological measures to reduce the numbers of growing tolerant bacteria suggests an approach to further shorten TB chemotherapy. Our finding that the same pump mediates both growth and tolerance suggests that the identification of more potent specific inhibitors with a dual bacterial killing mechanism is possible. Indeed, some are already being tested against Rv1258c (Sangwan et al., 2008; Sharma et al., 2010). Ultimately, the best assessment of whether targeting these newly discovered tolerant bacteria is the key to truly short course chemotherapy will come from clinical trials using efflux pump inhibitors like VER.
Finally, the tolerance mechanisms and counter strategies that we describe for mycobacteria may be relevant to other intracellular pathogens. Indeed, macrophage-induced tolerance to multiple drug classes is described for Legionella pneumophila, an agent of pneumonia (Barker et al., 1995). Thus, our findings may have relevance for Legionella as well as other recalcitrant intracellular pathogens that produce serious and often relapsing infections where tolerance is a barrier to therapy.
EXPERIMENTAL PROCEDURES
Bacterial strains and Methods
M. marinum strain M (ATCC BAA-535) and its fluorescent derivatives have been described (Cosma et al., 2006a; Davis and Ramakrishnan, 2009). A spontaneous INH resistant mutant identified in our laboratory (KA1), was found to have increased resistance to INH (MIC 64 µg/ml) and the katG locus was subsequently sequenced. Plasmid pBP10 (gift of D. Sherman, Seattle Biomed) was transformed into strain M to yield strain KA2, referred to in the text as Mm/pBP10. Mm were grown in standard media (see Supplemental Experimental Procedures) and prepared for experimental manipulations by growth to mid-log phase, followed by passage through a sterile 5µM filter to yield a single cell suspension. INH susceptibility of bacteria isolated from INH-treated larvae was verified by patching all outgrown colonies onto 7H10 agar containing 10, 20, and 40 µg/ml INH, and comparing growth to that of the parental strain. The INH MIC of three colonies was confirmed to be identical to strain M.
Mtb strain H37Rv was from D. Sherman (Seattle Biomed). Mtb strains JHU1258c-715, and JHU1258c-833 (harboring transposon insertions at positions 715 and 833, respectively, in the Rv1258c ORF), and the wild-type parent strain, CDC1551, were from W. R. Bishai and G. Lamichhane (Johns Hopkins University) (Lamichhane et al., 2003). Mtb were grown to mid log phase in standard medium prior to infection.
Zebrafish infections
Wild-type AB zebrafish were from our laboratory stock, and PU.1 morphant embryos have been described (Clay et al., 2007).. Larvae were infected via caudal vein injections at 36–48 hours post-fertilization as described (Davis and Ramakrishnan, 2009).
Microscopy
Wide field fluorescence microscopy was performed using a Nikon E600 equipped with a Nikon D-FL-E fluorescence unit with a 100W Mercury lamp. Wide field fluorescence images were captured using a CoolSnap HQ CCD camera (Photometrics) with MetaMorph 7.1 (Molecular Devices).
Fluorescent Pixel Count (FPC)
Quantification of infection with fluorescent Mm using images of individual embryos was performed using custom-made MATLAB software developed in-house. Briefly, in each image, the number of pixels with a fluorescence intensity greater than the brightest pixel observed in images of control uninfected embryos is counted. This count represents the total fluorescent area (in pixels) for each infected larva.
Calculation of Early Bactericidal Activity
FPC counts were log10 transformed and for each larva the difference between each day’s measurement and the initial burden was calculated. EBAs over defined intervals were calculated by taking the mean and SD of the Δ log10 FPC values calculated for individual larvae.
Macrophage Growth and Infection
J774A.1 and THP-1 macrophages were grown in DMEM and RPMI, respectively, supplemented with 10% FBS and 1% L-glutamine. THP-1 cells were differentiated with phorbol 12-myristate 13-acetate for 48 hours prior to infection. 1×105 J774A.1 or 5×105 THP-1 macrophages were infected at an MOI of 1.5 for 2–3 hours at 33°C (for Mm) or an MOI of 1 for 2–3 hours at 37°C (for Mtb). Cells were washed with medium and then 20 (for Mm) or 6 (for Mtb) µg/ml STM was added (t=0) for the duration of the intracellular growth. Medium was changed daily. Macrophages were lysed with 0.1% Triton X-100 for intracellular growth quantization. To lyse macrophages and release bacteria for subsequent tolerance assessment, each well was first washed once with 1× PBS, and once with diH2O, with the latter wash being removed immediately. Then, 200µl of diH2O was added and the cells incubated for 15 minutes to lyse macrophages. Finally, 800µl of 1.25× concentrated 7H9 medium (see Supplemental Experimental Procedures) was added and the wells scraped gently with a pipette tip to release all release all macrophages. CFU were enumerated from triplicate wells on supplemented 7H10 agar. For determination of antibiotic killing, the percent survival was calculated by dividing the CFU for each well by the mean CFU present prior to treatment.
Statistics
Statistical analyses were performed using Prism 5.01 (GraphPad). For data sets requiring log10 transformation prior to ANOVA, embryos with no detectable fluorescence above background, or with no detectable CFU were assigned a value of 0.9, with 1 being the limit of detection, prior to log10 transformation. A Mann Whitney rank test was used when values in one group were all below the limit of detection. Post test P values are as follows: *, P<0.05; **, P<0.01; ***P<0.001.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D.R. Sherman for the gift of pBP10, advice and the use of the BSL-3 facilities at Seattle BioMed, and the members of his laboratory, especially R. Liao and J. Pang for help in using the BSL-3 facilities, and M. Nixon for advice in analyzing the unstable plasmid data, W.R. Bishai and G. Lamichhane for providing the Mtb Rv1258c mutants through the Tuberculosis Animal Research and Gene Evaluation Taskforce (NIH/NIAID N01 AI30036), E. Nuermberger and P. Donald for sharing knowledge and insights about TB drugs, B. Cormack and M. Troll for discussion, J. M. Davis for technical advice, J. Ray and H. Volkman for figure design and preparation, and S. Phillips and T. Pecor for technical assistance. This work was supported by a Gates Foundation TB drug accelerator grant, a University of Washington Royalty Research Fund Grant, a Burroughs Wellcome Foundation Pathogenesis of Infectious Diseases award and NIH grants RO1 AI036396, RO1 AI54503 and U54AI057141 to LR, NIH grant T32 AI55396 to KNA, a Levinson Emerging Scholars Program Grant, a Mary Gates Endowment Grant, and a Washington NASA Space Grant to KW, a Pfizer Fellowship in Infectious Diseases and NIH grant K08 AI076620 to LEC, and NIH grants 5R21 AI073328-02 and 5R21 AI078189-02 to PHE. LR is a recipient of the NIH Director’s Pioneer Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akira M, Sakatani M, Ishikawa H. Transient radiographic progression during initial treatment of pulmonary tuberculosis: CT findings. J Comput Assist Tomogr. 2000;24:426–431. doi: 10.1097/00004728-200005000-00014. [DOI] [PubMed] [Google Scholar]
- Amaral L, Martins M, Viveiros M. Enhanced killing of intracellular multidrug-resistant Mycobacterium tuberculosis by compounds that affect the activity of efflux pumps. J Antimicrob Chemother. 2007;59:1237–1246. doi: 10.1093/jac/dkl500. [DOI] [PubMed] [Google Scholar]
- Aubry A, Chosidow O, Caumes E, Robert J, Cambau E. Sixty-three cases of Mycobacterium marinum infection: clinical features, treatment, and antibiotic susceptibility of causative isolates. Arch Intern Med. 2002;162:1746–1752. doi: 10.1001/archinte.162.15.1746. [DOI] [PubMed] [Google Scholar]
- Barker J, Scaife H, Brown MR. Intraphagocytic growth induces an antibiotic-resistant phenotype of Legionella pneumophila. Antimicrob Agents Chemother. 1995;39:2684–2688. doi: 10.1128/aac.39.12.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry CE, 3rd, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, Schnappinger D, Wilkinson RJ, Young D. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol. 2009;7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger AE, Besra GS, Ford ME, Mikusova K, Belisle JT, Brennan PJ, Inamine JM. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc Natl Acad Sci U S A. 1996;93:11919–11924. doi: 10.1073/pnas.93.21.11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrowitz ID. Reversible roentgenographic progression in the initial treatment of pulmonary tuberculosis. Am Rev Respir Dis. 1980;121:735–742. doi: 10.1164/arrd.1980.121.4.735. [DOI] [PubMed] [Google Scholar]
- Canetti G. American rev. ed. edn. New York: Springer Publishing Company; 1955. The tubercle bacillus in the pulmonary lesion of man; histobacteriology and its bearing on the therapy of pulmonary tuberculosis. [Google Scholar]
- Clay H, Davis J, Beery D, Huttenlocher A, Lyons S, Ramakrishnan L. Dichotomous Role of the Macrophage in Early Mycobacterium marinum Infection of the Zebrafish. Cell Host and Microbe. 2007;2:29–39. doi: 10.1016/j.chom.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangeli R, Helb D, Sridharan S, Sun J, Varma-Basil M, Hazbon MH, Harbacheuski R, Megjugorac NJ, Jacobs WR, Jr, Holzenburg A, et al. The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol Microbiol. 2005;55:1829–1840. doi: 10.1111/j.1365-2958.2005.04510.x. [DOI] [PubMed] [Google Scholar]
- Connolly LE, Edelstein PH, Ramakrishnan L. Why is long-term therapy required to cure tuberculosis? PLoS Med. 2007;4:e120. doi: 10.1371/journal.pmed.0040120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma CL, Davis JM, Swaim LE, Volkman H, Ramakrishnan L. Current Protocols in Microbiology. John Wiley and sons, Inc; 2006a. Zebrafish and Frog Models of Mycobacterium marinum Infection. [DOI] [PubMed] [Google Scholar]
- Cosma CL, Humbert O, Ramakrishnan L. Superinfecting mycobacteria home to established tuberculous granulomas. Nat Immunol. 2004;5:828–835. doi: 10.1038/ni1091. [DOI] [PubMed] [Google Scholar]
- Cosma CL, Klein K, Kim R, Beery D, Ramakrishnan L. Mycobacterium marinum Erp is a virulence determinant required for cell wall integrity and intracellular survival. Infect Immun. 2006b;74:3125–3133. doi: 10.1128/IAI.02061-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg AM., Jr Macrophage turnover, division and activation within developing, peak and "healed" tuberculous lesions produced in rabbits by BCG. Tuberculosis (Edinb) 2003;83:251–260. doi: 10.1016/s1472-9792(03)00048-9. [DOI] [PubMed] [Google Scholar]
- Davis JM, Ramakrishnan L. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell. 2009;136:37–49. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rossi E, Ainsa JA, Riccardi G. Role of mycobacterial efflux transporters in drug resistance: an unresolved question. FEMS Microbiol Rev. 2006;30:36–52. doi: 10.1111/j.1574-6976.2005.00002.x. [DOI] [PubMed] [Google Scholar]
- Decostere A, Hermans K, Haesebrouck F. Piscine mycobacteriosis: a literature review covering the agent and the disease it causes in fish and humans. Vet Microbiol. 2004;99:159–166. doi: 10.1016/j.vetmic.2003.07.011. [DOI] [PubMed] [Google Scholar]
- Donald PR, Diacon AH. The early bactericidal activity of anti-tuberculosis drugs: a literature review. Tuberculosis (Edinb) 2008;88 Suppl 1:S75–S83. doi: 10.1016/S1472-9792(08)70038-6. [DOI] [PubMed] [Google Scholar]
- Donald PR, McIlleron H. Antituberculosis drugs. Saunders Elsevier; 2009. [Google Scholar]
- Duits LA, Rademaker M, Ravensbergen B, van Sterkenburg MA, van Strijen E, Hiemstra PS, Nibbering PH. Inhibition of hBD-3, but not hBD-1 and hBD-2, mRNA expression by corticosteroids. Biochem Biophys Res Commun. 2001;280:522–525. doi: 10.1006/bbrc.2000.4157. [DOI] [PubMed] [Google Scholar]
- Ehrchen J, Steinmuller L, Barczyk K, Tenbrock K, Nacken W, Eisenacher M, Nordhues U, Sorg C, Sunderkotter C, Roth J. Glucocorticoids induce differentiation of a specifically activated, anti-inflammatory subtype of human monocytes. Blood. 2007;109:1265–1274. doi: 10.1182/blood-2006-02-001115. [DOI] [PubMed] [Google Scholar]
- Fontan PA, Aris V, Alvarez ME, Ghanny S, Cheng J, Soteropoulos P, Trevani A, Pine R, Smith I. Mycobacterium tuberculosis sigma factor E regulon modulates the host inflammatory response. J Infect Dis. 2008;198:877–885. doi: 10.1086/591098. [DOI] [PubMed] [Google Scholar]
- Garton NJ, Waddell SJ, Sherratt AL, Lee SM, Smith RJ, Senner C, Hinds J, Rajakumar K, Adegbola RA, Besra GS, et al. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 2008;5:e75. doi: 10.1371/journal.pmed.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill WP, Harik NS, Whiddon MR, Liao RP, Mittler JE, Sherman DR. A replication clock for Mycobacterium tuberculosis. Nat Med. 2009;15:211–214. doi: 10.1038/nm.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta AK, Katoch VM, Chauhan DS, Sharma R, Singh M, Venkatesan K, Sharma VD. Microarray analysis of efflux pump genes in multidrug-resistant Mycobacterium tuberculosis during stress induced by common anti-tuberculous drugs. Microb Drug Resist. 2010;16:21–28. doi: 10.1089/mdr.2009.0054. [DOI] [PubMed] [Google Scholar]
- Ibrahim M, Truffot-Pernot C, Andries K, Jarlier V, Veziris N. Sterilizing activity of R207910 (TMC207)-containing regimens in the murine model of tuberculosis. Am J Respir Crit Care Med. 2009;180:553–557. doi: 10.1164/rccm.200807-1152OC. [DOI] [PubMed] [Google Scholar]
- Jindani A, Dore CJ, Mitchison DA. Bactericidal and sterilizing activities of antituberculosis drugs during the first 14 days. Am J Respir Crit Care Med. 2003;167:1348–1354. doi: 10.1164/rccm.200210-1125OC. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol. 2010;8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koul A, Vranckx L, Dendouga N, Balemans W, Van den Wyngaert I, Vergauwen K, Gohlmann HW, Willebrords R, Poncelet A, Guillemont J, et al. Diarylquinolines are bactericidal for dormant mycobacteria as a result of disturbed ATP homeostasis. J Biol Chem. 2008;283:25273–25280. doi: 10.1074/jbc.M803899200. [DOI] [PubMed] [Google Scholar]
- Lamichhane G, Zignol M, Blades NJ, Geiman DE, Dougherty A, Grosset J, Broman KW, Bishai WR. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2003;100:7213–7218. doi: 10.1073/pnas.1231432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria: an update. Drugs. 2009;69:1555–1623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PT, Stenger S, Tang DH, Modlin RL. Cutting edge: vitamin D-mediated human antimicrobial activity against Mycobacterium tuberculosis is dependent on the induction of cathelicidin. J Immunol. 2007;179:2060–2063. doi: 10.4049/jimmunol.179.4.2060. [DOI] [PubMed] [Google Scholar]
- Louw GE, Warren RM, Gey van Pittius NC, McEvoy CR, Van Helden PD, Victor TC. A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob Agents Chemother. 2009;53:3181–3189. doi: 10.1128/AAC.01577-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RP, Nguyen L, Gatfield J, Visconti K, Nguyen K, Schnappinger D, Ehrt S, Liu Y, Heifets L, Pieters J, et al. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2005;102:12200–12205. doi: 10.1073/pnas.0505446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamolova GV, Turapov O, Malkin J, Woltmann G, Barer MR. Resuscitation-promoting factors reveal an occult population of tubercle Bacilli in Sputum. Am J Respir Crit Care Med. 2010;181:174–180. doi: 10.1164/rccm.200905-0661OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Elias EJ, Timm J, Botha T, Chan WT, Gomez JE, McKinney JD. Replication dynamics of Mycobacterium tuberculosis in chronically infected mice. Infect Immun. 2005;73:546–551. doi: 10.1128/IAI.73.1.546-551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L, Thompson CJ. Foundations of antibiotic resistance in bacterial physiology: the mycobacterial paradigm. Trends Microbiol. 2006;14:304–312. doi: 10.1016/j.tim.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Paramasivan CN, Sulochana S, Kubendiran G, Venkatesan P, Mitchison DA. Bactericidal action of gatifloxacin, rifampin, and isoniazid on logarithmic- and stationary-phase cultures of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2005;49:627–631. doi: 10.1128/AAC.49.2.627-631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon-Garcia S, Martin C, Thompson CJ, Ainsa JA. Role of the Mycobacterium tuberculosis P55 efflux pump in intrinsic drug resistance, oxidative stress responses, and growth. Antimicrob Agents Chemother. 2009;53:3675–3682. doi: 10.1128/AAC.00550-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich AR. The Pathogenesis of Tuberculosis. 2nd edn. Springfield, IL: Charles C. Thomas; 1946. [Google Scholar]
- Rivas-Santiago B, Hernandez-Pando R, Carranza C, Juarez E, Contreras JL, Aguilar-Leon D, Torres M, Sada E. Expression of cathelicidin LL-37 during Mycobacterium tuberculosis infection in human alveolar macrophages, monocytes, neutrophils, and epithelial cells. Infect Immun. 2008;76:935–941. doi: 10.1128/IAI.01218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde KH, Abramovitch RB, Russell DG. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe. 2007;2:352–364. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Rook GA, Steele J, Ainsworth M, Leveton C. A direct effect of glucocorticoid hormones on the ability of human and murine macrophages to control the growth of M. tuberculosis. Eur J Respir Dis. 1987;71:286–291. [PubMed] [Google Scholar]
- Sacchettini JC, Rubin EJ, Freundlich JS. Drugs versus bugs: in pursuit of the persistent predator Mycobacterium tuberculosis. Nat Rev Microbiol. 2008;6:41–52. doi: 10.1038/nrmicro1816. [DOI] [PubMed] [Google Scholar]
- Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. Tuberculosis drug resistance mutation database. PLoS Med. 2009;6:e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, et al. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kumar M, Nargotra A, Koul S, Khan IA. Piperine as an inhibitor of Rv1258c, a putative multidrug efflux pump of Mycobacterium tuberculosis. J Antimicrob Chemother. 2010;65:1694–1701. doi: 10.1093/jac/dkq186. [DOI] [PubMed] [Google Scholar]
- Siddiqi N, Das R, Pathak N, Banerjee S, Ahmed N, Katoch VM, Hasnain SE. Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a tap-like efflux pump. Infection. 2004;32:109–111. doi: 10.1007/s15010-004-3097-x. [DOI] [PubMed] [Google Scholar]
- Tobin DM, Ramakrishnan L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol. 2008;10:1027–1039. doi: 10.1111/j.1462-5822.2008.01133.x. [DOI] [PubMed] [Google Scholar]
- Tobin DM, Vary JC, Jr, Ray JP, Walsh GS, Dunstan SJ, Bang ND, Hagge DA, Khadge S, King MC, Hawn TR, et al. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell. 2010;140:717–730. doi: 10.1016/j.cell.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng YL, Ambrose KD, Zughaier S, Zhou X, Miller YK, Shafer WM, Stephens DS. Cationic antimicrobial peptide resistance in Neisseria meningitidis. J Bacteriol. 2005;187:5387–5396. doi: 10.1128/JB.187.15.5387-5396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viveiros M, Portugal I, Bettencourt R, Victor TC, Jordaan AM, Leandro C, Ordway D, Amaral L. Isoniazid-induced transient high-level resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2002;46:2804–2810. doi: 10.1128/AAC.46.9.2804-2810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman HE, Clay H, Beery D, Chang JC, Sherman DR, Ramakrishnan L. Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant. PLoS Biol. 2004;2:e367. doi: 10.1371/journal.pbio.0020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman HE, Pozos TC, Zheng J, Davis JM, Rawls JF, Ramakrishnan L. Tuberculous granuloma induction via interaction of a bacterial secreted protein with host epithelium. Science. 2010;327:466–469. doi: 10.1126/science.1179663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner DF, Mizrahi V. Tuberculosis chemotherapy: the influence of bacillary stress and damage response pathways on drug efficacy. Clin Microbiol Rev. 2006;19:558–570. doi: 10.1128/CMR.00060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner D, Zhou X, Chancey ST, Pohl J, Shafer WM, Stephens DS. Human antimicrobial peptide LL-37 induces MefE/Mel-mediated macrolide resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2010;54:3516–3519. doi: 10.1128/AAC.01756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.