Abstract
Summary: The relationship between genes and proteins is a dynamic relationship that changes across time and differs in different cells. The study of these differences can reveal various insights into biological processes and disease progression, especially with the aid of proper tools for network visualization. Toward this purpose, we have developed TVNViewer, a novel visualization tool, which is specifically designed to aid in the exploration and analysis of dynamic networks.
Availability: TVNViewer is freely available with documentation and tutorials on the web at http://sailing.cs.cmu.edu/tvnviewer.
Contact: epxing@cs.cmu.edu
1 INTRODUCTION
Biological relationships, such as those between proteins, between transcription factors and DNA binding sites, and between cells, differ across different tissues and change over time. Recent studies have shown that changes in network architecture from the cell cycle and in response to diverse stimuli are quite significant (Luscombe et al., 2004). Despite the dynamic nature of these interactions, the general scientific paradigm has often been to consider these relationships as static entities. However, many recent studies have advanced the field through insight from network dynamics across time or tissue in a variety of species including: human blood leukocyte response (Calvano et al., 2005), rice regulatory hierarchies of gene expression (Jiao et al., 2009), temporal interaction in Caenorhabditis elegans (Dupuy et al., 2007) and correlated changes in gene expression between mouse tissues (Keller et al., 2008). The study of network dynamics has the potential to produce crucial discoveries in gene regulation, cell cycle, and cancer progression.
Representing biological relationships visually through networks is considered the best way to demonstrate the interplay between different genes or proteins. Network representations of such data range from the small and simple networks to large, complex networks that represent thousands of genes. The potential for network visualization to aid in our fundamental understanding of biological relationships has resulted in an explosion of software platforms and visualization toolkits (Pavlopoulos et al., 2008; Suderman and Hallett, 2007). These software applications include popular tools such as Cytoscape (Shannon et al., 2003), Osprey (Breitkreutz et al., 2003), VisANT (Hu et al., 2009), Graphle (Huttenhower et al., 2009) and many more. While some tools are specific to biology, others such as Medusa (Hooper and Bork, 2005) are general purpose and can be customized to other domains. Each network visualization tool approaches the problem from a different angle; advantages and disadvantages between them are varied. However, despite the complex array of software available, there remains the unmet need for software to explore dynamic networks (Pavlopoulos et al., 2008; Suderman and Hallett, 2007). To explore dynamic networks, researchers must awkwardly create a series of static networks and manually inspect them to identify rewiring behavior.
We have developed TVNViewer, a web site built for the exploration and analysis of networks that change over time and space. Both circle and force-directed layouts are available for users to scroll through a series of networks in real time, providing a natural way to quickly identify network rewiring. Additionally, TVNViewer gives users access to analysis tools describing how the degree of the nodes and descriptors in the network change over time. Although TVNViewer was developed with biological interactions in mind, it can be used for any type of dynamic network.
2 TVNVIEWER OVERVIEW
TVNViewer is built for the visualization of small to moderate datasets of up to 500 nodes for gene–gene interactions, or if genes are grouped into a descriptor category (such as a GO category annotation), TVNViewer can handle up to 5000 nodes classified by up to 100 descriptors. TVNViewer accepts a series of up to 50 networks in text or xml format; detailed instructions on how to prepare files are available through TVNViewer's import wizard. TVNViewer supports three visualization paradigms: a gene–gene interaction paradigm, a two-tiered gene–gene interaction paradigm and a descriptor paradigm where nodes are grouped according to a descriptor category. Each visualization paradigm provides a slightly different way to explore the network; examples of each paradigm are available by running the examples on the TVNViewer web site. TVNViewer can be used through a temporary session to upload data, or users also have the option to create a free user account on the server in order to save data for future access.
3 TVNVIEWER VISUALIZATION TOOLS
There are many different views and controls available to users through TVNViewer, all of which are documented online. Additionally, we have provided online video tutorials. Here, we outline a few of the views and analysis tools available.
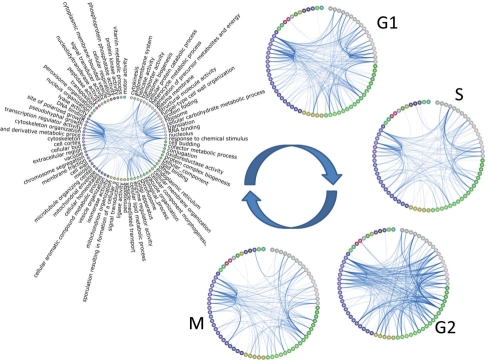
One way that users can use TVNViewer to see the dynamic rewiring of their networks is directly through the circle view. The circle view automatically clusters nodes according to connectivity, reducing the number of cross-circle connections to enhance the visualization's interpretability. Nodes are colored according to this clustering to give a natural representation of subgraphs that may exist. From this initial time point, users can scroll through the different graphs in the series, quickly moving through the networks to observe the rewiring taking place. The user has complete control over the node size, edge thickness, font size, presence of the node labels and the number of edges displayed in the graph. These can be adjusted according to the user's visualization preference. TVNViewer provides users with a direct link to download a .pdf or .png file of any view directly to their desktop, enabling the production of figures such as Figure 1.
Fig. 1.
TVNViewer dynamic network visualization. This figure was created from TVNViewer's visualization tools. A simulated dataset is shown at five stages with distinct differences in the relationships between nodes. In the TVNViewer, the user can use these types of visualizations to find and highlight how the network changes.
In addition to the circle view, the user can also explore the series of networks in a force-directed layout view. Again, the users can scroll through each time point in the series to observe the dynamics happening in their network series. The force-directed layout can also be stopped at any time and the nodes can be moved around in a user preference manner. The user can allow the network to update while scrolling through the time-series to see how the network structure is affected by the rewiring, or they can fix the location of the nodes and view only how the edges between nodes change.
In addition to exploring the network visualization directly, we provide two sets of analysis tools. These tools emphasize how the gene degree of different nodes changes over time. Users can examine how the overall degree distribution of the network changes over time. The other set of analysis tools includes two different views (a linear plot and a stacked plot) that users can use to explore how the degree of certain nodes or descriptors changes over time. Such a view may be useful to identify certain genes that may act as a transcription factor at some stages of development, but which fall back to other roles during other times.
In conclusion, TVNViewer provides a visualization framework for creating fine-tuned figures that highlight how networks change over time or space. Additionally, users can use the web site to explore the dynamics of the network and also to analyze how the degree of different nodes in the network changes over time. Examples and further documentation are available from the TVNViewer web site: http://sailing.cs.cmu.edu/tvnviewer.
4 IMPLEMENTATION NOTES
The TVNViewer web site is implemented using Adobe ActionScript, Apache and MySQL; TVNViewer runs on all major browsers with the Adobe Flash plug-in. TVNViewer's ActionScript code is built off of the Flare library, an open-source web visualization project (flare.prefuse.org). Thus, TVNViewer takes advantage of a recent, powerful visualization library with real-time animation and dynamically updated views.
ACKNOWLEDGEMENTS
We would like to recognize Ankur Parik and Dr Wei Wu for their helpful comments throughout the development of TVNViewer.
Funding: National Institutes of Health (1R01GM087694, 1R01GM093156, 0015699); National Science Foundation (DBI-0640543, IIS-0713379); Defense Advanced Research Projects Agency (Z931302); Alfred P. Sloan Research Fellowship.
Conflict of Interest: None declared.
REFERENCES
- Breitkreutz B.J, et al. Osprey: a network visualization system. Genome Biol. 2003;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvano S.E, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- Dupuy D., et al. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat. Biotechnol. 2007;25:663–668. doi: 10.1038/nbt1305. [DOI] [PubMed] [Google Scholar]
- Hooper S.D., Bork P. Medusa: a simple tool for interaction graph analysis. Bioinformatics. 2005;21:4432–4433. doi: 10.1093/bioinformatics/bti696. [DOI] [PubMed] [Google Scholar]
- Hu Z., et al. VisANT 3.5: multi-scale network visualization, analysis, and inference based on the gene ontology. Nucleic Acids Res. 2009;37:W115–W121. doi: 10.1093/nar/gkp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C., et al. Graphle: Interactive exploration of large, dense graphs. BMC Bioinformatics. 2009;10:417. doi: 10.1186/1471-2105-10-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., et al. A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat. Genet. 2009;41:258–263. doi: 10.1038/ng.282. [DOI] [PubMed] [Google Scholar]
- Keller M.P, et al. A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res. 2008;18:706–716. doi: 10.1101/gr.074914.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe N.M, et al. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos G.A, et al. A survey of visualization tools for biological network analysis. BioData Min. 2008;1:12. doi: 10.1186/1756-0381-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suderman M., Hallett M. Tools for visually exploring biological networks. Bioinformatics. 2007;23:2651–2659. doi: 10.1093/bioinformatics/btm401. [DOI] [PubMed] [Google Scholar]