Abstract
The KRAS gene is the most common locus for somatic gain-of-function mutations in human cancer. Germline KRAS mutations were shown recently to be associated with developmental disorders, including Noonan syndrome (NS), cardio-facio-cutaneous syndrome (CFCS), and Costello syndrome (CS). The molecular basis of this broad phenotypic variability has in part remained elusive so far. Here, we comprehensively analyzed the biochemical and structural features of ten germline KRAS mutations using physical and cellular biochemistry. According to their distinct biochemical and structural alterations, the mutants can be grouped into five distinct classes, four of which markedly differ from RAS oncoproteins. Investigated functional alterations comprise the enhancement of intrinsic and guanine nucleotide exchange factor (GEF) catalyzed nucleotide exchange, which is alternatively accompanied by an impaired GTPase-activating protein (GAP) stimulated GTP hydrolysis, an overall loss of functional properties, and a deficiency in effector interaction. In conclusion, our data underscore the important role of RAS in the pathogenesis of the group of related disorders including NS, CFCS, and CS, and provide clues to the high phenotypic variability of patients with germline KRAS mutations.
Keywords: Noonan syndrome, NS, gain of function, GAP resistance, KRAS, Ras isoforms, Ras mutations
Introduction
RAS proteins (HRAS, KRAS 4A, KRAS 4B, and NRAS) are central signal transduction molecules, which act as molecular switches through cycling between an active, GTP-bound, and an inactive, GDP-bound state [Vetter and Wittinghofer, 2001]. The intrinsic functions of RAS proteins, their GDP/GTP exchange and GTP-hydrolysis, are extremely slow. Guanine nucleotide exchange factors (GEFs) accelerate the exchange of bound GDP for the cellular abundant GTP [Guo et al., 2005], whereas GTPase activating proteins (GAPs) terminate RAS signaling by stimulation of the GTP hydrolysis reaction [Scheffzek and Ahmadian, 2005]. In its GTP-bound form, RAS interacts with and regulates a spectrum of functionally diverse downstream effectors including RAF kinases, phosphatidylinositol 3-kinase (PI3K), and RALGDS [Herrmann, 2003]. In the past years, considerable progress has been achieved in understanding the functions and underlying mechanisms of RAS proteins. Comprehensive structural studies resulted in determination of more than 50 structures (Supp. Tables S1 and S2), and provided a deep insight into the three-dimensional fold, the consequences of nucleotide binding and hydrolysis, the principles of regulation by GEFs and GAPs, and the specificity of effector binding [Fiegen et al., 2006]. These three classes of interacting proteins predominantly bind to two highly mobile regions, designated as switch I (residues 30–37) and switch II (residues 60–74) (Fig. 1) [Sprang, 1997; Vetter and Wittinghofer, 2001].
Figure 1.
Relative positions of amino acids in KRAS altered in patients with NS, CFCS, and CS. A: Secondary structure elements and conserved motifs of RAS. The α-helices and β-strands are illustrated as cylinders and arrows, respectively. The G-domain of RAS also consists of five conserved motifs (G1–G5; gray boxes) that are responsible for specific and tight nucleotide binding and hydrolysis. Bold lines indicate the position of specific RAS signatures including the hypervariable region (HVR), which is polybasic in KRAS 4B. Amino acids investigated in this study are indicated by arrows. The isoprenylation site of the protein is at the cysteine of the C-terminal CaaX motif. B, C: Solvent accessible surfaces of HRAS molecules are shown in the inactive GDP-bound state (B) and the active GTP-bound state (C). For clarity, structures are illustrated in three different views. Therefore, central panels are rotated 90° around the vertical axes to the right (left panel) and to the left (right panel). Amino acids altered in patients with NS, CFCS, or CS are color coded. Dashed arrows depict critical residues buried within the hydrophobic core of the protein.
Since their discovery as proto-oncogenes 35 years ago, somatic RAS mutations have been found to be highly prevalent in a variety of human cancers [Barbacid, 1990; Bos, 1989; Der, 1989; Kranenburg, 2005]. The majority of gain-of-function mutations affect amino acid residues G12, G13, and Q61 [Der et al., 1986; Malumbres and Barbacid, 2003; Seeburg et al., 1984] (www.sanger.ac.uk/genetics/CGP/cosmic/) triggering RAS accumulation in the active, GTP-bound state by impairing intrinsic GTPase activity, and conferring resistance to GAPs [Ahmadian, 2002; Ahmadian et al., 1999; Bos et al., 2007].
Recently, germline mutations in HRAS, NRAS, and KRAS genes have been identified in patients with various developmental disorders including Noonan syndrome (NS; MIM# 163950), Costello syndrome (CS; MIM# 218040), and cardio-faciocutaneous syndrome (CFCS; MIM# 115150) that share several phenotypic abnormalities, such as craniofacial dysmorphism, hair and skin abnormalities, cardiac defects, cognitive impairment, and postnatal growth deficiency [Schubbert et al., 2007a]. Moreover, these disorders have reportedly been associated with cancer (e.g., juvenile myelomonocytic leukemia in patients with NS and rhabdomyosarcoma in patients with CS).
HRAS point mutations affecting amino acids at positions 12, 13, and 117 or duplication at position 37 have been associated with CS [Denayer et al., 2008; Estep et al., 2006; Gremer et al., 2010; Gripp et al., 2006; Sol-Church et al., 2006]. NRAS mutations at positions 50 and 60 have been recently shown to enhance stimulus-dependent MAPK activation and account for rare cases of NS [Cirstea et al., 2010]. In contrast, the phenotypic spectrum caused by germline KRAS mutations at amino acid positions K5, V14, Q22, P34, I36, T58, G60, V152, D153, and F156 is remarkably broad and comprises NS, CFC, and, more rarely, a phenotype consistent with CS [Carta et al., 2006; Kratz et al., 2007; Lo et al., 2009; Nava et al., 2007; Niihori et al., 2006; Schubbert et al., 2006, 2007b; Zenker et al., 2007].
The pathophysiological mechanism underlying these clinically related syndromes is most likely a dysregulated signal flow through the RAS/MAPK pathway [Gelb and Tartaglia, 2006; Kratz et al., 2007; Tidyman and Rauen, 2009]. To gain insight into the underlying mechanisms, we set out to investigate 10 different germline KRAS mutants (p.K5N, p.V14I, p.Q22E, p.Q22R, p.P34L, p.P34R, p.T58I, p.G60R, p.D153V, p.F156L). Results from our global biochemical and functional characterization described in this study provide strong evidence for the existence of distinct structural, mechanistic, and functional changes that can result in an overall enhancement of RAS signaling.
Materials and Methods
Mutation Nomenclature
Amino acid substitutions are named according to the literature, with the first methionine encoded by the ATG start codon that is designated as amino acid number “1” following the journal guidelines (www.hgvs.org/mutnomen).
Plasmids
KRAS cDNA was cloned in the pEYFP–c1 vector via Xho1 and BamH1. pEYFP–KRAS and ptacHRAS [Tucker et al., 1986] were used as template, respectively, to generate the KRAS mutations using a polymerase chain reaction (PCR)-based site-directed mutagenesis protocol as described [Ahmadian et al., 1997]. Neurofibromin 1 catalytic domain NF1–333 (amino acids 1198–1531) was cloned in pGEX-4T-1 via EcoRI and NotI Gene segments encoding RAF1-RBD, RALGDS-RBD, and SOS1 catalytic domain CDC25 were cloned in the pGEX vectors as described [Herrmann et al., 1995; Lenzen et al., 1998; Vetter et al., 1999].
Proteins and Fluorescent Nucleotides
Wild-type and mutant HRAS proteins were prepared from Escherichia coli using the ptac-expression system as described [Tucker et al., 1986]. The nucleotide-free form of RAS was prepared as described [Ahmadian et al., 2002] and the fluorescent derivatives of GDP, GTP, and GppNHp (mantGDP, mantGTP, and mantGppNHp) were synthesized according to Ahmadian et al. [2002]. RAS · mantGDP, RAS · mantGTP, and RAS · mantGppNHp were prepared as described [Gremer et al., 2008]. RAF1-RBD, RALGDS-RBD, the catalytic domains of SOS1 (CDC25), and of neurofibromin (NF1–333) were produced as glutathione S-transferase (GST) fusion proteins in E. coli. All proteins were purified as described previously [Ahmadian et al., 2002; Hemsath and Ahmadian, 2005].
Biochemical Methods
Various intrinsic and extrinsic biochemical properties of the RAS proteins were measured as described before [Ahmadian et al., 2002; Hemsath and Ahmadian, 2005]. The association of mantGDP and mantGppNHp (0.2 µM, respectively) to the nucleotide-free RAS proteins (0.3 µM) was measured in 30mM Tris pH 7.5, 5mM MgCl2, and 3mM DTE at 25°C using an Applied-Photophysics stopped flow apparatus. Dissociation of mantGDP from the RAS proteins (0.3 µM) in the presence of 40 µM GDP was measured in 30mM Tris pH 7.5, 10mM KPi, 5mM MgCl2, and 3mM dithioerythritol (DTE) at 25°C using a Fluoromax 4 (Horiba Jobin Yvon™) fluorimeter at 366nm (excitation wavelength) and 450nm (emission wavelength). Observed rate constants (kobs) of association and dissociation were obtained by single exponential fitting of the data.
GTP hydrolysis of the RAS proteins (1 µM RAS · GTP containing 6 nM [γ]32GTP) was analyzed in a 30mM Tris/HCl pH 7.5, 10mM MgCl2, 3mM DTE, pH 7.5 buffer at 25°C by determining the release of radioactive (32γ)Pi in a charcoal assay. The time courses monitoring the release of radioactive Pi were fitted using single exponential equations. Observed rate constants (kobs) were obtained by single exponential fitting of the data.
GEF-catalyzed mantGDP dissociation from RAS proteins (0.3 µM) was measured in 30mMTris/HCl pH 7.5, 10mMKPi, 5mMMgCl2, and 3mM DTE at 25°C in the presence of CDC25 (2 µM), the catalytic domain of SOS1 and 40 µM GDP using an Applied- Photophysics™ stopped flow apparatus. Observed rate constants (kobs) were obtained by single exponential fitting of the data.
For determination of NF1–333 GAP activity, GDP-bound to RAS mutants was exchanged with excess mantGTP in presence of EDTA to result in a load of higher than 95%. Free unbound nucleotides were removed by gel filtration, and the RAS · mantGTP was immediately snap frozen in liquid nitrogen to avoid unmonitored hydrolysis [Gremer et al., 2008]. GAP-stimulated GTPase reaction of RAS proteins (0.2 µM) was measured in 30mM Tris pH 7.5, 10mM MgCl2, and 3mM DTE at 25°C using a Hightech™ stopped-flow apparatus. The monitored reactions show an increase of fluorescence due to association of NF1–333 (2 µM), the catalytic domain of neurofibromin 1, with RAS · mantGTP, and a subsequent hydrolysis of mantGTP, described by a decrease of fluorescence. This decay was fitted by a single exponential.
Effector binding assay was performed in 30mM Tris/HCl pH 7.5, 100mM NaCl, 5mM MgCl2, and 3mM DTE at 25°C using a Fluoromax 4 fluorimeter in polarization mode. Increasing amounts of GST-tagged RAS binding domains (RBD) of RAS effectors were titrated to 0.3 µM mantGppNHp-bound RAS proteins resulting in an increase of polarization. For the calculation of the dissociation constant (Kd) for the RAS-Effector interaction the concentration dependent binding curve was fitted using a quadratic ligand binding equation.
Cell-Based Assays
Monkey kidney epithelial COS-7 cells were grown in DMEM supplemented with 10% fetal calf serum (FCS) and transiently transfected using DEAE-dextran as described [Herbrand and Ahmadian, 2006].
GST pull down and MEK1/2, ERK1/2, and AKT activation assays were performed as described [Cirstea et al., 2010]. Briefly, the levels of GTP-bound RAS were determined using GST-fused RAF1-RBD protein to pull down active GTP-bound RAS by glutathione beads from extracts of COS-7 cells transfected with the respective KRAS mutants. The beads were washed four times and subjected to SDS-PAGE (15% polyacrylamide). Bound RAS proteins were detected by Western blotting using monoclonal antibodies against RAS (anti-RAS antibody, BD Transduction Laboratories™, Sparks, MD), anti-RAS (clone RAS10, Upstate-Millipore, Lake Placid, NY). MEK1/2, ERK1/2, AKT, phospho-MEK1/2, phospho-ERK1/2 and phospho-AKT, respectively, were determined by Western blotting analysis of the same COS-7 cell lysates used for the RAS pull down assay and were detected using antibodies against MEK1/2 (Cell Signaling, Danvers, MA), ERK1/2 (Cell Signaling), AKT (Cell Signaling), phospho-MEK1/2 (Ser 217/221; Cell Signaling), phosphor-ERK1/2 (Thr202/Tyr204; Cell Signaling), phospho-AKT (Ser473; Cell Signaling).
Structural Analysis
Because no wild type KRAS (KRASwt) structure is available to date, the structures of HRAS were used in our study. The G-domains of HRAS and KRAS share 97% identity and are generally accepted to be very similar in structure and function [Ahmadian et al., 1997]. The differences between the active and inactive state of RAS were analyzed by comparison of the GDP-bound [Milburn et al., 1990] (Protein Data Bank [PDB] code 4Q21) and GTP-bound [Pai et al., 1990] (1CTQ) HRAS structure, respectively. These structures were selected because they represent RASwt protein and have high resolutions among GDP-or GTP-bound structures. The interactions of RAS with its binding partners were analyzed on the basis of HRAS structure in complexes with p120RASGAP [Scheffzek et al., 1997] (1WQ1), the GEF SOS1 [Margarit et al., 2003] (1NVV), and the downstream effectors, RAF1-RBD [Nassar et al., 1995] (1C1Y), PI3Kγ [Pacold et al., 2000] (1HE8), BYR2-RBD [Scheffzek et al., 2001] (1K8R), RALGDS [Huang et al., 1998] (1LFD), and PLCε [Bunney et al., 2006] (2C5L).
Results
Because HRAS and KRAS proteins share 97% amino acid sequence identity in their G-domain and are 100% identical in regions responsible for interactions, their structural and biochemical properties can be considered to be very similar (e.g., D153 in KRAS is E153 in HRAS) if not identical [Ahmadian et al., 1997]. Furthermore, for a comprehensive structure–function analysis, no KRASwt structure is available to date. Thus, respective KRAS mutations were analyzed using HRAS structures (Supp. Tables S1 to S3), and for practical reasons we generated these KRAS mutations (p.K5N, p.V14I, p.Q22R/Q22E, p.P34R/P34L, p.T58I, p.G60R, p.E153V, and p.F156L; Fig. 1A) in the context of both the HRAS gene in the Escherichia coli expression system and the KRAS gene in the eukaryotic expression system. Purified mutant RAS proteins were comprehensively characterized using advanced physical and cellular biochemistry. As controls, we used RASwt, a GTPase deficient mutant (RASG12V) and a self-activating (“fast-cycling”) mutant (RASF28L) [Reinstein et al., 1991]. All data are summarized in Table 1 and Supp. Table S4.
Table 1.
Summary of Biochemical Properties of the RAS Proteins Grouped into Five Classes
| Classes | RAS mutant |
mant-GppNHp association |
Intrinsic mantGDP dissociation |
GEF-catalyzed mantGDP dissociation |
Intrinsic GTPase |
GAP-stimulated GTPase |
RAF-RBD binding |
|---|---|---|---|---|---|---|---|
| Controls | wt | 1.3a | 0.00003a | 0.09a | 0.0093a | 9.3a | 0.22b |
| G12V | ≈ | ≈ | ≈ | ↓ | |||
| F28L | ↑ | ↑ | ≈ | ↓ | |||
| A | K5N | ⇩ | ≈ | ≈ | ≈ | ≈ | ≈ |
| T58I | ≈ | ↑ | ≈ | ≈ | ≈ | ||
| E153V | ↓ | ≈ | ≈ | ≈ | ≈ | ||
| B | V14I | ↓ | ⇧ | ⇧ | ≈ | ≈ | |
| C | Q22R | ↓ | ≈ | ≈ | ≈ | ⇩ | ≈ |
| D | Q22E | ↓ | ⇧ | ⇧ | ≈ | ⇩ | |
| F156L | ↓ | ⇧ | ≈ | ⇩ | |||
| E | P34L | ≈ | ≈ | ≈ | ≈ | ||
| P34R | ≈ | ≈ | ≈ | ≈ | |||
| G60R | ↓ | ≈ | ↓ |
Based on our biochemical data, RAS mutants are grouped into different functional classes: (A) no major changes; (B) increase in intrinsic and catalyzed nucleotide exchange; (C) decrease in GAP-stimulated GTPase; (D) increase in intrinsic and catalyzed nucleotide exchange and decreased in GAP-stimulated GTPase; (E) defective interaction with GAPs and effectors and also with GEFs in the case of RASG60R. RASwt, RASG12V, and RASF28L were included as controls. Black arrows outlines effects that favor increased signaling, gray arrows those impaired signaling, such as lower affinity of effectors. ≈≥ 2-fold, ↓or ↑ = 3–15-fold, ⇩or ⇧ = 15–40-fold,  or
or  > 40-fold relative to RASwt values. Relative values are shown in Table S4.
> 40-fold relative to RASwt values. Relative values are shown in Table S4.
Observed rate constants (kobs) in sec−1.
Dissociations constants (Kd) in µM.
Germline RAS Mutations Are Located at Permissible Structural Sites
Overall location and spatial orientation of the mutated amino acids according to the nucleotide-bound forms were deduced from the structures of HRAS in the inactive, GDP-bound state (Fig. 1B) and in the active, GTP-bound state (Fig. 1C), respectively. We inspected solvent accessible areas of considered residues (Supp. Table S3) and found that only T58 and G60 undergo significant conformational rearrangements between the active and inactive state. These differences are not unexpected as these residues are nearby or part of the switch II region (Fig. 1). An interesting exception is P34, which is almost equally solvent exposed in both states although it is part of switch I (Fig. 1B and C; Supp. Table S3). The absolute solvent accessible area of P34 is relatively large in contrast to T58 and G60 (Fig. 1B and C; Supp. Table S3).
To assess if the mutations directly interfere with intermolecular interactions, we analyzed structures of RAS in complexes with regulators and effectors (Supp. Table S2). Remarkably, only P34 is clearly located within the interacting interface contacting GAPs, GEFs, and effectors (Supp. Fig. S1, yellow areas; Supp. Table S3). Other investigated residues are either on its edge or buried within the protein and are thus not directly participating in the interaction with RAS binding partners (Supp. Fig. S1; Supp. Table S3). Interacting interfaces (Supp. Fig. S1, yellow areas) are rather distinct between the complexes with SOS1 compared with GAP and effectors. Among the RAS mutants, three residues, P34, T58, and G60, contact the CDC25 domain of SOS1 (Supp. Table S3). As mentioned above, G60 is solvent exposed in the GDP-bound state (Fig. 1B; Supp. Table S3), which should be also true for an arginine side chain in the case of RASG60R (Supp. Fig. S1A). In this scenario a large, positively charged side chain would interfere sterically with the CDC25 binding. Alternatively, it is also possible that an arginine within the highly conserved DxxG60QE motif (part of the switch II; Fig. 1A), may interfere with the nucleotide dissociation itself. This motif has been recently implicated to play a critical role in GEF-mediated nucleotide exchange reactions [Gasper et al., 2008].
Finally, we calculated the vicinity of investigated amino acid residues to the nucleotides (Supp. Table S3) to explore a possible direct impact of the various mutations on GDP/GTP binding and GTP hydrolysis, respectively. Only two residues, V14 and G60, are involved in direct interaction with the nucleotide. Because V14 contacts the nucleotide with backbone atoms, only the substitution of G60 can directly affect nucleotide binding and hydrolysis as its Cα atoms faces the γ-phosphate in the GTP state.
Germline RAS Mutants Accumulate in the GTP-Bound State
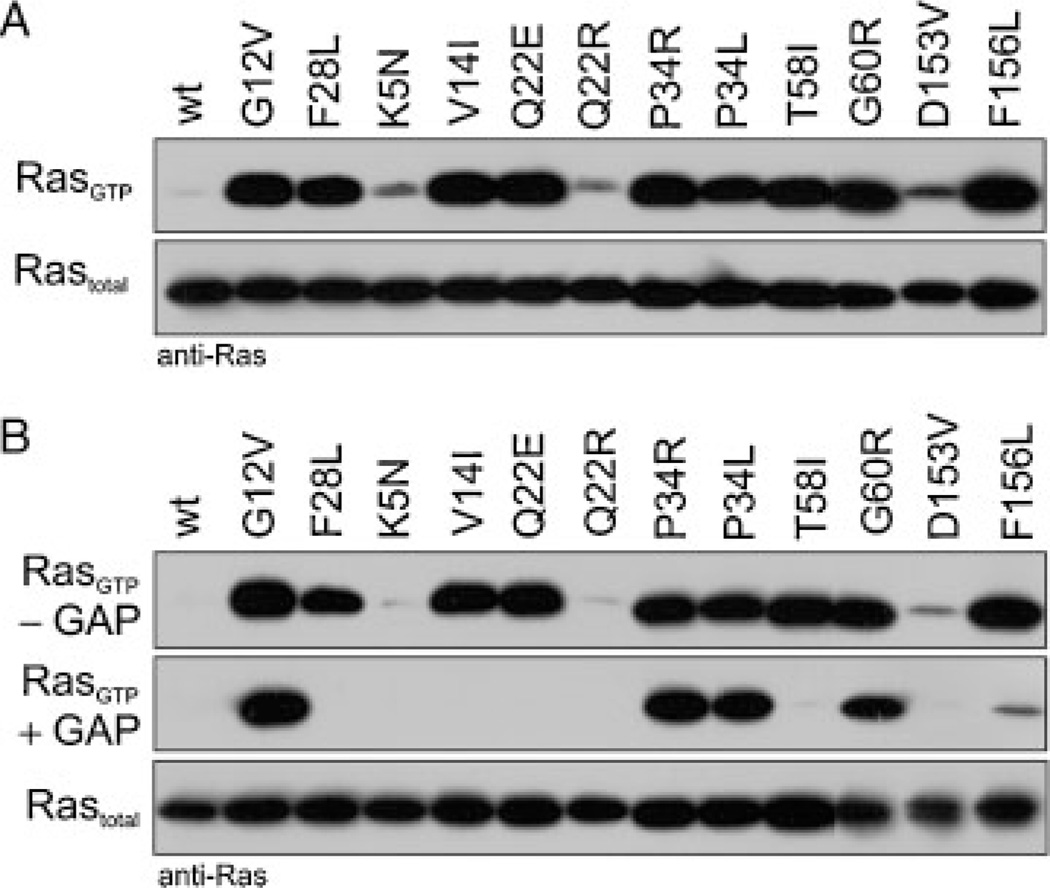
To gain insights into the regulatory cycle of the RAS mutants in cells we transiently transfected COS-7 cells with plasmids expressing KRASwt and KRAS mutant proteins. We determined the amount of active, GTP-bound RAS in the presence of serum using GST-fusion proteins of the RBD of RAF1 (GST–RAF1–RBD) immobilized on glutathione sepharose (GST pull-down assay described in Materials and Methods section). Fig. 2A shows that the majority of the RAS mutants exhibit a tremendously high level of activation compared to RASwt. We next repeated these experiments under serum starved conditions to exclude RAS activation by serum-containing stimuli. The majority of the RAS mutants remarkably remained in a hyperactive state except for p.K5N, p.Q22R, and p.D153V (Fig. 2B, upper panel). Two major reasons for the high level of GTP-bound active RAS mutants have to be considered: an increased GDP/GTP exchange (“fast cycling”) or a reduction of intrinsic or/and GAP-stimulated GTP hydrolysis.
Figure 2.
Cellular levels of the active, GTP-bound forms of germline KRAS mutants. Pull-down experiments of GTP-bound KRAS proteins (RASGTP) were performed in COS-7 cells transiently expressing either KRASwt or germline KRAS mutants in the presence (A) and in the absence of serum (B). Irrespective of culture conditions almost all KRAS mutants showed an increased GTP-bound level. Purified RAS-GAP, which was added to the cleared cell lysates proved the GAP sensitivity of the mutants (B, middle panel). GAP resistant mutants, RASP34L, RASP34R, and RASG60R, resided in the active state comparable to oncogenic RASG12V. Total amounts of recombinant RAS are shown for equal expression and loading. Anti-RAS antibodies used in these experiments were anti-RAS (RAS10 clone, Upstate-Millipore™, mutants p.G60R, p.D153V, p.F156L) and anti-RAS (BD Transduction Laboratories™, wt and all other mutants), because some mutations modified the RAS epitope recognized by the respective antibodies. Additional information is given in Supp. Fig. S2.
It is important to note that point mutations may lead to changes in RAS (epitope) recognition by the antibody. Therefore, we repeated these experiments using two different anti-RAS antibodies (Fig. 2 and Supp. Fig. S2), and found that the p.P34L and p.P34R mutants are not recognized by anti-RAS antibody clone RAS10 from Upstate-Millipore, but can be detected with the antibody anti-RAS obtained from BD Transduction Laboratories ™, and that the mutant p.D153V is much better recognized by the BD Transduction Laboratories™ anti-RAS (Fig. 2 and Supp. Fig. S2). These observations indicate that the affinity of antibodies to the proteins can be impaired by point mutation.
Increase in Nucleotide Exchange of RASV14I, RASQ22E, and RASF156L
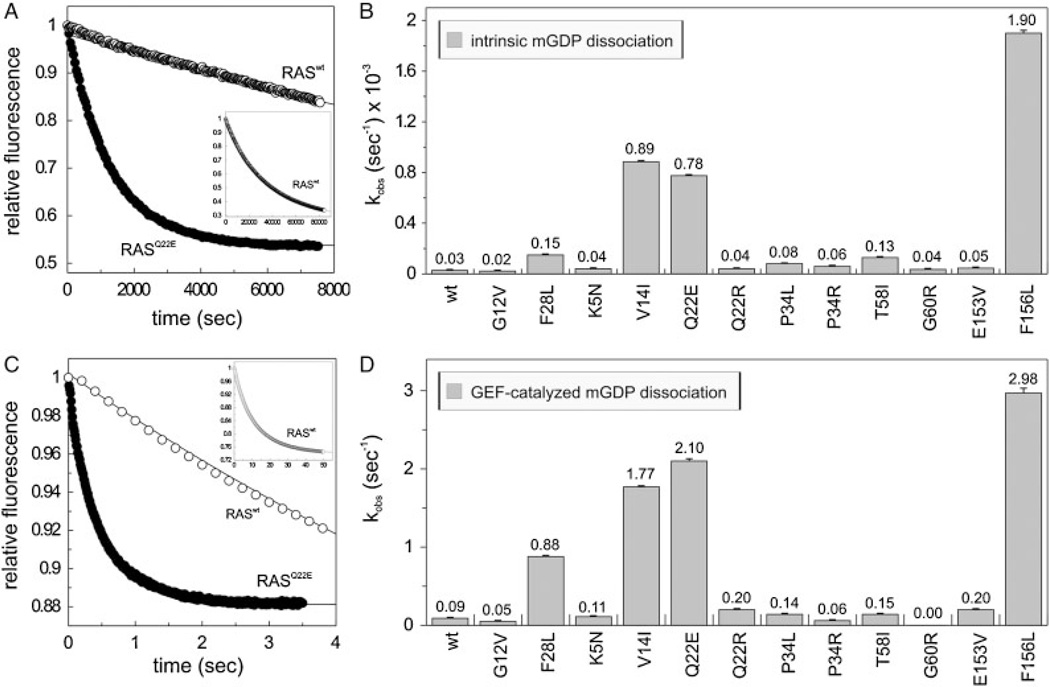
To examine the GDP/GTP exchange of the mutants, we measured both intrinsic and GEF-catalyzed nucleotide dissociation (Fig. 3). The most significant effect was an increase in dissociation of the fluorescently labeled GDP (mantGDP) from RAS mutants by almost 30-fold in the case of p.V14I and p.Q22E, and more than 60-fold in p.F156L (Fig. 3B). The fact that these mutants release mantGDP even faster than the already established “fast cycling” p.F28L [Reinstein et al., 1991] leads, most likely, to a GEF-independent activation of these mutants in cells (Fig. 2B). For completeness, we also measured the intrinsic functions of the RAS mutants concerning nucleotide association (Supp. Fig. S3). Interestingly and in contrast to the oncogenic p.G12V and the “fast cycling” p.F28L the association rates of mantGDP or a nonhydrolyzable fluorescently labeled GTP analog (mantGppNHp) with all other RAS mutants are slower then that of wild-type reaching a maximum 20-fold difference in the case of p.K5N (Supp. Fig. S3). However, as with RASwt, no preference for the nucleotide bound state was observed, and thus, the difference in association kinetics would probably have no major functional consequence due to the high affinity of guanine nucleotides to RAS and due to their high concentration in the cell.
Figure 3.
Modified nucleotide exchange properties of the RAS mutants. Intrinsic (A, B) and GEF-catalyzed (C, D) mantGDP dissociation from the RAS proteins (0.2 µM) in the presence of 40 µM GDP (A) or of 40 µM GDP and 2 µM CDC25 (C). On the panels A and C the respective time-dependent reactions of RASwt and a representative RAS mutant (p.Q22E) are shown. On the panels B and D the observed rate constant (kobs) of all RAS proteins are illustrated. RASwt, RASG12V, and RASF28L were included as controls. The insets (in A and C) show the complete time course of the mantGDP dissociation from RASwt. Standard errors of five to seven independent measurements are shown.
The dissociation of mantGDP from RASwt catalyzed by the catalytic domain of SOS1 (CDC25) [Pechlivanis et al., 2007] is three orders of magnitude faster than in the absence of this GEF (Fig. 3) under the conditions used. The effect of the germline RAS mutations on the CDC25-mediated dissociation was measured under the same conditions. Interestingly, differences in GEF-catalyzed nucleotide exchange reactions (Fig. 3C and D) correlate with the intrinsic dissociations of mantGDP from RAS proteins (Fig. 3A and B). This suggests that the observed differences in the acceleration of nucleotide exchange between wild-type and mutant RAS are caused primarily by structural changes of RAS itself, and not by altered RAS–GEF interactions. An exception is RASG60R that was virtually unresponsive toward CDC25 under our experimental condition, which is most likely due to the lack of a RAS–CDC25 interaction. Besides G60, the residues P34 and T58 also contact the CDC25 domain of SOS1 (Supp. Table S3), but obviously only the substitution of G60 to arginine abolishes the formation of the Ras–CDC25 complex, as no acceleration of nucleotide exchange by SOS1 was observed for the p.G60R mutant (Fig. 3C and D).
Profound GAP Insensitivity of the RASP34 and RASG60 Mutants
Mutants that do not exhibit a fast nucleotide exchange but accumulate in an active, GTP-bound form are strongly suggestive of having an impaired GTP hydrolysis activity. Therefore, we investigated the GAP sensitivity of the RAS mutants in cells. For this purpose, the catalytic domain of neurofibromin (NF1–333) [Ahmadian et al., 1997] was added to the cell lysates before performing the GST pull-down assay. The middle panel of Figure 2B shows that RASP34L, RASP34R, and RASG60R are GAP resistant and locked in the active state in a fashion similar to oncogenic RASG12V. Conversely, these results indicate that GAP-sensitive mutants RASV14I, RASQ22E, RAST58I, and RASF156L most likely have an increased GDP/GTP exchange explaining their accumulation in the active state (Fig. 2B, upper panel).
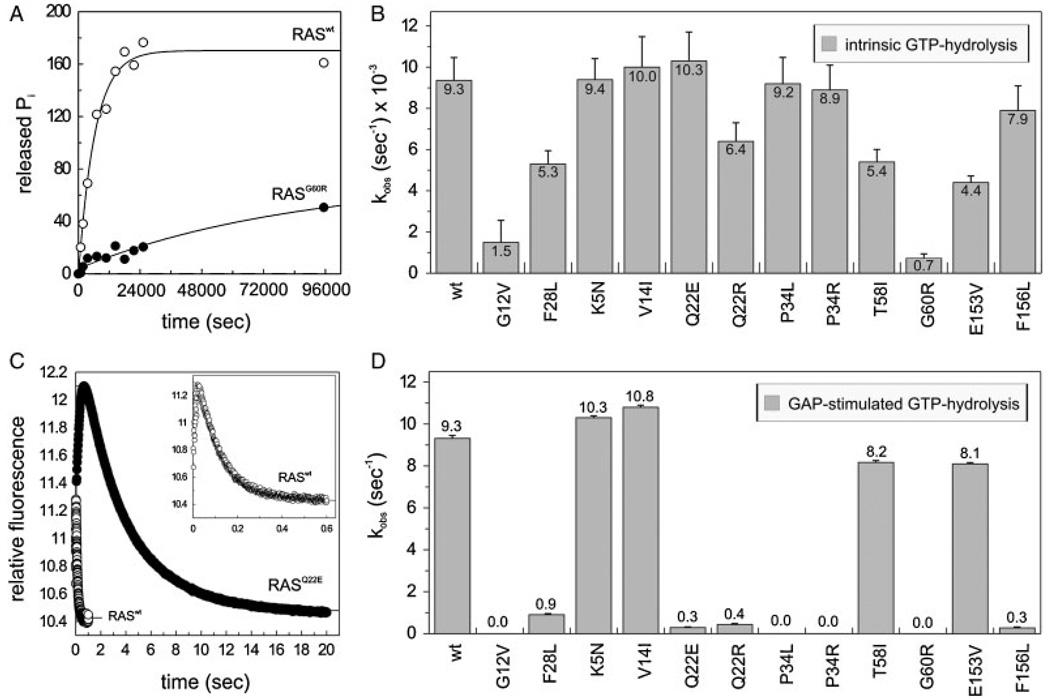
Next we examined the capabilities of RAS mutants to hydrolyze GTP in the absence and in the presence of a RAS–GAP in vitro. Intrinsic GTP hydrolysis was drastically reduced by the substitution of G60 by arginine, even more than the decrease caused by the oncogenic mutation p.G12V (Fig. 4A and B). All other RAS mutations altered the intrinsic GTP hydrolysis only marginally. A different situation was obtained for the GAP-stimulated GTPase activity after adding the catalytic domain of neurofibromin 1 (NF1–333). Six of 10 RAS mutants, p.Q22E, p.Q22R, p.P34L, p.P34R, p.G60R, and p.F156L revealed considerable reduction in GAP-stimulated GTPase rates (Fig. 4D) compared to Raswt. Among them, p.Q22E and p.F156L also showed faster intrinsic and GEF-catalyzed nucleotide dissociation as described above (Fig. 3B and D). The most severe impairment of the GAP-stimulated GTP hydrolysis is caused by the mutations p.P34L, p.P34R, and p.G60R, which is comparable to that of the oncogenic mutation p.G12V. Earlier mutational studies of RAS also showed that substitution of P34 by arginine abolished the GAP-stimulated GTP hydrolysis reaction [Chung et al., 1993; Stone et al., 1993]. The other RAS mutants, p.K5N, p.V14I, p.T58I, and p.E153V exhibited similar kobs values of stimulated GTP hydrolysis as obtained for RASwt (Fig. 4D). However, a recent report discussed that RASV14I exhibits a much lower GTP-hydrolysis in the presence of RAS-specific GAPs [Schubbert et al., 2006]. This discrepancy is possibly due to the different method used for the investigation of the GAP sensitivity. In contrast to our study, the former report used GST-fusion proteins of the respective RAS mutants and GAP proteins. In addition, unlike the nitrocellulose filter binding assay used by Schubbert et al. [2006], our studies are based on time-resolved fluorescence measurements using single turnover stopped-flow techniques in solution.
Figure 4.
GAP insensitivity of the RAS mutants. (A, B) Intrinsic γ32P-GTP hydrolysis reaction rates were measured for individual RAS proteins (1 µM). (C, D) GAP-stimulated GTPase reaction of the RAS proteins (0.2 µM) was measured in the presence of 2 µM NF1–333. On panels A and C, the respective time-dependent reactions of RASwt and a representative RAS mutant (p.G60R in A, p.Q22E in C) are shown. RASwt, RASG12V, and RASF28L were included as controls. The inset in panel C shows the complete and more detailed time course of GAP activity on RASwt. Standard errors of five to seven independent measurements are shown.
Moderate Gain of Signal Transduction Through the Germline RAS Mutants
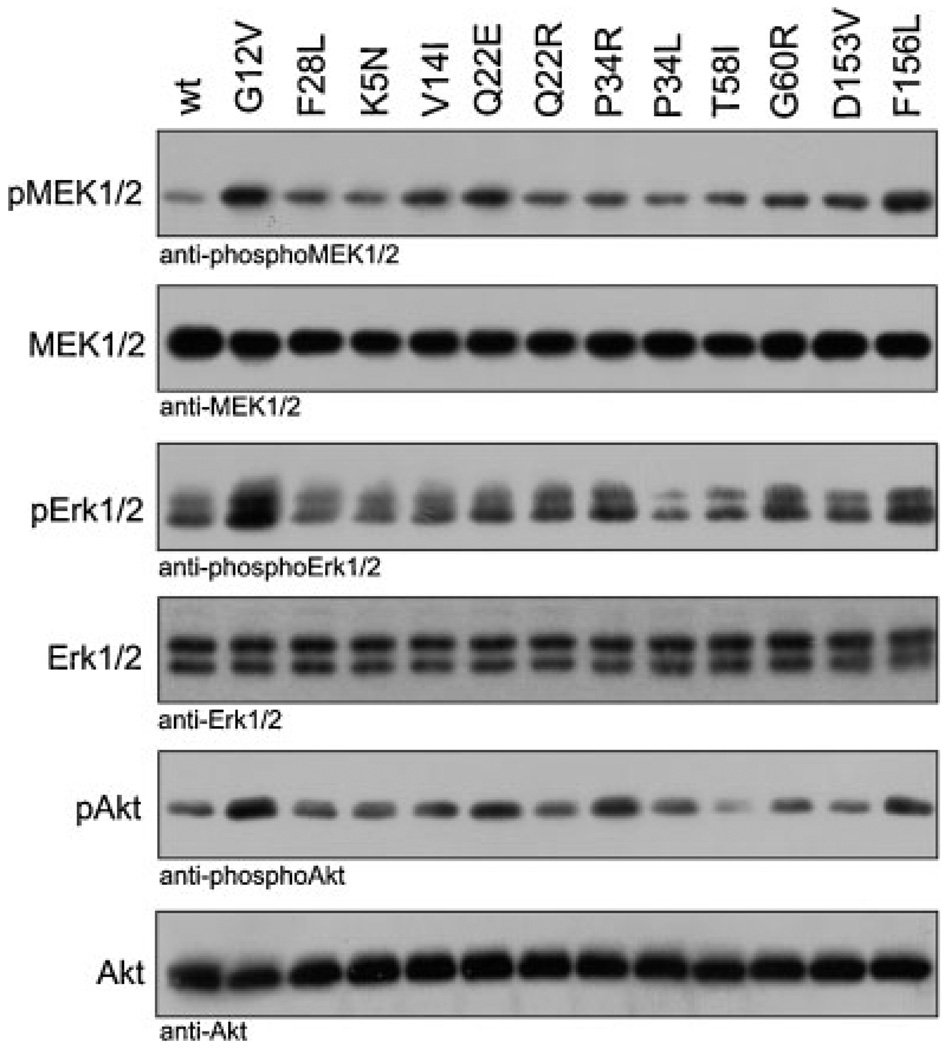
Accumulation of the KRAS mutants in their active state as a consequence of increased nucleotide exchange and impaired interaction with GAPs would predict a sustained activation of effectors and cellular signal transduction. To examine whether the elevated GTP-bound state of these proteins is correlated with an increased downstream signaling, we measured levels of phosphorylated MEK1/2, ERK 1/2, and AKT in COS-7 cells transiently transfected with the KRAS mutants. To avoid any upstream propagation by extracellular stimuli seen in experiments performed in the presence of serum (Supp. Fig. S4), we analyzed the potential of the germline KRAS mutants themselves to activate signaling pathways under serum-free culture conditions (Fig. 5). Given the abnormal biochemical properties that resulted in massive accumulation in the GTP-bound, active forms as shown above (Fig. 2B, upper panel), a strongly increased downstream signaling was expected. Surprisingly, most germline KRAS mutants induced only moderately increased phosphorylation levels of downstream signaling proteins. KRASV14I, KRASQ22E, and particularly KRASF156L, but also perceivably KRASP34R and KRASG60R showed increased levels of phosphorylated MEK1/2 (pMEK1/2), ERK1/2 (pERK1/2), and AKT (pAKT) compared to RASwt but less than RASG12V (Fig. 5). These results emphasize the ability of the majority of the germline KRAS mutants to activate downstream effectors under serum-free conditions to moderate degree. Expression of the KRAS mutants in presence of serum (Supp. Fig. S4) leads to overall enhanced MEK1/2 phosphorylation (e.g., p.K5N, p.V14I, p.Q22E, p.T58I, p.D153V, p.F156L) and enhanced ERK1/2 phosphorylation (e.g., p.V14I, p.Q22E, p.Q22R, p.F156L). These findings suggest that increased downstream signaling is a consistent feature of germline KRAS mutations, but this effect remains stimulus-dependent in some mutants, although it is constitutive in others.
Figure 5.
Increased downstream signaling activity of the germline RAS mutants under serum-free conditions. KRAS mutants, transiently transfected in COS-7 cells were analyzed for the phosphorylation level of MEK1/2 (pMEK1/2), ERK1/2 (pERK1/2), and AKT (pAKT) under serum-free culture conditions. The amounts of total RAS, MEK1/2, ERK1/2, and AKT in the cleared cell lysates as well as RASwt, RASG12V, and RASF28L were included as controls. Results of experiments in the presence of serum are shown in Supp. Fig. S4.
Significant Loss of Effector Binding Affinity
Our data show that most KRAS mutants accumulate in the active, GTP-bound form to an extent that can be similar to oncogenic RASG12V, but are disabled to equally activate downstream pathways, suggesting that an interaction with the downstream effectors might be impaired. Therefore, we set out to explore the impact of the investigated patient mutations on the interactions with the RAS-binding domains of two well-studied effectors RAF1 kinase and RALGDS. For this purpose, we established a fluorescence polarization-based assay (see Materials and Methods section), which enabled us to determine the equilibrium dissociation constants (Kd) of RAS · effector complexes (Fig. 6 and Supp. Fig. S5).
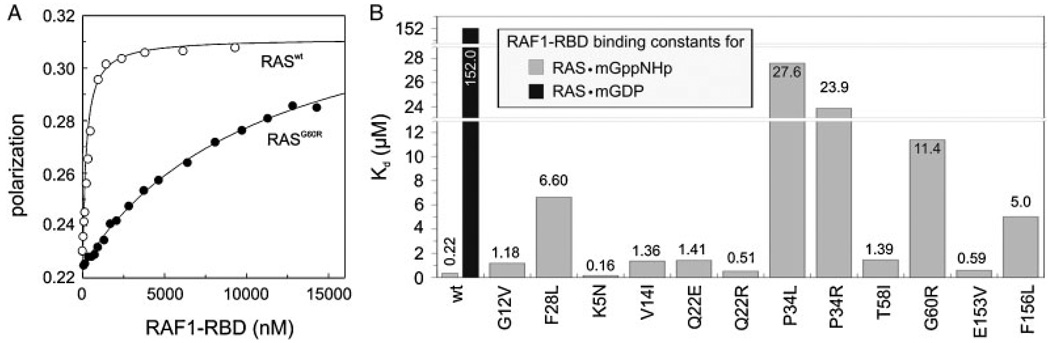
Figure 6.
Significant loss of RAF1 binding affinity for RAS mutants. Binding of RAF1-RBD (increasing concentrations as indicated) to mantGppNHp-bound RAS (0.2 µM) was measured using fluorescence polarization. On the panel A, the respective concentration-dependent measurements of RASwt and a representative RAS mutant (p.G60R) are shown. On panel B, the dissociation constants (Kd) of all RAS proteins are illustrated. RASwt, RASG12V, and RASF28L were included as controls. Data obtained with another RAS effector, RALGDS are shown in Supp. Fig. S5.
Downstream effectors bind with a much higher affinity to the GTP-bound form of RAS than to its GDP-bound form [Herrmann et al., 1995]. We quantified binding constants by fluorescence polarization for the interaction of RAS with the RBD of RAF1 (RAF1–RBD) (Fig. 6B) or with the RBD of RALGDS (RALGDS–RBD) (Supp. Fig. S5). Because RAS effectors, including RAF1, RALGDS, and PI3K share an overlapping interactive region on GTP-bound RAS with GAPs (Supp. Figs. S1B, S1C, and S1D) [Vetter and Wittinghofer, 2001], it is not surprising that most RAS mutations, which interfered with the GAP activity (Fig. 4D), also interfered with RAF1 and RALGDS binding (Fig. 6 and Supp. Fig. S5). The strongest reduction in binding affinity is observed with RAS mutations at position P34, G60, and F156. Taken together, our studies show that the remarkable increase in RAS activation (Fig. 2B, upper panel) due to GAP resistance or reduction of GAP interaction (Figs. 2B and 4D), which is most prominent in the case of P34 and G60 mutations, is at least in part compensated by another functional impairment, namely, the significant loss of interaction with downstream effectors of up to 125-fold (Kd value of 27.6 µM for KRASP34L divided by 0.22 µM for KRASwt) (Fig. 6B).
Discussion
For over 3 decades, the biochemical effects exhibited by cancer-associated RAS mutations have been studied in great detail. In contrast, only limited information is available on the newly discovered germline mutations of RAS. The data presented herein represent the most comprehensive biochemical and structural analysis of these novel RAS mutants to date.
The key phenomenon in RAS biology is its nucleotide-dependent interaction with different proteins of the signal transduction machinery, which is controlled by the GDP/GTP exchange and the GTP hydrolysis reactions. Any change of these functions or an impairment of the interaction of RAS with its binding partners can affect the fine-tuned balance of RAS regulation and its activity in cells. It is now clear that aberrant RAS function in the developing embryo leads to an abnormal progression of developmental programs. Understanding the mechanisms by which aberrant RAS disturbs normal development represents an important scientific goal. The results of this study confirm that germline KRAS mutations generally confer a milder gain-of-function phenotype than cancer-associated mutations at positions G12, G13, or Q61. Moreover, we show that germline KRAS mutations caused multifaceted effects that cannot simply be explained to result from one of the underlying mechanisms fine-tuning RAS functions. To gain insights into the structural alterations caused by the germline KRAS mutations, we inspected the environment of the respective residue and compared them with RASwt to explain their functional properties (Supp. Fig. S7, and “Assessement of possible structural consequences of RAS mutants” in the online Supporting Information). As summarized below and in Table 1 and Supp. Table S4, our data strongly suggest the existence of five partially interrelated mechanistic classes of KRAS mutants with altered signal transduction:
Class A groups the mutants KRASK5N, KRAST58I, and KRASD153V, which do not show major biochemical alterations compared to wild-type KRAS in vitro. All three mutants, especially p.T58I, are in a higher activated state and show a higher downstream signaling compared to RASwt indicating that mutation at these positions do impact RAS function but could not be monitored by the current tools of RAS biochemistry. KRASD153V expressing cells showed a slightly higher GTP-bound level and an increase in MEK1/2 phosphorylation compared to KRASwt but no difference regarding pERK1/2 and pAKT levels. The reason for this observation is not understood yet.
On the other hand, we have recently shown in a similar situation with NRAST50I identified in NS patients, that the residue T50 does not play a functional role, either in nucleotide binding and hydrolysis or in contacting protein partners of NRAS, but rather in the interaction with membrane lipids [Cirstea et al., 2010]. By inspecting such a RAS/membrane model [Abankwa et al., 2008], it is rather tempting to speculate that the E153 (in HRAS)/D153 (in KRAS) side chain may directly influence RAS interaction with the membrane. Moreover, we superposed HRAS with a recently published KRAS mutant structure (Supp. Table S1), which clearly showed that there is no significant difference between the E153 and D153 positions (Supp. Fig. S6).
Class B represented by KRASV14I, showed a dramatic increase, both in intrinsic and GEF-catalyzed nucleotide exchange as the probable major cause for its accumulation in the GTP-bound state and increased downstream signaling. In contrast to a previous study, where KRASV14I exhibited a significantly lower GTP-hydrolysis in the presence of RAS-specific GAPs in vitro [Schubbert et al., 2006], we did not observe any changes in the intrinsic and GAP-stimulated GTP-hydrolysis reactions (Fig. 4) and an only mild decrease in effector binding affinity.
Class C is represented by KRASQ22R, and characterized by an impaired GAP-stimulated GTP hydrolysis while its intrinsic functions including the intrinsic GTP hydrolysis reaction (Fig. 4) remained unaffected and its interaction with effectors is virtually functional (Fig. 6). Consistent with our results, a KRASQ22K mutant, which is physiologically homologous to p.Q22R, has been shown to transform NIH-3T3 fibroblasts [Tsukuda et al., 2000], an effect that is presumably caused by accumulation of RAS in its active state. The underlying pathogenetic mechanism is most likely due to a surface exposed guanidinium group of the arginine which prevents GAP binding (Supp. Fig. S7C) but does not interfere with effector binding (Fig. 6).
Class D comprises the mutants KRASQ22E and KRASF156L. The members of this class are characterized by an increase in intrinsic and catalyzed nucleotide exchange in combination with the resistance to GAPs, but still with a functional interaction with effectors. These effects which cause a profound activation of the MAPK and PI3K/AKT pathways are not directly affecting nucleotide binding and hydrolysis (Figs. 3 and 4) because Q22 and F156 are not directly involved in the coordination of the active center. F156 substitution by leucine creates a cavity within the hydrophobic core causing loss of contact with surrounding residues (Supp. Fig. S7E), which lead to an overall reduction of the nucleotide binding affinity [Xu et al., 1998], an increase in the cellular level of GTP-bound RAS and subsequent activation of the transforming potential of RAS [Quilliam et al., 1995].
All mutations that cause faster nucleotide dissociation (KRASV14I, KRASQ22E, KRASF156L) in comparison to RASwt affect amino acids that are either barely (V14, Q22) or not at all (F156) exposed to the solvent (Fig. 1B and C and Supp. Table S3). It implicates that disturbed integrity of RAS structure is responsible for the alteration of its intrinsic property as the substitutions of buried amino acids by smaller side chains very likely affect the internal dynamics of the proteins.
Class E is represented by the mutants KRASP34L, KRASP34R, and KRASG60R and is characterized by a defective GAP sensitivity and a strongly reduced interaction with effectors. Although these mutants are locked in a hyperactivated state, which is rather comparable to the oncogenic RASG12V, their ineffectiveness for downstream signaling in turn causes only a mild gain-of-function phenotype. Accordingly, class E mutants are able to activate downstream pathways as shown by ERK1/2 and AKT phosphorylation (Fig. 5). A similar case has been recently reported in a germline HRAS mutant associated with CS [Gremer et al., 2010]. Hereby, E37 duplication in the switch I region of HRAS impairs both binding of GAP and effector proteins. Therefore, this mutant can also be assigned as a class E member. Although KRASP34L and KRASP34R do not respond to GAP they are in principle able to hydrolyze GTP intrinsically (Fig. 4). This strongly suggests that the respective amino acid substitutions either interfere with GAP binding or with the positioning of the catalytic arginine of GAP in the active site. P34 is invariant in RAS and RHO proteins [Eberth et al., 2005] and any substitution of P34 has been suggested to affect interaction with the binding partner of RAS [Chung et al., 1993; Stone et al., 1993].
The mutation of KRAS G60 to arginine has most severe consequences, namely, an overall impairment of almost all biochemical and functional properties. Its substitution by a large and charged amino acid like arginine in KRAS or glutamate in NRAS [Cirstea et al., 2010] seems to corrupt the switch regions including the critical catalytic Q61, affect nucleotide binding, GTP hydrolysis, and impairs intermolecular interaction with regulators and effectors. Previous studies have shown that a conservative mutation of G60 to alanine impairs the normal GTPase function of RAS and Gα [Ford et al., 2005; Sung et al., 1996]. G60A mutation of HRAS dramatically affects intrinsic and GAP-stimulated GTP hydrolysis without major changes on its interaction with effector proteins [Hwang et al., 1996]. Structural analysis of HRASG60A showed that its switch I region adopts an open conformation [Ford et al., 2005]. However, G60 substitution by arginine (KRAS) or glutamate (NRAS) may affect both switch regions, which in turn may be the reason for loss of intrinsic and extrinsic functions.
Interestingly, certain RAS mutations such as p.P34L, p.P34R, or p.G60R are compromised in their interaction with effectors as evidenced by the inability to bind efficiently RAF1–RBD or RALGDS–RBD (Fig. 6 and Supp. Fig. S5). This is surprising considering that enhanced downstream signaling is the primary cause of the developmental diseases. At the same time, these mutations have the most severe effect on the GAP-mediated GTPase reaction, in a range that is quite similar to the oncogenic mutation prototype p.G12V. It is likely that the impairment of effector interactions damps the consequences of GAP resistance of these mutants on downstream signal flow. This is in contrast to RAS proteins with oncogenic mutations at the positions G12 or Q61, which contribute to potent transforming properties [Der et al., 1884; Seeburg et al., 1886]. Our results therefore provide an explanation for the lower levels of activated KRAS signaling exerted by germline mutations compared to the classical oncogenic mutations. The fact that the majority of investigated amino acids of RAS are neither involved in contacts with interacting partners nor with the nucleotide also suggests that the effects of changes at these sites are milder compared to oncogenic mutations and may at least in part explain why these alterations are tolerated in the germline and are generally not associated with tumor development in affected individuals [Karnoub and Weinberg, 2008; Quinlan and Settleman, 2009].
The diversity of functional consequences of germline KRAS mutations is paralleled by a remarkably wide phenotypic spectrum associated with mutations in this gene and its tempting to assume a causal relation between certain genotypes and phenotypic expressions. There is indeed a tendency towards an association of more severe phenotypes (CFC/CS) with mutations that proved to have stronger effects on ERK1/2 phosphorylation in our experiments (p.Q22E, p.Q22R, p.P34R, p.G60R, p.F156L) (Fig. 5). In contrast, patients harboring the mutations p.V14I, p.P34L, p.D153V tend to have less severe physical and mental handicaps and are more commonly classified as having NS, the less severe form among this group of developmental disorders [Aoki et al., 2008]. However, the number of known patients with a proven KRAS mutation is still too small to delineate clear genotype–-phenotype correlations.
In conclusion, we describe and classify in detail the functional properties of a spectrum of germline mutations of KRAS that have been previously identified as a cause of developmental syndromes. Our studies reveal several new mechanisms by which germline KRAS mutations contribute to human disease and lead to disturbed embryonic development.
Acknowledgments
We thank Dorothee Vogt and Patricia Stege for expert technical assistance and Katja T. Koessmeier for critical reading of the manuscript.
Grant sponsor: The German Research Foundation; Grant number: DFG AH 92/5-1; Grant sponsor: The NGFNplus program of the German Ministry of Science and Education (BMBF); Grant number: 01GS08100; Grant sponsors: The Research Committee of the Medical Faculty of the Heinrich-Heine University Düsseldorf; Max-Planck Society; E-Rare project NSEuroNet.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Abankwa D, Hanzal-Bayer M, Ariotti N, Plowman SJ, Gorfe AA, Parton RG, McCammon JA, Hancock JF. A novel switch region regulates H-ras membrane orientation and signal output. EMBO J. 2008;27:727–735. doi: 10.1038/emboj.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian MR. Prospects for anti-ras drugs. Br J Haematol. 2002;116:511–518. doi: 10.1046/j.0007-1048.2001.03314.x. [DOI] [PubMed] [Google Scholar]
- Ahmadian MR, Hoffmann U, Goody RS, Wittinghofer A. Individual rate constants for the interaction of Ras proteins with GTPase-activating proteins determined by fluorescence spectroscopy. Biochemistry. 1997;36:4535–4541. doi: 10.1021/bi962556y. [DOI] [PubMed] [Google Scholar]
- Ahmadian MR, Wittinghofer A, Herrmann C. Fluorescence methods in the study of small GTP-binding proteins. Methods Mol Biol. 2002;189:45–63. doi: 10.1385/1-59259-281-3:045. [DOI] [PubMed] [Google Scholar]
- Ahmadian MR, Zor T, Vogt D, Kabsch W, Selinger Z, Wittinghofer A, Scheffzek K. Guanosine triphosphatase stimulation of oncogenic Ras mutants. Proc Natl Acad Sci USA. 1999;96:7065–7070. doi: 10.1073/pnas.96.12.7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Niihori T, Narumi Y, Kure S, Matsubara Y. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum Mutat. 2008;29:992–1006. doi: 10.1002/humu.20748. [DOI] [PubMed] [Google Scholar]
- Barbacid M. Ras oncogenes—their role in neoplasia. Eur J Clin Invest. 1990;20:225–235. doi: 10.1111/j.1365-2362.1990.tb01848.x. [DOI] [PubMed] [Google Scholar]
- Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Brooks BR, Bruccoleri RE, Olafson D, States DJ, Swaminathan S, Karplus M. Charmm—a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- Buhrman G, de Serrano V, Mattos C. Organic solvents order the dynamic switch II in Ras crystals. Structure. 2003;11:747–751. doi: 10.1016/s0969-2126(03)00128-x. [DOI] [PubMed] [Google Scholar]
- Buhrman G, Wink G, Mattos C. Transformation efficiency of RasQ61 mutants linked to structural features of the switch regions in the presence of Raf. Structure. 2007;15:1618–1629. doi: 10.1016/j.str.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunney TD, Harris R, Gandarillas NL, Josephs MB, Roe SM, Sorli SC, Paterson HF, Rodrigues-Lima F, Esposito D, Ponting CP, Gierschik P, Pearl LH, Driscoll PC, Katan M. Structural and mechanistic insights into ras association domains of phospholipase C epsilon. Mol Cell. 2006;21:495–507. doi: 10.1016/j.molcel.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Carta C, Pantaleoni F, Bocchinfuso G, Stella L, Vasta I, Sarkozy A, Digilio C, Palleschi A, Pizzuti A, Grammatico P, Zampino G, Dallapiccola B, Gelb BD, Tartaglia M. Germline missense mutations affecting KRAS isoform B are associated with a severe Noonan syndrome phenotype. Am J Hum Genet. 2006;79:129–135. doi: 10.1086/504394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HH, Benson DR, Schultz PG. Probing the structure and mechanism of Ras protein with an expanded genetic code. Science. 1993;259:806–809. doi: 10.1126/science.8430333. [DOI] [PubMed] [Google Scholar]
- Cirstea IC, Kutsche K, Dvorsky R, Gremer L, Carta C, Horn D, Roberts AE, Lepri F, Merbitz-Zahradnik T, Konig R, Kratz CP, Pantaleoni F, Dentici ML, Joshi VA, Kucherlapati RS, Mazzanti L, Mundlos S, Patton MA, Silengo MC, Rossi C, Zampino G, Digilio C, Stuppia L, Seemanova E, Pennacchio LA, Gelb BD, Dallapiccola B, Wittinghofer A, Ahmadian MR, Tartaglia M, Zenker M. A restricted spectrum of NRAS mutations causes Noonan syndrome. Nat Genet. 2010;42:27–29. doi: 10.1038/ng.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denayer E, Parret A, Chmara M, Schubbert S, Vogels A, Devriendt K, Frijns JP, Rybin V, De Ravel TJ, Shannon K, Cools J, Scheffzek K, Legius E. Mutation analysis in Costello syndrome: functional and structural characterization of the HRAS p.Lys 117Arg mutation. Hum Mutat. 2008;29:232–239. doi: 10.1002/humu.20616. [DOI] [PubMed] [Google Scholar]
- Der CJ. The ras family of oncogenes. Cancer Treat Res. 1989;47:73–119. doi: 10.1007/978-1-4613-1599-5_4. [DOI] [PubMed] [Google Scholar]
- Der CJ, Finkel T, Cooper GM. Biological and biochemical properties of human rasH genes mutated at codon 61. Cell. 1986;44:167–176. doi: 10.1016/0092-8674(86)90495-2. [DOI] [PubMed] [Google Scholar]
- Eberth A, Dvorsky R, Becker CFW, Beste A, Goody RS, Ahmadian MR. Monitoring the real-time kinetics of the hydrolysis reaction of guanine nucleotide-binding proteins. Biol Chem. 2005;386:1105–1114. doi: 10.1515/BC.2005.127. [DOI] [PubMed] [Google Scholar]
- Estep AL, Tidyman WE, Teitell MA, Cotter PD, Rauen KA. HRAS mutations in Costello syndrome: detection of constitutional activating mutations in codon 12 and 13 and loss of wild-type allele in malignancy. Am J Med Genet A. 2006;140:8–16. doi: 10.1002/ajmg.a.31078. [DOI] [PubMed] [Google Scholar]
- Fiegen D, Dvorsky R, Ahmadian MR. Structural principles of Ras interaction with regulators and effectors. In: Der CJ, editor. Proteins and cell regulation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2006. [Google Scholar]
- Ford B, Hornak V, Kleinman H, Nassar N. Structure of a transient intermediate for GTP hydrolysis by ras. Structure. 2006;14:427–436. doi: 10.1016/j.str.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Ford B, Skowronek K, Boykevisch S, Bar-Sagi DB, Nassar N. Structure of the G60A mutant of Ras. J Biol Chem. 2005;280:25697–25705. doi: 10.1074/jbc.M502240200. [DOI] [PubMed] [Google Scholar]
- Franken SM, Scheidig AJ, Krengel U, Rensland H, Lautwein A, Geyer M, Scheffzek K, Goody RS, Kalbitzer HR, Pai EF, Wittinghofer A. Three-dimensional structures and properties of a transforming and a nontransforming glycine-12 mutant of p21H-ras. Biochemistry. 1993;32:8411–8420. doi: 10.1021/bi00084a005. [DOI] [PubMed] [Google Scholar]
- Gasper R, Thomas C, Ahmadian MR, Wittinghofer A. The role of the conserved switch II glutamate in guanine nucleotide exchange factor-mediated nucleotide exchange of GTP-binding proteins. J Mol Biol. 2008;379:51–63. doi: 10.1016/j.jmb.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Gelb BD, Tartaglia M. Noonan syndrome and related disorders: dysregulated RAS-mitogen activated protein kinase signal transduction. Hum Mol Genet. 2006;15:R220–R226. doi: 10.1093/hmg/ddl197. [DOI] [PubMed] [Google Scholar]
- Gremer L, De Luca A, Merbitz-Zahradnik T, Dallapiccola B, Morlot S, Tartaglia M, Kutsche K, Ahmadian MR, Rosenberger G. Duplication of Glu37 in the switch I region of HRAS impairs effector/GAP binding and underlies Costello syndrome by promoting enhanced growth factor-dependent MAPK and AKT activation. Hum Mol Genet. 2010;19:790–802. doi: 10.1093/hmg/ddp548. [DOI] [PubMed] [Google Scholar]
- Gremer L, Gilsbach B, Ahmadian MR, Wittinghofer A. Fluoride complexes of oncogenic Ras mutants to study the Ras-RasGAP interaction. Biol Chem. 2008;389:1163–1171. doi: 10.1515/BC.2008.132. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Lin AE, Stabley DL, Nicholson L, Scott CI, Doyle D, Aoki Y, Matsubara Y, Zackai EH, Lapunzina P, Gonzalez-Meneses A, Holbrook J, Agresta CA, Gonzalez IL, Sol-Church K. HRAS mutation analysis in Costello syndrome: genotype and phenotype correlation. Am J Med Genet A. 2006;140:1–7. doi: 10.1002/ajmg.a.31047. [DOI] [PubMed] [Google Scholar]
- Guo Z, Ahmadian MR, Goody RS. Guanine nucleotide exchange factors operate by a simple allosteric competitive mechanism. Biochemistry. 2005;44:15423–15429. doi: 10.1021/bi0518601. [DOI] [PubMed] [Google Scholar]
- Hall BE, Bar-Sagi D, Nassar N. The structural basis for the transition from Ras-GTP to Ras-GDP. Proc Natl Acad Sci USA. 2002;99:12138–12142. doi: 10.1073/pnas.192453199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsath L, Ahmadian MR. Fluorescence approaches for monitoring interactions of Rho GTPases with nucleotides, regulators, and effectors. Methods. 2005;37:173–182. doi: 10.1016/j.ymeth.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Herbrand U, Ahmadian MR. p190-RhoGAP as an integral component of the Tiam1/Rac1-induced downregulation of Rho. Biol Chem. 2006;387:311–317. doi: 10.1515/BC.2006.041. [DOI] [PubMed] [Google Scholar]
- Herrmann C. Ras-effector interactions: after one decade. Curr Opin Struct Biol. 2003;13:122–129. doi: 10.1016/s0959-440x(02)00007-6. [DOI] [PubMed] [Google Scholar]
- Herrmann C, Martin GA, Wittinghofer A. Quantitative analysis of the complex between p21ras and the Ras-binding domain of the human Raf-1 protein kinase. J Biol Chem. 1995;270:2901–2905. doi: 10.1074/jbc.270.7.2901. [DOI] [PubMed] [Google Scholar]
- Huang L, Hofer F, Martin GS, Kim SH. Structural basis for the interaction of Ras with RalGDS. Nat Struct Biol. 1998;5:422–426. doi: 10.1038/nsb0698-422. [DOI] [PubMed] [Google Scholar]
- Hwang MCC, Sung YJ, Hwang YW. The differential effects of the Gly-60 to Ala mutation on the interaction of H-Ras p21 with different downstream targets. J Biol Chem. 1996;271:8196–8202. doi: 10.1074/jbc.271.14.8196. [DOI] [PubMed] [Google Scholar]
- Ito Y, Yamasaki K, Iwahara J, Terada T, Kamiya A, Shirouzu M, Muto Y, Kawai G, Yokoyama S, Laue ED, Wälchli M, Shibata T, Nishimura S, Miyazawa T. Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein. Biochemistry. 1997;36:9109–9119. doi: 10.1021/bi970296u. [DOI] [PubMed] [Google Scholar]
- Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kigawa T, Yamaguchi-Nunokawa E, Kodama K, Matsuda T, Yabuki T, Matsuda N, Ishitani R, Nureki O, Yokoyama S. Selenomethionine incorporation into a protein by cell-free synthesis. J Struct Funct Genomics. 2002;2:29–35. doi: 10.1023/a:1013203532303. [DOI] [PubMed] [Google Scholar]
- Klink BU, Goody RS, Scheidig AJ. A newly designed microspectrofluorometer for kinetic studies on protein crystals in combination with x-ray diffraction. Biophys J. 2006;91:981–992. doi: 10.1529/biophysj.105.078931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranenburg O. The KRAS oncogene: past, present, and future. Biochim Biophys Acta. 2005;1756:81–82. doi: 10.1016/j.bbcan.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Kratz CP, Niemeyer CM, Zenker M. An unexpected new role of mutant Ras: perturbation of human embryonic development. J Mol Med. 2007;85:223–231. doi: 10.1007/s00109-006-0135-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis PJ, Domaille PJ, Campbell-Burk SL, Van Aken T, Laue ED. Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry. 1994;33:3515–3531. doi: 10.1021/bi00178a008. [DOI] [PubMed] [Google Scholar]
- Krengel U. Ph.D. thesis. Heidelberg: 1991. Struktur und Guanosintriphosphathydrolysemechanismus des C-terminal verkürzten menschlichen Krebsproteins P21-H-RAS. [Google Scholar]
- Krengel U, Schlichting I, Scherer A, Schumann R, Frech M, John J, Kabsch W, Pai EF, Wittinghofer A. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell. 1990;62:539–548. doi: 10.1016/0092-8674(90)90018-a. [DOI] [PubMed] [Google Scholar]
- Lenzen C, Cool RH, Prinz H, Kuhlmann J, Wittinghofer A. Kinetic analysis by fluorescence of the interaction between Ras and the catalytic domain of the guanine nucleotide exchange factor Cdc25Mm. Biochemistry. 1998;37:7420–7430. doi: 10.1021/bi972621j. [DOI] [PubMed] [Google Scholar]
- Lo FS, Lin JL, Kuo MT, Chiu PC, Shu SG, Chao MC, Lee YJ, Lin SP. Noonan syndrome caused by germline KRAS mutation in Taiwan: report of two patients and a review of the literature. Eur J Pediatr. 2009;168:919–923. doi: 10.1007/s00431-008-0858-z. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:708. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- Margarit SM, Sondermann H, Hall BE, Nagar B, Hoelz A, Pirruccello M, Bar-Sagi D, Kuriyan J. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 2003;112:685–695. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- Milburn MV, Tong L, deVos AM, Brunger A, Yamaizumi Z, Nishimura S, Kim SH. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- Nassar N, Horn G, Herrmann C, Scherer A, McCormick F, Wittinghofer A. The 2.2 A crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature. 1995;375:554–560. doi: 10.1038/375554a0. [DOI] [PubMed] [Google Scholar]
- Nava C, Hanna N, Michot C, Pereira S, Pouvreau N, Niihori T, Aoki Y, Matsubara Y, Arveiler B, Lacombe D, Pasmant E, Parfait B, Baumann C, Heron D, Sigaudy S, Toutain A, Rio M, Goldenberg A, Leheup B, Verloes A, Cavé H. Cardio-facio-cutaneous and Noonan syndromes due to mutations in the RAS/MAPK signalling pathway: genotype–phenotype relationships and overlap with Costello syndrome. J Med Genet. 2007;44:763–771. doi: 10.1136/jmg.2007.050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niihori T, Aoki Y, Narumi Y, Neri G, Cave H, Verloes A, Okamoto N, Hennekam RCM, Gillessen-Kaesbach G, Wieczorek D, Kavamura MI, Kurosawa K, Ohashi H, Wilson L, Heron D, Bonneau D, Corona G, Kaname T, Naritomi K, Baumann C, Baumann C, Matsumoto N, Kato K, Kure S, Matsubara Y. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat Genet. 2006;38:294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- Pacold ME, Suire S, Perisic O, Lara-Gonzalez S, Davis CT, Walker EH, Hawkins PT, Stephens L, Eccleston JF, Williams RL. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103:931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, Wittinghofer A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechlivanis M, Ringel R, Popkirova B, Kuhlmann J. Prenylation of Ras facilitates hSOS1-promoted nucleotide exchange, upon Ras binding to the regulatory site. Biochemistry. 2007;46:5341–5348. doi: 10.1021/bi602353k. [DOI] [PubMed] [Google Scholar]
- Quilliam LA, Zhong S, Rabun KM, Carpenter JW, South TL, Der CJ, Campbellburk S. Biological and structural characterization of a Ras transforming mutation at the phenylalanine-156 residue, which is conserved in all members of the Ras superfamily. Proc Natl Acad Sci USA. 1995;92:1272–1276. doi: 10.1073/pnas.92.5.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan MP, Settleman J. Isoform-specific ras functions in development and cancer. Future Oncol. 2009;5:105–116. doi: 10.2217/14796694.5.1.105. [DOI] [PubMed] [Google Scholar]
- Reinstein J, Schlichting I, Frech M, Goody RS, Wittinghofer A. p21 with a phenylalanine 28-leucine mutation reacts normally with the GTPase activating protein GAP but nevertheless has transforming properties. J Biol Chem. 1991;266:17700–17706. [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR. GTPase activating proteins: structural and functional insights 18 years after discovery. Cell Mol Life Sci. 2005;62:3014–3038. doi: 10.1007/s00018-005-5136-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Grünewald P, Wohlgemuth S, Kabsch W, Tu H, Wigler M, Wittinghofer A, Herrmann C. The Ras-Byr2RBD complex: structural basis for Ras effector recognition in yeast. Structure. 2001;9:1043–1050. doi: 10.1016/s0969-2126(01)00674-8. [DOI] [PubMed] [Google Scholar]
- Scheidig AJ, Burmester C, Goody RS. The pre-hydrolysis state of p21(ras) in complex with GTP: new insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins. Structure. 1999;7:1311–1324. doi: 10.1016/s0969-2126(00)80021-0. [DOI] [PubMed] [Google Scholar]
- Scheidig AJ, Franken SM, Corrie JE, Reid GP, Wittinghofer A, Pai EF, Goody RS. X-ray crystal structure analysis of the catalytic domain of the oncogene product p21H-ras complexed with caged GTP and mant dGppNHp. J Mol Biol. 1995;253:132–150. doi: 10.1006/jmbi.1995.0541. [DOI] [PubMed] [Google Scholar]
- Scheidig AJ, Sanchez-Llorente A, Lautwein A, Pai EF, Corrie JE, Reid GP, Wittinghofer A, Goody RS. Crystallographic studies on p21(H-ras) using the synchrotron Laue method: improvement of crystal quality and monitoring of the GTPase reaction at different time points. Acta Crystallogr. 1994;D50:512–520. doi: 10.1107/S090744499301443X. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Bollag G, Lyubynska N, Nguyen H, Kratz CP, Zenker M, Niemeyer CM, Molven A, Shannon K. Biochemical and functional characterization of germ line KRAS mutations. Mol Cell Biol. 2007b;27:7765–7770. doi: 10.1128/MCB.00965-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007a;7:563. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Zenker M, Rowe SL, Boll S, Klein C, Bollag G, van der Burgt I, Musante L, Kalscheuer V, Wehner LE, Nguyen H, West B, Zhang KYJ, Sistermans E, Rauch A, Niemeyer CM, Shannon K, Kratz CP. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- Schweins T, Scheffzek K, Assheuer R, Wittinghofer A. The role of the metal ion in the p21ras catalysed GTP-hydrolysis: Mn2+ versus Mg2+ J Mol Biol. 1997;266:847–856. doi: 10.1006/jmbi.1996.0814. [DOI] [PubMed] [Google Scholar]
- Seeburg PH, Colby WW, Capon DJ, Goeddel DV, Levinson AD. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature. 1984;312:71–75. doi: 10.1038/312071a0. [DOI] [PubMed] [Google Scholar]
- Sol-Church K, Stabley DL, Nicholson L, Gonzalez IL, Gripp KW. Paternal bias in parental origin of HRAS mutations in Costello syndrome. Hum Mutat. 2006;27:736–741. doi: 10.1002/humu.20381. [DOI] [PubMed] [Google Scholar]
- Sondermann H, Soisson SM, Boykevisch S, Yang SS, Bar-Sagi D, Kuriyan J. Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell. 2004;19:393–405. doi: 10.1016/j.cell.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Spoerner M, Herrmann C, Vetter IR, Kalbitzer HR, Wittinghofer A. Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc Natl Acad Sci USA. 2001;98:4944–4949. doi: 10.1073/pnas.081441398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprang SR. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- Stieglitz B, Bee C, Schwarz D, Yildiz O, Moshnikova A, Khokhlatchev A, Herrmann C. Novel type of Ras effector interaction established between tumour suppressor NORE1A and Ras switch II. EMBO J. 2008;27:1995–2005. doi: 10.1038/emboj.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JC, Colleton M, Bottorff D. Effector domain mutations dissociate p21(Ras) effector function and GTPase-activating protein-interaction. Mol Cell Biol. 1993;13:7311–7320. doi: 10.1128/mcb.13.12.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YJ, Hwang MCC, Hwang YW. The dominant negative effects of H-Ras harboring a Gly to Ala mutation at position 60. J Biol Chem. 1996;271:30537–30543. doi: 10.1074/jbc.271.48.30537. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Williams RL, Rabbitts TH. Tumour prevention by a single antibody domain targeting the interaction of signal transduction proteins with RAS. EMBO J. 2007;26:3250–3259. doi: 10.1038/sj.emboj.7601744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidyman WE, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev. 2009;19:230–236. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong LA, de Vos AM, Milburn MV, Kim SH. Crystal structures at 2.2 A resolution of the catalytic domains of normal ras protein and an oncogenic mutant complexed with GDP. J Mol Biol. 1991;217:503–516. doi: 10.1016/0022-2836(91)90753-s. [DOI] [PubMed] [Google Scholar]
- Tsukuda K, Tanino M, Soga H, Shimizu N, Shimizu K. A novel activating mutation of the K-ras gene in human primary colon adenocarcinoma. Biochem Biophys Res Commun. 2000;278:653–658. doi: 10.1006/bbrc.2000.3839. [DOI] [PubMed] [Google Scholar]
- Tucker J, Sczakiel G, Feuerstein J, John J, Goody RS, Wittinghofer A. Expression of p21 proteins in Escherichia coli and stereochemistry of the nucleotide-binding site. EMBO J. 1986;5:1351–1358. doi: 10.1002/j.1460-2075.1986.tb04366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter IR, Linnemann T, Wohlgemuth S, Geyer M, Kalbitzer HR, Herrmann C, Wittinghofer A. Structural and biochemical analysis of Ras-effector signaling via RalGDS. FEBS Lett. 1999;45:175–180. doi: 10.1016/s0014-5793(99)00555-4. [DOI] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A. Signal transduction—the guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- Xu JA, Baase WA, Baldwin E, Matthews BW. The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect. Protein Sci. 1998;7:158–177. doi: 10.1002/pro.5560070117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenker M, Lehmann K, Schulz AL, Barth H, Hansmann D, Koenig R, Korinthenberg R, Kreiss-Nachtsheim M, Meinecke P, Morlot S, Mundlos S, Quante AS, Raskin S, Schnabel D, Wehner LE, Kratz CP, Horn D, Kutsche K. Expansion of the genotypic and phenotypic spectrum in patients with KRAS germline mutations. J Med Genet. 2007;44:131–135. doi: 10.1136/jmg.2006.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]