Abstract
Circadian clocks are present in most organisms and provide an adaptive mechanism to coordinate physiology and behavior with predictable changes in the environment. Genetic, biochemical, and cellular experiments have identified more than a dozen component genes and a signal transduction pathway that support cell-autonomous, circadian clock function. One of the hallmarks of biological clocks is their ability to reset to relevant stimuli while ignoring most others. We review recent results showing intracellular and intercellular mechanisms that convey this robust timekeeping to a variety of circadian cell types.
Introduction
Clocks need to be robust. How useful would a watch be if it was reset by eating lunch or walking to work? Instead, watches are built to withstand and ignore shocks to keep accurate time. However, because no watch keeps perfect time, they need adjusting. The same concepts are true for biological clocks -- they are remarkably robust to perturbation and yet entrain to daily environmental cues such as light and dark. In this review, we explore the underlying basis of the robustness in the mammalian circadian clock to perturbation. Here, robustness is defined as the ability to sustain daily oscillations with an accurate period length day after day. Other studies have highlighted that metrics of robustness depend on how we define circadian performance and at what level {PMIDs: 17158515; 17482552). In oscillatory systems, after perturbation, changes in amplitude, period length, and phase are indicators of robustness, or its inverse, sensitivity. These could relate to the adaptive features of the system to its environment. For instance, jet lag is a byproduct of robust circadian timekeeping, in essence ignoring temporal displacement. Jet lag diminishes, however, due to the system’s sensitivity toward light cues; a change in light pattern resets the circadian phase. The ability to maintain robust circadian rhythms in the face of perturbations and uncertainty (e.g. temporal displacement or day length) is a long-recognized and critical property of living systems. Of course, though cellular in nature, physiological disruptions are products of both intercellular and intercellular processes and robustness needs to be considered and measured at multiple levels. In this review, we highlight how this robustness arises from both intracellular and intercellular processes.
When considering the mechanisms underlying reliable circadian rhythmicity, the plasma membrane naturally divides the processes into intracellular and intercellular. Both processes can differ between cell types. Intracellular processes include the genetic regulatory architecture of the cell. What do we mean by that? We define genetic architecture as the suite of transcriptional and post-transcriptional mechanisms that support normal circadian rhythms. This includes core clock genes such as the E-box activators, Clock and Bmal1, and their transcriptional repressors, the Period and Cryptochrome genes. In addition, it includes more recently identified factors such as Rev-erb-alpha and Rora that function in the clock to regulate levels of Clock and Bmal1. Intercellular processes include those signaling pathways from one circadian cell which impact oscillations in another. To date, neuropeptides and perhaps gap junctions have been implicated. We will discuss how this architecture changes upon genetic perturbation, and the implications of dynamic network analysis for future study. Finally, we will discuss how recent advances have allowed the exploration of these genetic architectures for the underlying principles that convey robustness.
Intracellular mechanisms for robustness
Circadian cells not only respond to environmental change – they anticipate it
The circadian clock pathway is remarkably similar to two other adaptive pathways, the dioxin and hypoxia pathways (reviewed in (1)). While the dioxin response pathway helps mammals to sense and metabolize environmental contaminants, the hypoxia pathway helps mammals respond to low oxygen conditions. Like the dioxin and hypoxia pathways, the circadian clock senses inputs such as light and food. However, in contrast to the dioxin and hypoxia pathways, which are reactive to changes in the environment, the circadian clock anticipates environmental change. All three pathways use bHLH PAS proteins in their signal transduction. As DNA-binding transcription factors, these proteins share remarkably similar response elements, the dioxin response element, a hypoxia response element, and the E-box.
To establish the roles of key players in these pathways, researchers in the circadian, hypoxia, and dioxin signaling fields generated and studied the effect of specific gene knockouts on these pathways. In addition to establishing the roles of these key players in their respective pathways, knockout of many of these genes often resulted in mice with early lethality (reviewed in (1)). In contrast, loss of circadian clock genes (e.g. knockout mice for Bmal1, Clock, Npas2, Per1, Per2, Per3, Cry1, Cry2) had no effect on survival at birth (2–6). Not only were these mice alive, deficits in circadian regulation of locomotor activity were relatively minor for all but Bmal1 null mice. How could this be? Circadian clocks are important -- almost all organisms have them -- how could dispensing with key clock components have relatively minor effects on behavior and physiology? Answers to these questions lie in the genetics, biochemistry, and cell biology of the clock, all critical features of its robustness.
Genetic duplication in the mammalian circadian core loop
The concept of redundancy dominated early thinking on the genetic robustness of the clock. Initial work stemmed from the observation that for every fly circadian clock gene there were typically two or three mammalian paralogs, descendants of a common ancestor of this fly gene (7). Realizing this, several labs created compound homozygous, null mice to test the redundancy hypothesis. First up for these studies were the Cryptochrome and Period genes. While Cry1−/− mice had a short period length for locomotor activity in constant darkness, Cry2−/− mice ran with a long period suggestive that CRY1 and CRY2 proteins are not functionally identical (8). However, Cry1/Cry2 double knockout mice had a much more severe phenotype -- arrhythmicity (8) -- indicating their importance in the clock. The same was true for the Per genes. While Per1, Per2, or Per3 null mice had minor locomotor activity phenotypes in constant darkness, Per1/Per2 double knockout mice were arrhythmic (5, 9). Analysis of Npas2 and Clock, homologs of fly Clock, took longer, but the result was the same. Both Npas2 and Clock knockout mice had a mild shortening of locomotor periodicity (3, 10). The compound homozygote Npas2/Clock double-knockout mice were again arrhythmic in constant conditions (11). Note: this is condition dependent –Clock mutant mice, which, in contrast to the knockout, retain an altered protein version of Clock, are arrhythmic in constant darkness, but become rhythmic in constant light {PMIDs: 9160755, 9160756, 12465885}. Analysis of Bmal1/Bmal2 compound null mice was preempted by the observation that Bmal1 knockout mice were themselves arrhythmic (2). The essentiality of Bmal1 may in part be explained by its role in regulating its paralogBmal2 – when Bmal1 levels are low, Bmal2 levels are also low (2, 12). Thus, the clock was by and large tolerant to deletion of its individual components with redundancy as a likely explanation.
Biochemical mechanism and genetic architecture
But it wasn’t that simple. As more biochemical details of the clock were discovered, a surprisingly complex genetic architecture was revealed. To get an oscillation, the clock requires an activator, a repressor, and some mechanism(s) to delay the repressor’s action on the activator. Many components of the mammalian clock are transcriptional activators and repressors (Figure 1). Two families of activators, Clock/Npas2 and Bmal1/Bmal2, heterodimerize to bind E-box sequences present in two families of repressors, Cry1/Cry2 and Per1/Per2/Per3 (13–14). As message levels of these repressors build up, their proteins are translated and form physical complexes with members of the casein kinase 1 family in the cytoplasm (6, 15–21). After a delay, this repressor complex translocates to the nucleus where it interacts with the activator complex to repress their transcription. This results in lower levels of their own messages. Later in the cycle, these repressor proteins are actively degraded by Fbxl3 and Btrc in a proteasome-dependent fashion, thus relieving repression and allowing repressor message levels to accumulate and begin another cycle (22–25). This biochemical mechanism is called the core loop.
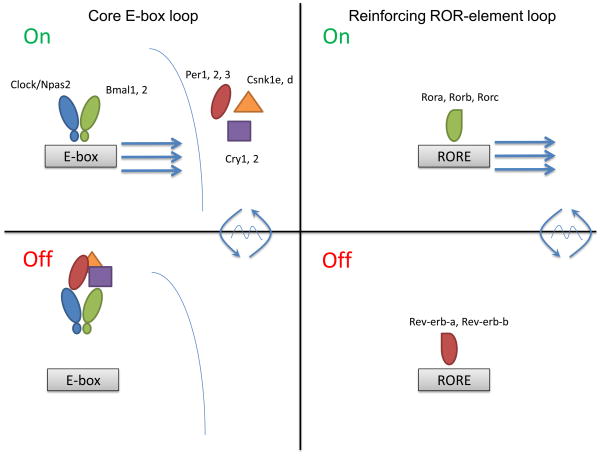
Figure 1.
Biochemical engines and regulatory architectures. The two primary biochemical engines of the clock are the E-box and RORE regulators. (Top) A simplified diagram of these engines. In the upper panels, a theoretical “100% on” state is depicted. Clock and Bmal1 bind to E-box elements recruiting the basal PolII machinery and activating transcription. Rora, Rorb, and Rorc likewise bind to their response elements, the RORE, to activate transcription from structural genes containing these elements. (Bottom) In the lower panels, a theoretical “100% off” state is depicted. In the case of the E-box, the repressor complex (Cryptochrome, Period, and casein kinase 1) has translocated to the nucleus, bound to the Clock/Bmal1 complex, and repressed the complex possibly by disrupting their DNA-binding potential. In the case of the RORE, Rev-erb-alpha and Rev-erb-beta displace the Ror factors to actively repress transcription by recruiting histone deacetylaces.
But is it one loop or a series of them {PMID: 12620213}? Inherent in this simple mechanism is the capacity for each factor in the clock to regulate the others. For example, Bmal1-knockout mice have lower levels of Per1, Per2, Cry1, and Cry2, consistent with its role as an activator of these genes (2). Conversely, mice deficient in Cry1 and Cry2 have higher levels of Per1 and Per2 (8). One consequence of “the core loop” structure, therefore, is the capacity for mutual regulation of gene expression of all components by all components. Although the biochemical mechanism may constitute a loop, the consequence on gene regulatory architecture is a network of E-box loops regulating many core clock genes (Figure 1). In this sense, it may be helpful to think of the core loop as an important motif in the general architecture of the clock network in a cell.
Evidence for a stabilizing secondary intracellular loop
The circadian clock regulatory structure, however, is composed of more than just the core E-box loop. Schibler and colleagues recognized an ROR element (RORE) in the Bmal1 promoter (26). This element was identified earlier as a target of two classes of transcription factors, the Rors (activators) and Rev-erbs (repressors). Schibler’s group tested a knockout mouse for Rev-erb-alpha and found that, as predicted, Bmal1 message levels were high (26). In addition, Rev-erb-alpha had an E-box element in its promoter and a high amplitude circadian pattern to its mRNA expression driven by the core loop (27). Later, our group and others showed that Rora, an activator of the ROR element, positively activated Bmal1 expression in vitro (28–30). Moreover, Rora deficient mice had lower Bmal1 levels in the suprachiasmatic nucleus (SCN) of the hypothalamus, a population of about 20,000 neurons critical for circadian rhythms in vertebrates. Mice lacking Rora also had short period circadian locomotor activity, a behavior driven by the SCN (28, 30). Because of this mutual regulation, similar to that seen in the core loop, this structure was termed a secondary loop (Figure 1). A similar secondary loop was identified earlier in the fly system, though the fly genes were not related to those found in mammals.
The role of the secondary loop
The secondary loop reinforces the primary loop by regulating core loop components – this reinforcement confers additional robustness. Mice deficient in Rev-erb-alpha were short period length and have a greater dispersion in period length and lower amplitude than wild type littermates (26). A confounding phenotype of the Rorasg/sg mutation, ataxia, made it impossible to directly link its similar phenotypes to a deficit in circadian clock function (28). Flies or mice deficient in these components have disruptions in gene expression and behavior, but their phenotypes are not as severe as components in the primary loop. Thus, this secondary transcription-translation loop stabilizes, but is not required for, circadian rhythm generation. Moreover, rather than play a direct role in rhythm generation, secondary loop factors seem to act by regulating the expression of components of the core loop.
Architecture from loops
At nearly the same time, Hiroki Ueda and colleagues began to probe the genetic regulatory architecture of the clock for additional layers of regulation using sequence informatics. They built sophisticated computational models of the E-box, RORE, and D-box (promoter elements bound by PAR bZip factors that were identified as regulators of clock output). In their seminal study, the Ueda group showed that activators and repressors that bind these different elements were mutually ‘wired’ in a web-like genetic architecture (31). The core, E-box loop as well as the ROR and DBP loops were repeated motifs in this genetic architecture (Figure 2). Importantly, in cell-based assays, they went on to show that many of these elements were functional. Thus, while the clock may be composed of core and secondary loops, in action these loops regulate the expression of multiple core clock genes both independently and coordinately. Describing the function of clocks, then, requires an understanding of the higher order genetic architecture in addition to the aforementioned biochemical mechanisms.
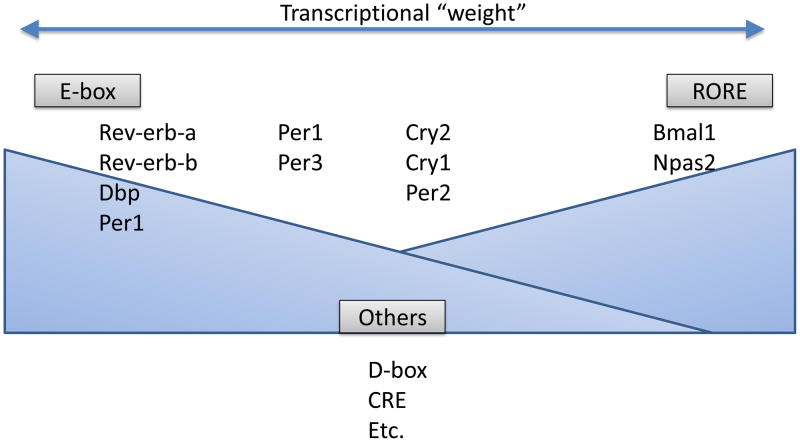
Figure 2.
The contribution of response elements to clock gene expression is depicted. Certain genes are dominated by E-box input, e.g. Rev-erb-alpha or DBP. Other genes are dominated by RORE input, e.g. Bmal1. Many genes integrate input from both elements, e.g. Cry1 or Per2. Other elements also contribute – the CRE and D-box elements are notable examples. In this model, like a synaptic weight, all genes have the potential to get input from all transcriptional complexes, but do so to a greater or lesser extent depending on the complex.
Probing the genetic architecture for characteristics underlying robustness
But how do you study genetic regulatory architecture? Unlike buildings, it’s not easy to see. To get at this issue, our group applied sensitivity analysis and a technique called Gene Dosage Network Analysis (32). In this approach, an individual gene is depleted by RNAi in a dose-dependent fashion generating an “allelic series” of gene dosage. These cells are then put in functional assays to determine the impact of this depletion on clock function. In addition, message levels for other components of the clock system were investigated by RT PCR. If all clock components are truly wired to all clock components, changing any one of them has the capacity to regulate the levels of all others. We did this analysis, and most clock components were found to regulate most others (32) (Figure 2). The finding that several genes are upregulated following knockdown of their paralogs suggests that the clock network utilizes active compensatory mechanisms rather than simple redundancy to confer robustness and maintain function. But there was more. By taking this systematic approach and by collecting experimental data at scale, we were able to investigate this perturbation data for general principles. We found two: paralog compensation and proportional response.
Paralog compensation: it takes (at least) two, baby
Paralog compensation occurs when one paralog is upregulated in response to deletion of another. This phenomenon has been observed at a genome scale. Pilpel and colleagues analyzed the yeast knockout panel and found that paralogous genes were more likely to respond to a specific knockout than knockouts of unrelated genes (33). It has also been observed in vertebrates (34). In perhaps the most famous case, deletion of the master regulator of myogenesis, Myod1, unexpectedly caused little to no phenotype in muscle development (35). Upon investigation, researchers found that it’s paralog, Myf-5, was upregulated in response to deletion of Myod1 (35). When the double knockout was made, the mice failed to develop normal skeletal muscle and died soon after birth (36). Paralog compensation, rather than simple redundancy, provides a mechanism to ensure gene function upon challenge (i.e. gene loss) and, therefore, conferred robustness.
Our investigation of the circadian system likewise found several examples of paralog compensation, particularly those associated with repressors. For example, depletion of Per1 by RNAi resulted in up regulation of its paralogs Per2 and Per3. Likewise, depletion of Cry1 resulted in up regulation of its paralog Cry2. Finally, down regulation of Rev-erb-beta resulted in up regulation of its paralog Rev-erb-alpha. In all three cases, this paralog compensation was unidirectional. In other words, depletion of Per2 did not produce up regulation of Per1 and Per3, nor did Per1 and Per2 respond to depletion of Per3. Thus, paralog compensation imparts robustness against some, but not all, perturbations of the genetic architecture. This helps explain how forward genetics has revealed some, but not many, genes in the circadian gene network.
How does directional gene compensation work? It is possible that the paralogs are not wired in exactly the same way; without the right regulatory element, a gene can’t respond. Indeed, while Per1, Per2, and Per3 have E-box elements in their promoters (what genes don’t?), their levels respond differently in Clock and Bmal1 mutant mice (37). Furthermore, they are differentially regulated by light and by the CREB system (38–39). It is also possible that genes are present at vastly different protein concentrations or activity levels. For example, depletion of a less abundant gene may have little or no effect on a more abundant gene, even if it’s wired to respond. This may explain tissue-specific robustness. For example, Npas2 and Clock are expressed at similarly low levels in the SCN. Loss of one or the other yields a modest circadian phenotype, perhaps due to paralog compensation, whereas loss of both abolishes circadian behavior (11). In contrast, Npas2 is expressed at high levels in the neocortex, where it has been speculated to play a more important role in circadian timing than Clock (40). In liver, Npas2 levels are elevated in response to Clock knockout; however, the liver clock is arrhythmic (3, 51). Interestingly, there is no evidence for cell-autonomous circadian timing in these brain areas (41), suggesting that cell-specific changes in the relative abundance of clock genes may not always support circadian clock function. This needs further examination, especially given the recent report that Per1 and Per2 are not always found in the same cells, at least in brain (42). Thus, tissue specific expression and other perturbations change the genetic architecture of the clock and consequently its function.
The principle of proportionality
Perturbation of the clock by Gene Dosage Network Analysis showed that the oscillator can produce three types of proportional responses: linear, inverse, and nonlinear. For example, depletion of Bmal1 resulted in strictly linear transcriptional responses of Rev-erb-alpha and Rev-erb-beta levels: deplete Bmal1 levels by 50%, Rev-erb-alpha levels drop by 50%. These responses can also be sub-linear, but proportional: deplete Clock levels by 50%, Rev-erb-alpha levels drop 25%. Responses can be disproportionate: deplete Cry1 and Cry2 levels by 50%, and Per2 and Rorc levels increase by 11 times and seven times respectively. This is by no means a clock-specific principle. Similar proportionality has been observed in both yeast and higher eukaryotic systems (34). Thus, this network sensitivity analysis uncovered how simple principles of paralog compensation and proportionality were built into the circadian oscillator.
Intercellular mechanisms for robustness
Hiding weaknesses in a network
One of the most remarkable recent discoveries is that cell-cell interactions can make circadian oscillations more robust against a wide variety of perturbations including genetic mutations. The success of genetics in circadian biology depended on easily measured changes in the stable, repeatable daily rhythms of locomotor activity. In mammals, forward genetics revealed a role for the Clock gene in the determination of circadian periodicity of locomotor activity in mice (43). The discovery of Clock came quickly (the first Clock heterozygote was mouse #25 in the mutagenesis screen for altered circadian locomotor patterns). So it’s surprising that the subsequent 15 years of mutagenesis screens yielded only one additional central regulator of circadian periodicity, the Fbxl3 gene (44–46). Forward genetics in flies was more productive and offered candidates that, when knocked out in mice, revealed roles for genes including Per1, Per2, Cry1, and Cry2. The regulatory architecture in mice appeared to protect against loss of key clock components, where flies, which lack clock gene paralogs, had more severe phenotypes.
More recently, studies of the cellular basis for the behavioral deficits in these mice revealed that the coupled SCN network is more resistant to genetic perturbations than single SCN neurons. For example, the Clock mutation produces arrhythmicity in the firing patterns of dispersed SCN neurons, but a long period in SCN slices and behavior (47). Similarly, dispersed SCN neurons from Cry1−/− or Per1−/− mice exhibit weak circadian rhythms in PER2::LUC activity but high amplitude rhythms in SCN slices (48). These results are consistent with analyses of chimeric, Clock-mutant mice which showed that lengthening of circadian period and loss of amplitude correlated with a decreasing proportion of wild-type to Clock/Clock cells. Strikingly, the circadian pacemaker mechanism does not suddenly break down with the introduction of some Clock-mutant cells (49). Thus, cellular network interactions can protect against some genetic deficits. This provides a warning to geneticists who give up when they see no behavioral phenotype in their knockout mouse; cellular communication could mask the loss of function that may be apparent in single cells.
The same idea holds for peripheral clocks. Imaging analysis of liver, lung, cornea, or fibroblasts from Cry1−/− mice showed an arrhythmic phenotype, while behavioral rhythms or those from SCN slices were short period length (48). Lung and fibroblasts from Per1 knockout mice are arrhythmic, while behavioral analysis of the same mice show short period length (48). While Rorasg/sg mice are short period length in locomotor behavior, fibroblasts from these mice are arrhythmic (50). In contrast to ClockΔ19/Δ19 mice, which have a mutation that causes exon skipping and loss of 51 amino acids in the Clock protein and are long period for locomotor activity, Clock−/− mice, with no expressed exons, have short period length behavioral rhythms. Lung and liver samples from these knockout mice are arrhythmic (51). Thus, behavioral rhythms and intact SCN slices from circadian clock gene knockout mice are protected against the more severe phenotypes seen in cell autonomous preparations from their periphery.
Sloppy clocks made precise
The idea that single cells are unstable circadian clocks made robust by network interactions also holds for wild-type SCN neurons. We recently found that fully isolated SCN neurons can express circadian rhythms in gene expression or firing rate, but their rhythms are low amplitude and their periods are unreliable (52). Dispersed SCN neurons show about 10 times greater cycle-to-cycle period variability than those in the connected SCN explant (53). In SCN slices, blocking action potentials (and presumably intercellular communication) with tetrodotoxin (TTX) also results in sloppy circadian rhythms of gene expression in many neurons (54). Furthermore, individual cells in SCN slices that were arrhythmic in a primary TTX treatment can remain arrhythmic or regain rhythmicity in a subsequent TTX treatment (52). This ability to generate and lose rhythmicity appears to be common across multiple classes of cells in the SCN. This is reminiscent of how neurons that fire in rhythmic bursts can transition to tonic firing patterns with small changes in their various conductances depending on a combination of intrinsic and network properties (55). In circadian cells, perhaps small changes in the molecular clock (e.g. degradation rates of PER2) result in cells which fall off their limit cycle and become unstable.
Remarkably, Rat-1 fibroblasts are not unstable, but instead appear to oscillate with a relatively stable, cell-autonomous period for at least 15 days (56). This is similar to the precise circadian timing of individual cyanobacteria, which do not appear to share circadian timing information with each other (57). Among mammalian cells, SCN cells appear to be unique in their ability to synchronize to each other.
Intercellular signals that improve robustness
One of the signals responsible for the synchrony and precision among SCN cells has been identified -- vasoactive intestinal polypeptide (VIP). VIP is secreted in a circadian pattern from a subset of approximately 10% of SCN neurons (58). Loss of VIP or its receptor results in mice that have trouble entraining to a poorly lit day, one hour of light at dawn and another at dusk (59). In constant darkness, the locomotor activity of most of these mice is arrhythmic or comprised of multiple circadian periods (59–61). SCN from these mice match their behavioral phenotypes with no rhythm at the population level, although a subset of individual cells express desynchronized, unstable circadian rhythms in firing rate or gene expression. A minority of mice deficient in VIP signaling can express a shortened free-running period in behavior and in the SCN (59–61), suggesting additional signaling can coordinate the ensemble. These other coupling signals have not been identified, but candidates include other neuropeptides (e.g. gastrin releasing peptide or vasopressin) and communication through gap junctions. Even signals such as glucocorticoids or body temperature, acting indirectly or directly on SCN rhythmicity, could play a role in this robustness against the loss of the primary intercellular coupling factor, VIP (62–64).
How do intercellular signals stabilize intracellular circadian signals?
Ultimately, they need to act on the molecular oscillator. Although little is known about the specific transduction pathways, we and others have hypothesized that cell-cell signaling increases the amplitude of the circadian oscillator, reducing its sensitivity to perturbations (65). For example, robustness could result from amplification of transcription of critical clock genes (Fig. 3). In this respect, VIP increases cAMP (66–67), requires protein kinase A (PKA) to phase advance firing rate rhythms in the SCN (68), and can induce transcription of Per1 and Per2 in a CREB-dependent manner (69). Other transmitters, hormones, gap junctions, or even temperature changes in the body, could similarly entrain and augment Per gene transcription in local or global populations of circadian cells (70).
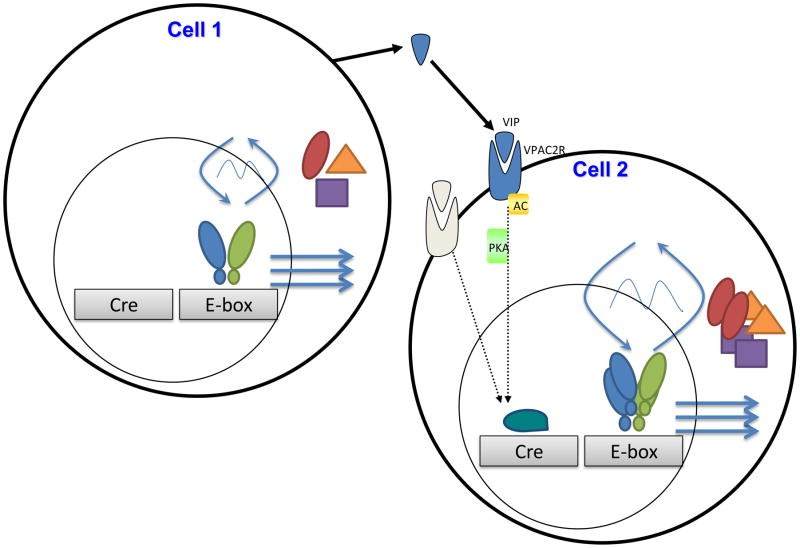
Fig. 3.
Schematic of putative network pathways to amplify the intracellular circadian oscillator. Because the loss of VIP or its receptor leads to fewer high amplitude circadian cells, we illustrate how intercellular VIP may increase the amplitude of intracellular gene expression rhythms. Cell 1 expresses the E-box based molecular oscillator shown in Fig. 1. Cell 2 expresses the same molecular oscillator and receives a signal (VIP) which acts through its receptor (VPAC2R) to activate second messengers (e.g. adenylatecyclase and protein kinase A) to ultimately enhance clock gene expression (e.g. phosphorylated CREB binding to the CRE promoter element on the clock gene or another interacting gene). The increased transcription results in more clock gene products at the time of their peak production and a higher amplitude oscillation. The same perturbation on these two cells would be predicted to have a larger effect on the oscillations of Cell 1. This highlights that intercellular signaling both increases robustness and provides points of sensitivity in the architecture underlying reliable circadian cycling. Future studies will consider how these pathways may change with age, may adjust to the seasons, or may be impacted by modern, non-circadian lifestyles.
Relevance of robust rhythmicity to reality
To be useful a circadian clock must keep accurate time so that it can control processes in anticipation of daily changes in physiology and the environment. It must also be adjustable to local time and, thus, seek a balance between robustness and adaptability. Therefore, there may be conditions when circadian oscillators are naturally less robust.
Neonatal SCN may be less robust
SCN from newborn rodents express circadian rhythms in gene expression, metabolism, and firing rate, but do not control daily rhythms in behavior for several more days to weeks depending on the model and behavior. Recently, Nishide et al. showed that SCN from postnatal day 6 mice phase shift more than 8 hours to stimuli that have no effect on adult SCN (71). It may be that developmental changes in molecular oscillator components or intercellular connectivity regulate the responsiveness of the circadian clock to perturbations. This would suggest that, for whatever reasons, neonatal rhythms are particularly susceptible to environmental disruptions.
Times to throw out a clock
Arctic reindeer and voles may turn off circadian oscillations as an adaptive advantage in their unusual lighting environments. Recently, Loudon and colleagues showed that fibroblasts from reindeer either have extremely low amplitude rhythms or have lost autonomous clock function altogether (72). It remains to be seen how changes in intracellular state, SCN connectivity, or their combined effects mediate some of the developmental or seasonal adaptations in mammalian circadian rhythms (73).
Future directions
Recent work has raised the possibility that inputs outside the core clock can influence or even substitute for its function. For example, cell metabolism (i.e. redox state) can be under circadian clock control and also regulate transcription of key clock genes (74–75). Similarly, cAMP levels oscillate with time of day and can shift circadian gene expression (76). In SCN context, intercellular signals likely modulate ATP and cAMP levels, a path from network effects to single-cell circadian oscillations. On a systems level, body temperature, under the control of the SCN, can influence circadian timing in peripheral tissues and in the SCN (62–63). How and whether these intracellular and intercellular signals protect the clock against environmental perturbations remains to be determined.
Traditional, ground-up modeling will also play an important role. Much of our recent progress has benefited from using cell autonomous models of the clock. These models are convenient in that they allow for high throughput analysis of clock gene perturbations, and indeed, even whole genome screening for clock modifiers. They are also more amenable to biochemical approaches than the SCN. Indeed, much of the biochemistry that fuels ground-up models of the clock comes from either the cell autonomous models or easily studied peripheral tissues such as the liver. As the field moves further, understanding the tissue specific biochemical parameters of the clock will become important. There isn’t one clock; there shouldn’t be one clock model.
Finally, the nascent field of network biology is just beginning for the clock. In that regard, the transcriptional translational feedback loop may have gotten us here, but its utility is waning in explaining the complexities of clock biology. As future studies uncover more about the clock network architecture, we may consider these biochemical loops as motifs in larger structures that explain the complexity of clock biology. While the circadian gene network has begun to give up its principles, the features that make it so robust will be the hardest nuts to crack
Research highlights
Circadian systems oscillate in a noisy environment and entrain to daily timing cues.
Their robustness against environmental and genetic perturbations depends on their biochemical and genetic architecture.
Paralog compensation, transcriptional response programs and balanced biochemical feedback loops help sustain daily cycling.
Robustness also arises from intercellular (e.g. neurotransmission) and systems-level signals (e.g. body temperature).
Acknowledgments
We acknowledge members of the Hogenesch and Herzog labs for comments on this manuscript. The National Institutes of Health funds JBH (5R01HL 097800-04 and 5R01NS 054794-05) and EH (MH63104 and GM078993)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- 1.McIntosh BE, Hogenesch JB, Bradfield CA. Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu Rev Physiol. 2010 Mar 17;72:625–45. doi: 10.1146/annurev-physiol-021909-135922. [DOI] [PubMed] [Google Scholar]
- 2.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000 Dec 22;103(7):1009–17. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Debruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM. A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron. 2006 May 4;50(3):465–77. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 4.Garcia JA, Zhang D, Estill SJ, Michnoff C, Rutter J, Reick M, Scott K, Diaz-Arrastia R, McKnight SL. Impaired cued and contextual memory in NPAS2-deficient mice. Science. 2000 Jun 23;288(5474):2226–30. doi: 10.1126/science.288.5474.2226. [DOI] [PubMed] [Google Scholar]
- 5.Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001 Jun 1;105(5):683–94. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 6.Okamura H, Miyake S, Sumi Y, Yamaguchi S, Yasui A, Muijtjens M, Hoeijmakers JH, van der Horst GT. Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science. 1999 Dec 24;286(5449):2531–4. doi: 10.1126/science.286.5449.2531. [DOI] [PubMed] [Google Scholar]
- 7.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002 Aug 29;418(6901):935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 8.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999 Apr 15;398(6728):627–30. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 9.Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001 May;30(2):525–36. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 10.Dudley CA, Erbel-Sieler C, Estill SJ, Reick M, Franken P, Pitts S, McKnight SL. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science. 2003 Jul 18;301(5631):379–83. doi: 10.1126/science.1082795. Epub 2003 Jul 3. [DOI] [PubMed] [Google Scholar]
- 11.DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007 May;10(5):543–5. doi: 10.1038/nn1884. Epub 2007 Apr 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi S, Hida A, McGuinness OP, Wasserman DH, Yamazaki S, Johnson CH. Circadian clock gene Bmal1 is not essential; functional replacement with its paralog, Bmal2. Curr Biol. 2010 Feb 23;20(4):316–21. doi: 10.1016/j.cub.2009.12.034. Epub 2010 Feb 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gekakis N, Saez L, Delahaye-Brown AM, Myers MP, Sehgal A, Young MW, Weitz CJ. Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science. 1995 Nov 3;270(5237):811–5. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 14.Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A. 1998 May 12;95(10):5474–9. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999 Jul 23;98(2):193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 16.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001 Dec 28;107(7):855–67. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 17.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000 May 12;288(5468):1013–9. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 18.Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006 Mar;38(3):312–9. doi: 10.1038/ng1745. Epub 2006 Feb 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akashi M, Tsuchiya Y, Yoshino T, Nishida E. Control of intracellular dynamics of mammalian period proteins by casein kinase I epsilon (CKIepsilon) and CKIdelta in cultured cells. Mol Cell Biol. 2002 Mar;22(6):1693–703. doi: 10.1128/MCB.22.6.1693-1703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eide EJ, Vielhaber EL, Hinz WA, Virshup DM. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon. J Biol Chem. 2002 May 10;277(19):17248–54. doi: 10.1074/jbc.M111466200. Epub 2002 Mar 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005 Apr;25(7):2795–807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salzberg HC. Group psychotherapy screening scale: a validation study. Int J Group Psychother. 1969 Apr;19(2):226–8. doi: 10.1080/00207284.1969.11507784. [DOI] [PubMed] [Google Scholar]
- 23.Shirogane T, Jin J, Ang XL, Harper JW. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005 Jul 22;280(29):26863–72. doi: 10.1074/jbc.M502862200. Epub 2005 May 24. [DOI] [PubMed] [Google Scholar]
- 24.Reischl S, Vanselow K, Westermark PO, Thierfelder N, Maier B, Herzel H, Kramer A. Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms. 2007 Oct;22(5):375–86. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- 25.Ohsaki K, Oishi K, Kozono Y, Nakayama K, Nakayama KI, Ishida N. The role of {beta}-TrCP1 and {beta}-TrCP2 in circadian rhythm generation by mediating degradation of clock protein PER2. J Biochem. 2008 Nov;144(5):609–18. doi: 10.1093/jb/mvn112. Epub 2008 Sep 8. [DOI] [PubMed] [Google Scholar]
- 26.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002 Jul 26;110(2):251–60. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 27.Triqueneaux G, Thenot S, Kakizawa T, Antoch MP, Safi R, Takahashi JS, Delaunay F, Laudet V. The orphan receptor Rev-erbalpha gene is a target of the circadian clock pacemaker. J MolEndocrinol. 2004 Dec;33(3):585–608. doi: 10.1677/jme.1.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004 Aug 19;43(4):527–37. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 29.Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005 May;12(5):441–8. doi: 10.1038/nsmb925. Epub 2005 Apr 10. [DOI] [PubMed] [Google Scholar]
- 30.Guillaumond F, Dardente H, Giguere V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005 Oct;20(5):391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 31.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005 Feb;37(2):187–92. doi: 10.1038/ng1504. Epub 2005 Jan 23. [DOI] [PubMed] [Google Scholar]
- 32.Baggs JE, Price TS, DiTacchio L, Panda S, Fitzgerald GA, Hogenesch JB. Network features of the mammalian circadian clock. PLoS Biol. 2009 Mar 10;7(3):e52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kafri R, Bar-Even A, Pilpel Y. Transcription control reprogramming in genetic backup circuits. Nat Genet. 2005 Mar;37(3):295–9. doi: 10.1038/ng1523. Epub 2005 Feb 20. [DOI] [PubMed] [Google Scholar]
- 34.Kafri R, Levy M, Pilpel Y. The regulatory utilization of genetic redundancy through responsive backup circuits. Proc Natl Acad Sci U S A. 2006 Aug 1;103(31):11653–8. doi: 10.1073/pnas.0604883103. Epub 2006 Jul 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to upregulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992 Oct 30;71(3):383–90. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 36.Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993 Dec 31;75(7):1351–9. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 37.Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch JB, Takahashi JS. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007 Feb 27;104(9):3342–7. doi: 10.1073/pnas.0611724104. Epub 2007 Feb 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yokota S, Yamamoto M, Moriya T, Akiyama M, Fukunaga K, Miyamoto E, Shibata S. Involvement of calcium-calmodulin protein kinase but not mitogen-activated protein kinase in light-induced phase delays and Per gene expression in the suprachiasmatic nucleus of the hamster. J Neurochem. 2001 Apr;77(2):618–27. doi: 10.1046/j.1471-4159.2001.00270.x. [DOI] [PubMed] [Google Scholar]
- 39.Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A. 2002 May 28;99(11):7728–33. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science. 2001 Jul 20;293(5529):506–9. doi: 10.1126/science.1060699. Epub 2001 Jul 5. [DOI] [PubMed] [Google Scholar]
- 41.Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Circadian rhythms in isolated brain regions. J Neurosci. 2002 Jan 1;22(1):350–6. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng HY, Alvarez-Saavedra M, Dziema H, Choi YS, Li A, Obrietan K. Segregation of expression of mPeriod gene homologs in neurons and glia: possible divergent roles of mPeriod1 and mPeriod2 in the brain. Hum Mol Genet. 2009 Aug 15;18(16):3110–24. doi: 10.1093/hmg/ddp252. Epub 2009 May 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994 Apr 29;264(5159):719–25. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007 May 11;316(5826):900–4. doi: 10.1126/science.1141194. Epub 2007 Apr 26. [DOI] [PubMed] [Google Scholar]
- 45.Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, O’neill J, Chesham JE, Brooker D, Lalanne Z, Hastings MH, Nolan PM. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007 May 11;316(5826):897–900. doi: 10.1126/science.1141138. Epub 2007 Apr 26. [DOI] [PubMed] [Google Scholar]
- 46.Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007 Jun 1;129(5):1011–23. doi: 10.1016/j.cell.2007.04.030. Epub 2007 Apr 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakamura W, Honma S, Shirakawa T, Honma K. Clock mutation lengthens the circadian period without damping rhythms in individual SCN neurons. Nat Neurosci. 2002 May;5(5):399–400. doi: 10.1038/nn843. [DOI] [PubMed] [Google Scholar]
- 48.Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ, 3rd, Takahashi JS, Kay SA. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007 May 4;129(3):605–16. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Low-Zeddies SS, Takahashi JS. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell. 2001 Apr 6;105(1):25–42. doi: 10.1016/s0092-8674(01)00294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008 Feb 29;4(2):e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeBruyne JP, Weaver DR, Reppert SM. Peripheral circadian oscillators require CLOCK. Curr Biol. 2007 Jul 17;17(14):R538–9. doi: 10.1016/j.cub.2007.05.067. [DOI] [PubMed] [Google Scholar]
- 52.Webb AB, Angelo N, Huettner JE, Herzog ED. Intrinsic, nondeterministic circadian rhythm generation in identified mammalian neurons. Proc Natl Acad Sci U S A. 2009 Sep 22;106(38):16493–8. doi: 10.1073/pnas.0902768106. Epub 2009 Sep 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herzog ED, Aton SJ, Numano R, Sakaki Y, Tei H. Temporal precision in the mammalian circadian system: a reliable clock from less reliable neurons. J Biol Rhythms. 2004 Feb;19(1):35–46. doi: 10.1177/0748730403260776. [DOI] [PubMed] [Google Scholar]
- 54.Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003 Nov 21;302(5649):1408–12. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- 55.Taylor AL, Goaillard JM, Marder E. How multiple conductances determine electrophysiological properties in a multicompartment model. J Neurosci. 2009 Apr 29;29(17):5573–86. doi: 10.1523/JNEUROSCI.4438-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004 Dec 29;14(24):2289–95. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amdaoud M, Vallade M, Weiss-Schaber C, Mihalcescu I. Cyanobacterial clock, a stable phase oscillator with negligible intercellular coupling. Proc Natl Acad Sci U S A. 2007 Apr 24;104(17):7051–6. doi: 10.1073/pnas.0609315104. Epub 2007 Apr 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001 Oct 19;916(1–2):172–91. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- 59.Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005 Apr;8(4):476–83. doi: 10.1038/nn1419. Epub 2005 Mar 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X, Waschek JA. Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am J Physiol Regul Integr Comp Physiol. 2003 Nov;285(5):R939–49. doi: 10.1152/ajpregu.00200.2003. Epub 2003 Jul 10. [DOI] [PubMed] [Google Scholar]
- 61.Brown TM, Colwell CS, Waschek JA, Piggins HD. Disrupted neuronal activity rhythms in the suprachiasmatic nuclei of vasoactive intestinal polypeptide-deficient mice. J Neurophysiol. 2007 Mar;97(3):2553–8. doi: 10.1152/jn.01206.2006. Epub 2006 Dec 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herzog ED, Huckfeldt RM. Circadian entrainment to temperature, but not light, in the isolated suprachiasmatic nucleus. J Neurophysiol. 2003 Aug;90(2):763–70. doi: 10.1152/jn.00129.2003. Epub 2003 Mar 26. [DOI] [PubMed] [Google Scholar]
- 63.Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol. 2002 Sep 17;12(18):1574–83. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- 64.Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000 Sep 29;289(5488):2344–7. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 65.Abraham U, Granada AE, Westermark PO, Heine M, Kramer A, Herzel H. Coupling governs entrainment range of circadian clocks. MolSyst Biol. 2010 Nov 30;6:438. doi: 10.1038/msb.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rea MA. VIP-stimulated cyclic AMP accumulation in the suprachiasmatic hypothalamus. Brain Res Bull. 1990 Dec;25(6):843–7. doi: 10.1016/0361-9230(90)90179-4. [DOI] [PubMed] [Google Scholar]
- 67.Vanecek J, Watanabe K. Melatonin inhibits the increase of cyclic AMP in rat suprachiasmatic neurons induced by vasoactive intestinal peptide. Neurosci Lett. 1998 Aug 7;252(1):21–4. doi: 10.1016/s0304-3940(98)00530-8. [DOI] [PubMed] [Google Scholar]
- 68.Meyer-Spasche A, Piggins HD. Vasoactive intestinal polypeptide phase-advances the rat suprachiasmatic nuclei circadian pacemaker in vitro via protein kinase A and mitogen-activated protein kinase. Neurosci Lett. 2004 Mar 25;358(2):91–4. doi: 10.1016/j.neulet.2003.12.114. [DOI] [PubMed] [Google Scholar]
- 69.Nielsen HS, Hannibal J, Fahrenkrug J. Vasoactive intestinal polypeptide induces per1 and per2 gene expression in the rat suprachiasmatic nucleus late at night. Eur J Neurosci. 2002 Feb;15(3):570–4. doi: 10.1046/j.0953-816x.2001.01882.x. [DOI] [PubMed] [Google Scholar]
- 70.Ko CH, Yamada YR, Welsh DK, Buhr ED, Liu AC, Zhang EE, Ralph MR, Kay SA, Forger DB, Takahashi JS. Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol. 2010 Oct 12;8(10):e1000513. doi: 10.1371/journal.pbio.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nishide SY, Honma S, Honma K. The circadian pacemaker in the cultured suprachiasmatic nucleus from pup mice is highly sensitive to external perturbation. Eur J Neurosci. 2008 May;27(10):2686–90. doi: 10.1111/j.1460-9568.2008.06231.x. Epub 2008 May 29. [DOI] [PubMed] [Google Scholar]
- 72.Lu W, Meng QJ, Tyler NJ, Stokkan KA, Loudon AS. A circadian clock is not required in an arctic mammal. Curr Biol. 2010 Mar 23;20(6):533–7. doi: 10.1016/j.cub.2010.01.042. Epub 2010 Mar 11. [DOI] [PubMed] [Google Scholar]
- 73.Ciarleglio CM, Axley JC, Strauss BR, Gamble KL, McMahon DG. Perinatal photoperiod imprints the circadian clock. Nat Neurosci. 2011 Jan;14(1):25–7. doi: 10.1038/nn.2699. Epub 2010 Dec 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009 May 1;324(5927):651–4. doi: 10.1126/science.1171641. Epub 2009 Mar 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009 May 1;324(5927):654–7. doi: 10.1126/science.1170803. Epub 2009 Mar 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008 May 16;320(5878):949–53. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]