Abstract
Motivation: Deciphering the complex mechanisms by which regulatory networks control gene expression remains a major challenge. While some studies infer regulation from dependencies between the expression levels of putative regulators and their targets, others focus on measured physical interactions.
Results: Here, we present Physical Module Networks, a unified framework that combines a Bayesian model describing modules of co-expressed genes and their shared regulation programs, and a physical interaction graph, describing the protein–protein interactions and protein-DNA binding events that coherently underlie this regulation. Using synthetic data, we demonstrate that a Physical Module Network model has similar recall and improved precision compared to a simple Module Network, as it omits many false positive regulators. Finally, we show the power of Physical Module Networks to reconstruct meaningful regulatory pathways in the genetically perturbed yeast and during the yeast cell cycle, as well as during the response of primary epithelial human cells to infection with H1N1 influenza.
Availability: The PMN software is available, free for academic use at http://www.compbio.cs.huji.ac.il/PMN/.
Contact: aregev@broad.mit.edu; nirf@cs.huji.ac.il
1 INTRODUCTION
Transcription regulation plays a major role in controlling gene expression and cell function. Despite intensive research, the topology and function of most regulatory circuits remain largely unknown. The increasing availability of large-scale datasets, such as genomics sequence, gene expression profiles and protein–DNA or protein–protein interaction data, provides an opportunity to automatically infer regulatory circuits on a genome-wide scale.
Three main types of approaches have been used to infer regulatory models from genomic data (Kim et al., 2009), each suffering from substantial limitations. Observational models, including Bayesian networks (Friedman, 2004) and their extensions (e.g.Hartemink et al., 2002; Segal et al., 2003; Zou et al., 2005), rely on dependencies between the expression profiles of regulators to those of their target genes. These can handle abundant expression data, but often fail to distinguish true regulation from co-expression (Amit et al., 2009; Kim et al., 2009; Segal et al., 2003;). Perturbational models associate targets to factors based on the effect of the factors' genetic manipulation on gene expression (Amit et al., 2009; Capaldi et al., 2008; Hu et al., 2007;). These identify functional effects, but may fail to correctly distinguish direct from indirect targets (Wagner et al., 2001). Finally, physical models associate regulatory factors with the targets whose promoters they bind (Breitkreutz et al., 2010; Harbison et al., 2004; Lee et al., 2002), or where a cis-regulatory site is present e.g.(Suzuki et al., 2009). These identify molecular targets, but some of those may be functionally silent (Capaldi et al., 2008).
Thus, a major challenge is to build a realistic, molecular and functional model of gene regulation that combines changes in gene expression with the underlying physical interactions. Previous attempts toward this goal have mostly introduced new hypotheses only at one level, while using the other level to support them. These include works that detect functional modules by integrating phenotypic and physical data (Ideker et al., 2002; Nariai et al., 2004; Peleg et al., 2010) or that reconstruct or annotate signaling pathways or binding events (Gao et al., 2004; Ourfali et al., 2007; Yeang et al., 2004; Yeger-Lotem et al., 2009). A notable exception (Kundaje et al., 2008) integrated binding, sequence and expression information in a Bayesian framework to identify both clusters of genes and their transcriptional regulators. However, this work did not consider upstream signaling pathways.
Here, we present Physical Module Networks (PMN), a novel probabilistic graphical method that learns transcriptional networks by combining gene expression profiles, protein–protein and protein-DNA binding data. The PMN model, based on Module Networks (Segal et al., 2003), discovers modules of co-expressed genes, sets of regulators that control their activity, and a path of physical interactions that connects the regulators to their target module (Fig. 1). The modules, regulators and paths are inferred simultaneously, resulting in the most probable physical model of gene regulation that underlies the observed data.
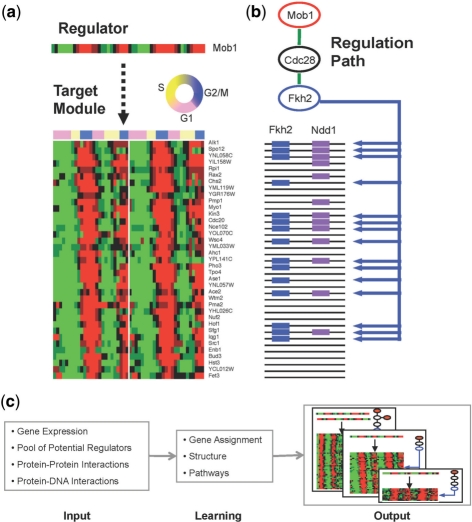
Fig. 1.
PMNs physical module. (a) Expression pattern of the Mob1 regulator (top) and its 37 target genes (bottom), during 2 cell cycles (two replicates are shown). (b) A physical pathway connecting Mob1 via Cdc28 to the transcription factor FKH2 that binds 15 of the module genes. (c) Learning procedure. Input: gene expression, a set of potential regulators and physical protein–protein and protein–DNA interactions. PMN Learner: an iterative optimization procedure that finds modules, their regulators and the physical pathways that explain the regulation. Output: the best configuration found by the learner.
Using synthetic data, we show that the addition of physical interactions to the simple Module Network (MN) model improves the model's precision, without compromising recall. We evaluate the biological power of the model in two yeast systems (gene perturbations and cell cycle), and a human dataset (response of epithelial cells to flu infection). In each case, the learned modules and pathways are biologically sound, and lead to novel insights, emphasizing the power of integrated probabilistic models.
2 THE PMN MODEL
A PMN (Fig. 1) consists of two components: an MN representing the relation in expression between a regulator and its targets and a Physical Interaction Graph providing a path of physical interactions between them.
2.1 Module Networks
An MN model (Segal et al., 2003) represents regulatory dependencies in expression profiles between genes. It relies on two major assumptions. First, it assumes that the expression level of a target gene can be predicted from the expression level of one or more regulators. Second, it assumes that genes are organized into modules, where all members of a module are regulated by the same factors and in the same way. For example (Fig. 1a), the expression values of all the genes in a G2/M module can be predicted by the level of the same regulator (Mob1), in the same way. In some cases, multiple regulators act combinatorially. This may be modeled in different ways, including a decision tree (Segal et al., 2003, 2005), linear regression (Lee et al., 2006), etc.
Formally, a Module Network is a Bayesian network which is defined over gene groups (modules) rather than over individual genes (Fig. 2a, Segal et al., 2005). Briefly, an MN model ℳ consists of three components. The first is the partition of all genes G in the domain into a set of modules 𝒞={M1,…, Mn}. The second is a structure 𝒮 that assigns for each module Mj a set of parents PaMj⊂G, which we term regulators of the genes in Mj. This structure induces a graph 𝒢ℳ with 𝒞 as vertices and edges {Mj→Mk:Mj∩PaMk≠∅}. A legal MN has induced an acyclic graph 𝒢ℳ. The third component is a set of template conditional probabilitiess P(Mj∣PaMj), each of which specifies a distribution over the values of a gene for each value assignment Val(PaMj) to the parent set. Together these components provide a concise description of the joint distribution over all genes in G, such that each gene depends on the parents of the module it belongs to; see (Segal et al., 2005).
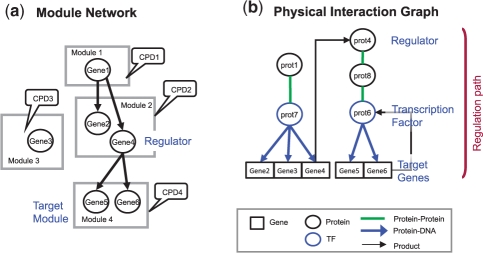
Fig. 2.
Consistency toy example. (a) MN over 4 modules. Gene 1 is the regulator of Module 2, Gene 4 is the regulator of Module 4. (b) Physical pathways connecting the regulators to the target genes. The pathways start from the regulator proteins (circles) and connect them via transcription factors to their target genes (rectangles).
2.2 Physical Interaction Graph
A Physical Interaction Graph (Figs 1b and 2b) describes possible interactions between proteins and genes. Formally, an interaction graph ℐ is a graph over a set of genes and proteins with three types of edges: protein–protein interactions (an undirected edge between two proteins), protein–DNA interactions (directed edge from a protein to a gene), and transcription interactions (directed edge from a gene to its protein product). Thus, each gene in the domain is represented by two nodes, one for the gene and one for its protein product. Protein nodes corresponding to transcription factors are marked according to prior biological knowledge. The interaction graph may contain genes whose expression was not measured (and thus do not appear in the MN).
2.3 Regulatory Paths
An MN ℳ and a Physical Interaction Graph ℐ are consistent, if we can explain how the state of the regulators in the MN reaches the target genes through physical interactions. More precisely, for each pair of regulator Xi and target module Mj in ℳ, there should be a consistent physical Regulation Path from Xi to Mj. A Regulation Path (Fig. 2b) is a sequence of nodes 〈v1,…, vn〉 in ℐ, where v1 is a protein node of the protein product of Xi and vn is a transcription factor (TF) that binds all the genes in Mj. If the protein v1 is a transcription factor, the path can be the trivial one, of length zero. The regulation path is partially directed, such that the edge between vℓ and vℓ+1 is either undirected or directed from vℓ to vℓ+1.
A PMN 𝒫=〈ℳ,ℐ〉 model has a consistent configuration if each regulator in the Module Network has a regulation path from its protein to the genes in its target module (Fig. 2).
3 LEARNING A PMN
Given input data, our goal is to find the configuration of the model—modules, regulators and regulation paths—that best describes the data. The input consists of (1) gene expression measurements, 𝒟X, (2) a pool of potential (‘candidate’) regulators R⊆G from which the algorithm can choose, and (3) protein–protein and protein–DNA interaction measurements (𝒟I), associated with P-values that represent their confidence. Following the literature on graphical models (Koller and Friedman, 2009), we pose the learning problem as a discrete optimization problem. We define a score function that captures how well a model decribes the observed data, and then search for the model that maximizes the score.
3.1 Model score
Formally, let 𝒫=〈ℳ,ℐ〉 be a PMN model which consists of an MN ℳ and a Physical Interaction Graph ℐ. We score how well a given model describes the data 𝒟=〈𝒟X, 𝒟I〉 using a Bayesian Score, which is derived from the posterior probability of the model. According to Bayes rule, we know that
where P(𝒫) is the prior of the model, P(𝒟|𝒫) is the likelihood of the model given the data. Thus, we can define a score
Note that we are ignoring the normalization constant, since it depends only on the data and thus does not change the relative score of different models.
The score must fulfill two major requirements. First, it must be decomposable into the two model components, allowing us to calculate the score of one component while the other is fixed, an important attribute for efficient learning. Second, we must ensure model consistency, by a high penalty on inconsistent configurations, such that they will not be chosen during the learning.
Under the following two constructions, the Bayesian score satisfies both decomposability and consistency. First, the prior has the following structure:
where C(ℳ,ℐ)=1 if ℳ and ℐ describe a consistent configuration, and 0 otherwise. Although this requirement limits the class of priors we can use, it ensures our model is both consistent and enables efficient learning. Second, we require that the likelihood of each component be independent from the other, given the model. That is,
This is a reasonable assumption since the gene expression and physical interaction observations are derived from independent sources.
If these assumptions are satisfied, the Bayesian score can be further developed for consistent pairs 〈ℳ,ℐ〉:
For inconsistent pairs the score is −∞.
The MN score Score(ℳ:𝒟X) is described by (Segal et al., 2005). Briefly, under several assumptions, it is decomposed to the score of the gene assignment and the score of the network structure, allowing efficient learning of each aspect, while the other is fixed.
The new contribution of our work is the Physical Interaction Graph score, Score(ℐ:𝒟I). This score is constructed from local scores of the individual edges in the graph. We make the simplifying assumption that the observations about edges of the interaction graph are independent of each other, given the underlying interaction network. This assumption is reasonable since the experimental measurement of each pair of edges is independent of the other pairs. Thus, each edge, and the evidence supporting it, contribute to the score independently of other edges in the graph.
Formally, we define E to be the set of edges that can appear in ℐ: all undirected edges between two protein nodes, and directed edges from a protein (TF) node to a gene node. (The transcription edge from a gene to its protein is assumed as given.) Let e be a (potential) edge in E, we define ℐe to be the indicator random variable with value 1 if e is in ℐ and 0 otherwise. The set of variables {ℐe:e∈E} defines uniquely the graph ℐ. We represent the evidence from various interaction screens about an edge e in 𝒟I as another indicator random variable de, such that de=1 if e was observed in 𝒟I and 0 otherwise.
The independence assumption implies that
where P(ℐe) is a prior belief in the edge e, and it can be set to be uniform, or to reflect other external biological knowledge, such as the quality of the experimental assay. and P(de|ℐe) is the probability to observe e given ℐe.
This definition of the score seems to involve all edges in E, which can be a very large number. This would be in contrast to the typical assumption that real interaction graphs are sparse. To avoid this problem, we rewrite the score as
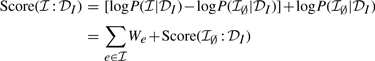 |
where Score(ℐ∅:𝒟I) is the score of the empty graph and
is the likelihood cost of adding e to the graph. Since Score(ℐ∅:𝒟I) is constant, we can ignore it when evaluating different ℐ. The remaining terms involve only the weights of the edges present in ℐ. We note that we choose a uniform prior P(ℐe)=0.001 so as to ensure that We<0 for all edges. It means that adding an edge always has a cost, and thus penalizing long pathways.
Finally, we choose the probability of the observation function. There is no clear mechanistic argument for a specific function, and for mathematical convenience we choose to use the following function
where ω=0.9 is a constant estimated from the data and pe is the P-value of e. That is, if the edge is not in the graph we assume that the probability to observe it in the data is its P-value. If the edge is in the graph, the probability to observe it has an exponential distribution which is dependent on its P-value. Note that under these assumptions, an edge which is not observed in the data may still be included in the graph, associated with a low prior probability.
3.2 Model optimization
The MN learning procedure (Segal et al., 2005) performs a coordinate-wise greedy optimization (Koller and Friedman, 2009), iterating between two optimization procedures. The first procedure keeps the partition of genes into modules fixed, and improves the score by modifyng the regulatory programs. This involves steps of adding and removing regulators to modules. The second procedure keeps the regulatory program fixed, and improves the score by moving genes from one module to another. Given the properties of the Bayesian score, each procedure can be implemented as a greedy hill-climbing search with local operations that can be evaluated relatively easily.
In learning PMNs, we maintain the same overall architecture, with an important distinction: at the same time that a change in ℳ is evaluated, we also need to change ℐ to maintain consistency, and determine what is the score of these changes. Thus, each local step in the MN learning procedure is accompanied by a step on the Physical Interaction Graph, and the impact of the proposed step on the score is the sum of these two components.
Specifically, when adding an edge from a regulator R to a module M in ℳ, we must ensure that there is a consistent regulatory path. If there is none, then we consider the addition of protein–DNA edges that will introduce at least one such path. If after such inclusion there is still no path, the step will not be taken. Recall that a regulatory path is a single path from the regulator to a transcription factor, and then branches into edges from the transcription factor to each of the genes in the module. Considering each transcription factor T seperately, the procedure searches for the heaviest path (using the weights We defined above) from R to T using the Bellman–Ford algorithm (Bellman, 1958). (Since all the weights are negative, finding the heaviest path is equivalent to finding the shortest path by Bellman–Ford.) It then evaluates the cost of edges from the TF to the genes in M. The total score for T is the sum of these two terms. The transcription factor T that maximizes the score is chosen, and the associated edges are added to ℐ.
When removing an edge from R to M, we may remove some edges in ℐ. The algorithm examines edges on the path from R to M, and then considers which ones can be removed, while still maintaining consistency of all other regulatory paths. This is done with all edges until no more edges can be removed.
When reassigning a gene from one module to another, a set of TF to gene edges has to be removed and others added. These depend on the regulatory paths choosen for each module.
Thus, the resulting procedure is essentially isomorphic to the MN procedure except for the additional book-keeping for maintaining changes to ℐ that accompany each change in ℳ. The initialization of the procedure is with an initial clustering of genes, and the procedure stops upon convergence.
4 EVALUATION WITH SYNTHETIC DATA
We first compared the performance of PMN on synthetic data to that of a standard MNs model, examining its robustness to noise and comparing its precision and recall in recovering regulatory interactions. We reasoned that in this comparison we can most directly evaluate the specific impact of the additional information (physical data) and constraints (e.g. consistency) in a PMN.
We generated a synthetic network over 312 genes, partitioned to 7 modules that are regulated by 10 genes (out of the 312 genes). The conditional probability distributions (CPDs) are taken from a Bayesian network test case (Beinlich et al., 1989). We sampled physical interaction observations from a set of real interactions (Batada et al., 2006; Herrgard et al., 2006; MacIsaac et al., 2006), covering 43% of the original network. The distributions of node degrees and of edge weights were similar to that of the original network. We built the synthetic Physical Interaction Graph by choosing 7 TF proteins as the ‘true’ TFs, and finding pathways that connect them to the regulators. (Data available online at http://www.compbio.cs.huji.ac.il/PMN/.)
We evaluated the models by three criteria: (i) the likelihood of a previously unseen set of gene expression samples (test set); (ii) the ability to choose the correct regulators for each gene (reconstruction); and (iii) the PMN's ability to reconstruct the correct regulation pathway by counting all the interactions that were selected for the model (this criteria is not applicable for MN). The latter two criteria were measured by recall (True Positive/(True Positive + False Negative)) and precision (True Positive/(True Positive + False Positive)).
We learned the MN model from 200 gene expression samples and used 10-fold cross validation to estimate its train and test scores. We learned the PMN model from the same train and test sample sets, in addition to a set of physical interaction observations. To simulate noise which is abundant in expression data, we generated data from different levels of smoothed distribution. We smoothed the distribution to degree α, by transforming each CPD, replacing each term P(X=xi|Pax) by a smoothed version
where we normalize the entries to sum to 1.
We find that the likelihood of a test set given the learned PMN is almost identical to the likelihood given the MN (Fig. 3a), indicating that both models have similar predictive power. Thus, the additional constraints in a PMN (by consistency requirements) do not compromise its predictive power.
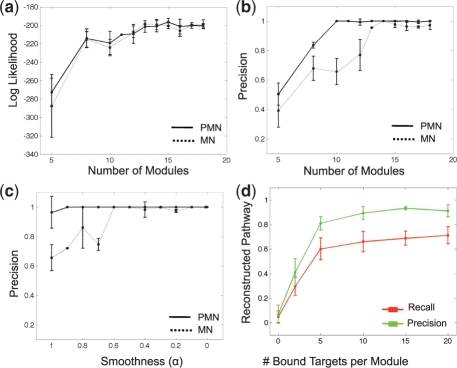
Fig. 3.
Performance on synthetic data. (a) Log likelihood of test samples, achieved by PMN (solid line) and MN (dashed line) as a function of module number. Plots show average over 10-fold cross validation experiments; error bars show 2 STD. (b) Precision rate of reconstructing regulator-target pairs, achieved by PMN and MN, as a function of number of modules. Plots as in (a). (c) Precision rate of reconstructing regulator-target pairs, achieved by PMN and MN, as a function of smoothness of the expression data. Plots as in (a). (d) Reconstructed pathways as a function of noise in the protein–DNA data. Plots show average precision and recall over 10-fold cross validation; error bars indicate 2 STD.
While both models have good recall, the PMN has substantially higher precision. Recall ranges between 80% and 100%, when learning with a sufficient number of modules. The PMN, however, chooses fewer false regulatory relations than the MN, resulting in a higher precision (Fig. 3b). This is likely due to the additional constraints on the choice of regulators in a PMN, which must be supported by both expression and physical interactions. Notably, when smoothness is introduced to the expression data, the performance of MN and PMN is similar in terms of test set likelihood, recall and precision (data not shown). With strong smoothing the observation values are nearly uniformly distributed, and the MN recall is higher compared to the PMN (data not shown). In this case, the train score is so low, that the addition of the physical interaction score prevents the algorithm from adding regulators, even if they are correct. Conversely, when the smoothing is lower, the PMN outperforms the MN in precision (Fig. 3c).
We next examined the contribution of protein–protein interactions and protein–DNA interactions to the correctness of the reconstructed pathways. First, introducing noise to the protein–protein interaction observations (by adding random edges, weighted by the average weight of edges in the interaction set), had little, if any, effect on the PMN's ability to correctly reconstruct regulator–target relationships and pathways. Second, increasing the number of protein–DNA interactions from the true TFs to their true modules, had a negligible effect on the accuracy of regulator–target relationships, but dramatically improved the accuracy of the reconstructed pathways (Fig. 3d), emphasizing the importance of the TF choice in the model.
5 A MAP OF THE YEAST RESPONSE TO GENE PERTURBATIONS
We next assessed the biological accuracy and insights in real data, addressing two capabilities of the PMN approach. First, we considered the reconstruction of correct regulation pathways, between known modules and their known regulators, as determined by gene perturbation in yeast (Hughes et al., 2000; Mnaimneh et al., 2004). Second, we assessed the biological performance of a full learning of PMN from yeast cell cycle expression data (Pramila et al., 2006), a relatively well characterized system which enables us to estimate the biological relevance of the inferred model.
In both cases, we collected observations for protein–protein and protein–DNA interactions from multiple sources (Batada et al., 2006; Herrgard et al., 2006; MacIsaac et al., 2006), generating a dataset of 18.1K protein–protein and 90.6K protein–DNA interactions over 5640 proteins. To avoid spurious over-representation of highly connected (‘hub’) proteins in reconstructed pathways (as they connect most protein pairs in the network in the most efficient way), we determined the confidence in each edge (P-value) according to the degree of the adjacent nodes in the observed network, such that edges that involve highly connected proteins are penalized. In addition, the P-value prefers edges that are supported by more than a single line of evidence. Formally, the P-value of an edge e with degree x of its heaviest adjacent node, was set to be:
where C=0 when the edge is supported by two or more lines of evidence, or C=1 otherwise. Unobserved edges were associated with a constant P-value which is higher than any observed edge. (See http://www.compbio.cs.huji.ac.il/PMN/ for additional information.)
To evaluate the ability of PMN to reconstruct regulation pathways, we used a test case in which the regulators and the modules are known. We used a set of expression profiles measured following single-gene knockouts (Hughes et al., 2000) and knockdowns (Mnaimneh et al., 2004). We defined each perturbed gene as a regulator, the genes that were significantly repressed as one target module, and those that were significantly induced as another target module. The threshold for significance was defined as more than a 2-fold increase or decrease, and one set (Hughes et al., 2000) was further filtered using a significance threshold presented in the original paper (P<0.01). We removed small target modules (less than 5 genes), resulting in a set of 827 regulator-module pairs, for 446 distinct regulators. We used the interaction data as described above.
For each of the 827 regulator-module pairs we reconstructed the most probable (primary) regulation pathway that starts with the regulator and ends with a TF that binds some of the target genes. For example, consider the simple pathway that was reconstructed from NDD1 (Fig. 4a), a component of the MCM1/NDD1/FKH transcription factor complex that activates G2/M transition genes (Koranda et al., 2000). The pathway correctly reconstructs the connection of NDD1 to FKH2, which in turn has evidence for binding 28% of the 60 target genes. We successfully reconstructed 660 primary pathways with an average length of 2.6 edges, including 48 pathways of size zero (directly from a perturbed TF to its targets). On average, the selected TFs had evidence for binding 20% of genes per target module. We failed to reconstruct pathways for the remaining 167 pairs, where the observed physical interactions did not have paths from the regulator proteins to a TF.
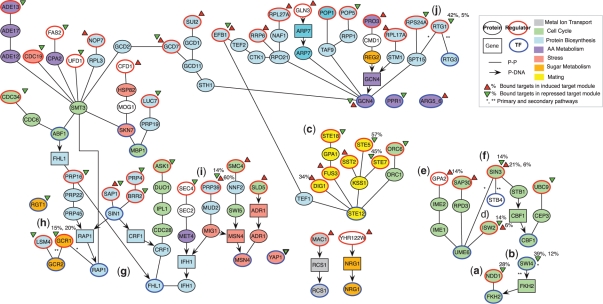
Fig. 4.
Yeast pathway map. Unification of sixty two selected pathways, reconstructed by PMN from gene perturbation data. The pathways are also available online at www.compbio.cs.huji.ac.il/PMN/. Proteins (circles) and genes (squares) are connected by protein–protein interactions (lines) and protein–DNA interactions (arrows). Triangles represent repressed (green) and induced (red) target modules. Numbers indicate the percentage of target genes bound by the TF in pathways which are mentioned in the text. Proteins were manually annotated with the following categories: Metal-ion transport (grey), cell-cycle (green), protein biosynthesis (light blue), amino-acid metabolism (purple), stress (pink), sugar metabolism (orange), mating (yellow) or other (white). (a–j) Pathways that are discussed in the text.
Long-term gene perturbation can cause drastic changes in the cell, and may thus affect the target genes in more than one way. To search for secondary effects, we removed the edges in the primary pathway, and repeated the pathway reconstruction procedure. For example, after removing the (trivial) primary pathway from SWI4 (Fig. 4b) to its targets genes (39% of the genes that were repressed after its perturbation are bound by SWI4), we detected a secondary pathway from SWI4 to FKH2 gene, which in turn binds 12% of the altered genes.
To statistically assess the reconstructed pathways we estimated their score's P-value empirically, by randomization of the input interactions. We used an edge-swapping algorithm (Maslov and Sneppen, 2002) to generate 50 randomized observation networks that maintain the node degrees of the original network, and then learned pathways for the same 827 regulator-module pairs using each of the 50 networks. We then assessed the significance of each pathway learned with the real network, based on the frequency of observing a path with that score or higher in the randomized networks. We found that 99 primary pathways and 6 secondary pathways were significant [above 2 STD (P<0.021)]. (Data available online at http://www.compbio.cs.huji.ac.il/PMN/).
These significant reconstructed pathways span a spectrum of cellular processes, including cell-cycle, mating, stress response, protein biosynthesis, amino-acid metabolism and metal ion transport (Fig. 4). To biologically interpret them, we annotated each pathway member according to the process in which it participates, and each target module with its enriched GO process terms (Fisher's exact test, corrected with FDR 5%). We then inspected the functional relevance of each pathway and its terminal TF to the module and regulator with which it was associated.
There are 6 coherent mating pathways, each consists of known members of the pheromone response, and ends with STE12, the pheromone response TF (Fig. 4c). Three start with STE18, STE5 and STE7, and end with repressed modules enriched for pheromone response genes, a fourth starts with DIG1, a STE12 repressor (Olson et al., 2000), and indeed ends with an induced module enriched for pheromone response genes. These provide a proof of concept.
We detected 10 coherent cell-cycle pathways, of which 5 are meiosis pathways that end with UME6, a TF that regulates both induction and repression of early meiotic genes (Steber and Esposito, 1995). Two of the pathways (Fig. 4d) start with an ISW2 knockout, a member of the ISWI chromatin remodeling complex, which is known to be recruited by UME6 (Goldmark et al., 2000). Another pathway starts from a GPA2 knockout (Fig. 4e), a G-protein that is coupled with the carbon sensor GPR1, through the early meiosis activators IME2 and IME1, to UME6, which binds a target module enriched for sporulation genes. This is consistent with induction of sporulation during carbon (and nitrogen) starvation.
Some of the reconstructed meiosis pathways lead to novel hypotheses. Most notably, the secondary pathway from SIN3 (Fig. 4f), a histone deacetylase component involved in meiosis regulation, which is connected to its target module through STB4. STB4 is a zinc-finger with an unknown function that interacts with SIN3 in a 2-hybrid assay (Kasten and Stillman, 1997), and binds 6% of the genes in the target module. We hypothesize that STB4 plays a role in induction of early meiosis genes.
Several pathways reflect the cellular response to stress following gene perturbations, and the coupling between reduction of cell growth (through repression of ribosomal functions) and changes to carbon metabolism during stress. Twelve pathways lead to ribosomal protein (RP) gene modules (Fig. 4g), ending with the known RP activators FHL1 or RAP1. Interesting exceptions are two pathways that start with the knockout of GCR1, a transcriptional activator of glycolysis genes (Fig. 4h): in the primary pathway GCR1 binds RAP1 leading to repression of both carbohydrate catabolism genes [known to be regulated by RAP1 and GCR1 (Mizuno et al., 2004)] and RP genes. In the secondary pathway GCR1 binds GCR2, a known co-activator, which binds 19 of the repressed genes, including 3 glycolysis genes. Coupling between RP gene regulation and sugar metabolism is also observed following perturbation of the splicing factor PRP39 (Fig. 4i), proposing a new molecular pathway.
Finally, 14 pathways reflect protein biosynthesis and amino acid metabolism, ending with GCN4, a major TF activator of amino acid metabolism genes. The pathways control 10 modules, both repressed and induced, that are enriched for amino acid metabolism genes. While statistically significant, these pathways likely reflect mostly indirect effects on amino acid metabolism by many of the genetic perturbations in the study, a fact noted in the original study (Hughes et al., 2000). In such cases, the reconstructed pathways may not reflect a ‘real’ transduction of a regulatory signal, although their intiation (perturbed gene) and endpoint (the TF GCN4) are correct. Indeed, when GCN4 is removed, other, more relevant pathways may be revealed, for example in the case of RTG1 (Fig. 4j). Despite these limitations, we concluded that PMN can recover known coherent pathways in the correct context, as well as reveal novel potential mechanisms.
6 RECONSTRUCTING A YEAST CELL CYCLE NETWORK
We assessed the reconstruction of a full PMN—modules, regulators and pathways—using the yeast cell cycle, a well-studied system that facilitates careful evaluation. We used expression profiles measured at 50 time points during 2 cell cycles from yeast cultures synchronized with α-factor (Pramila et al., 2006). We focused on the 594 top cyclic genes as defined in the original publication (PNM5 posterior > 0.999), 68 of which have a known regulatory role in cell cycle (by GO), and were designated as candidate regulators for the model to choose from. We augmented the physical interactions described above with protein–DNA interactions measured specifically during the yeast cell cycle (Horak et al., 2002).
The inferred network consists of 36 modules of average size 17.5 genes (±19.7 genes), 11 with a single regulator and 4 with two regulators, with regulation paths of average length of 2.5. Nine modules peak at G1 or G1/S (Fig. 5 right, pink or pink-yellow), of which one is enriched for cytokinesis genes (module #43), two for mitosis genes (#5, #44), and four for DNA replication genes (#36, #39, #44, #48). For example, module 36 (Fig. 5 left) is regulated by SWI4, via SWI6 and MBP1, which binds 59% of the module genes, consistent with the known role of the SBF (SWI4-SWI6) and MBF (SWI6-MBP1) complexes in regulating late G1 genes. Module 36 is also regulated by ACM1, an inhibitor of mitosis that is induced during G1/S, via a pathway consisting of (in order) CLN1, SIC1 (both G1/S transition regulators), and CDC14, a phosphatase required for mitotic exit. Although CDC14 does not seem to be directly connected to G1/S regulation, it is known to interact with SWI6-MBP1 complex that binds the module genes (Geymonat et al., 2004).
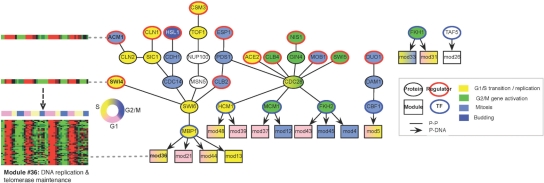
Fig. 5.
Yeast cell cycle map. Pathways reconstructed by PMN from the yeast cell cycle data. Left - An example module (# 36), indcued in G1 & S, is enriched for DNA replication and telomerase maintenance genes, and is regulated by ACM1 and SWI4. Right - all other pathways reconstructed. Modules (rectangles) are colored according to their peak activity phase, and proteins are colored according to the phase in which they are known to play a role.
Six modules peak at S and four at G2/M (Fig. 5, blue). For example, Module 33, enriched for mitosis genes, is active throughout S and G2/M, and is regulated by FKH1, a key TF of G2/M genes, which binds directly to 46% of the module genes. In another example, Module 45 (Figs 1a and b, 5 right) is regulated by MOB1, an essential component of the mitotic exit network, through a pathway that consist of (in order) CDC28, the catalytic subunit of the main cell cycle CDK, and FKH2, a major G2/M TF, which binds 43% of the module genes. The module genes are also enriched for binding of NDD1, an FKH2 transcriptional co-activator (Fig. 1b).
In some notable exceptions, biologically ‘incoherent’ regulators are chosen by the PMN. For instance, Module 13 (Fig. 5 right) peaks in S phase, is highly enriched for histones, and is regulated by ESP1 and HSL1, mitosis and budding regulators. This may reflect the fact that in some cases, induction of a regulator's mRNA precedes the time of its protein's activity (Amit et al., 2009; Ramsey et al., 2008). While the choice of regulator in the MN is based solely of the expression pattern, the physical interactions in the PMN may be able to reduce the number of such false cases.
Conversely, the selected TF is coherent with the target module in all cases where the TF binds more than one gene per module: MBP1 is chosen as the TF of 4 G1 and S modules; FKH1 for two S and G2/M modules; FKH2 for two G2/M modules; MCM1, a major cell cycle transcription and DNA replication regulator, for a G1 module; and HCM1, a transcription regulator of S phase genes, for two G1 and S modules. Besides FKH1, the TFs are not chosen as the regulators themselves (at the beginning of a pathway), because their own mRNA expression profiles do not correlate with their protein activity: some are induced in a different phase, and others do not show a cyclic pattern at all. Yet, the PMN correctly associated them with their target modules.
7 PATHWAY RECONSTRUCTION OF HUMAN–FLU INTERACTIONS
Infection of human cells with the H1N1 influenza virus results in substantial changes in the cells' transcription profile. A recent study (Shapira et al., 2009) measured these changes in human primary bronchial epithelial cells at 10 time point along 18 h following infection with wild type virus, attenuated virus, viral RNA or treatment with interferon. It also identified (with yeast two hybrid assays), potential interactions between each of the 10 viral proteins and 87 human proteins.
We used PMN to identify the potential mechanisms that lead from the viral proteins to changes in expression. This presents a special challenge, since the human physical network is much larger than in yeast, and is still poorly covered (Hart et al., 2006). We used 12 gene modules that were pre-defined (Shapira et al., 2009), and reconstructed the pathways from 10 viral genes to each cluster. We used human protein–protein and protein–DNA interactions collected from multiple published sources (Badis et al., 2009; Berger et al., 2008; Chang et al., 2008; Suzuki et al., 2009; Yosef et al., 2009), and human-viral protein interactions from the original paper. To reduce noise, we included only 32 TFs whose targets were enriched (Fisher's Exact Test, FDR 5%) in at least one cluster.
We ran the pathway reconstruction procedure six times, removing in each round the TF that was selected in the previous round, to reconstruct multiple secondary pathways. The first three rounds, selected only general TFs (TAF1, CREB1, NRF1 and SP1) for each of the 12 modules. Specific pathways emerged only after we removed these general factors.
One intriguing example is the pathway that connects the viral polymerase subunits NP and PB2 to the human TF Interferon Regulatory Factor 2 (IRF2) (Fig. 6a), a known regulator of interferon-dependent gene expression, which binds 16 out of 57 genes in cluster #2, which is induced by interferon. The initial study identified a novel role for the viral polymerase in perturbing host signaling (Shapira et al., 2009), but its relation to the transcriptional program and mode of action remained unknown. Our PMN analysis suggests that the polymerase subunits act through a pathway that includes apoptotic proteins TRAF1, API1 and p53, and impact interferon-dependent gene expression, thus raising testable mechanistic hypotheses. Other interesting pathways (Fig. 6b) connect the NA and HA viral proteins to three clusters. All three pathways consist of (in order) CREBP, VCAF, MLL and CREBBP, but end in three different TFs: NFKB1 that binds cluster #7 (5 out of 60 genes, induced only in presence of whole virus), E2F1 that binds cluster #5 (5 out of 55 genes, induced by virus or viral RNA) and the interferon-dependent factor IRF1 that binds cluster #4 (23 out of 139 genes, induced by interferon alone), all major regulators in host response pathways. This suggests a novel role and mechanism for additional viral proteins in modulating the host transcriptional response.
Fig. 6.
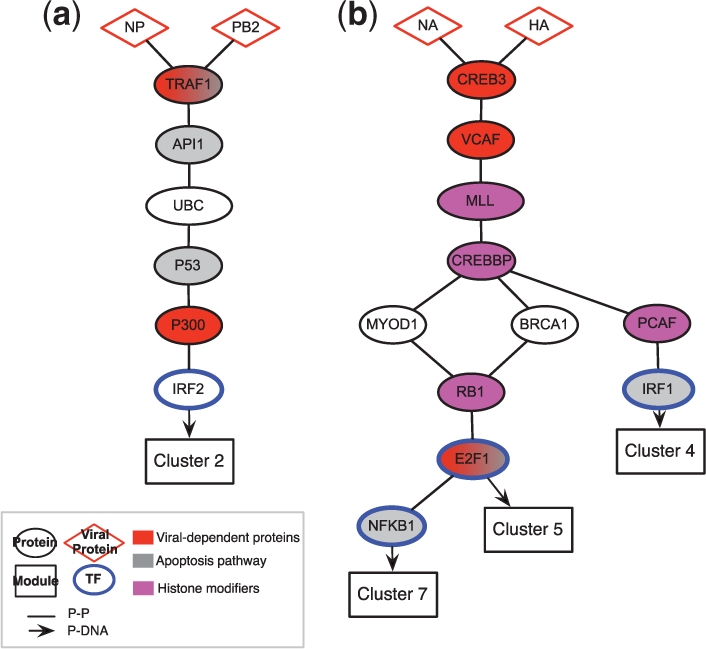
Viral infection in human host. Pathways reconstructed from H1N1 influenza virus proteins to responsive gene clusters. (a) Pathway connecting NP and PB2 viral proteins to cluster 2. (b) Pathway connecting NA viral protein to clusters 4 and 5 and HA viral protein to clusters 4 and 7. Color indicates protein categories (see legend).
8 DISCUSSION
Here, we presented PMNs, a probabilistic graphical method that learns transcriptional networks by combining phenotypic effects (changes in gene expression) with their underlying physical mechanism (protein–protein and protein–DNA interactions). We evaluated the model by comparing to a simple MN on synthetic data, and by reconstructing coherent pathways and regulation in yeast and human data.
A PMN builds and subtantially extends on MNs, a special case of a Bayesian network that is defined over a set of gene modules rather than single genes (Segal et al., 2003). The main drawback of a simple MN is that each regulator is chosen solely based on its expression pattern, limiting its ability to distinguish between true regulators and false positives. The PMN addresses this limitation by presenting additional constraints on the choice of regulators, requiring a physical pathway that connects it to some of the target genes. Indeed, as we have shown on synthetic data, the added physical interactions increase the model's precision without compromising recall. Furthermore, the PMN presents a new hypothesis for the regulation mechanism. We showed that it can determine coherent pathways and regulatory mechanisms that connect genetic perturbations to target genes (in yeast and human data), as well as coherent modules, regulators and pathways in the yeast cell-cycle system.
Notably, adding physical interactions also carries disadvantages, especially when interaction data are very partial and noisy. PMN may miss a true regulator if a full regulation path is missing (hurting recall), the reconstructed pathways are sensitive to noise in the interaction measurements, and certain types of interactions (protein modifications, chromatin occupancy, cellular localization), as well as alternative splicing, are not modeled in the simple interaction graph. Nevertheless, PMN's success in several realistic cases, in yeast and human, suggest that it provides an important advance toward reconstructing models of regulatory circuits in eukaryotic cells.
ACKNOWLEDGEMENTS
We thank Gal Elidan for his support in the implementation process.
Funding: US-Israel Binational Foundation (BSF) grant (to N.F. and A.R., in part).
Conflict of Interest: none declared.
REFERENCES
- Amit I., et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G., et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batada N., et al. Stratus not altocumulus: a new view of the yeast protein interaction network. PLoS Biol. 2006;4:e317. doi: 10.1371/journal.pbio.0040317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinlich I., et al. The ALARM monitoring system: a case study with two probabilistic inference techniques for belief networks. In Proceedings of the 2nd European Conference on AI in Medicine. 1989;38:247–256. [Google Scholar]
- Bellman R. On a routing problem. Q. Appl. Math. 1958;16:87–90. [Google Scholar]
- Berger M., et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz A., et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldi A., et al. Structure and function of a transcriptional network activated by the MAPK Hog1. Nat. Genet. 2008;40:1300–1306. doi: 10.1038/ng.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., et al. Computational identification of the normal and perturbed genetic networks involved in myeloid differentiation and acute promyelocytic leukemia. Genome Biol. 2008;9:R38. doi: 10.1186/gb-2008-9-2-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N. Inferring cellular networks using probabilistic graphical models. Science. 2004;303:799–805. doi: 10.1126/science.1094068. [DOI] [PubMed] [Google Scholar]
- Gao F., et al. Defining transcriptional networks through integrative modeling of mRNA expression and transcription factor binding data. BMC Bioinformatics. 2004;5:31. doi: 10.1186/1471-2105-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M., et al. Clb6/Cdc28 and Cdc14 regulate phosphorylation status and cellular localization of Swi6. Mol. Cell Biol. 2004;24:2277–2285. doi: 10.1128/MCB.24.6.2277-2285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark J., et al. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- Harbison C., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart G., et al. How complete are current yeast and human protein-interaction networks? Genome Biol. 2006;7:120. doi: 10.1186/gb-2006-7-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartemink A.J., et al. Combining location and expression data for principled discovery of genetic regulatory network models. Pac. Symp. Biocomput. 2002:437–449. [PubMed] [Google Scholar]
- Herrgard M., et al. Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in saccharomyces cerevisiae. Genome Res. 2006;16:627–635. doi: 10.1101/gr.4083206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak C., et al. Complex transcriptional circuitry at the G1/S transition in saccharomyces cerevisiae. Genes Dev. 2002;16:3017–3033. doi: 10.1101/gad.1039602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., et al. Genetic reconstruction of a functional transcriptional regulatory network. Nat. Genet. 2007;39:683–687. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- Hughes T., et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Ideker T., et al. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18(Suppl. 1):S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- Kasten M., Stillman D. Identification of the Saccharomyces cerevisiae genes STB1-STB5 encoding Sin3p binding proteins. Mol. Gen. Genet. 1997;256:376–386. doi: 10.1007/s004380050581. [DOI] [PubMed] [Google Scholar]
- Kim H., et al. Transcriptional regulatory circuits: predicting numbers from alphabets. Science. 2009;325:429–432. doi: 10.1126/science.1171347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller D., Friedman N. Probabilistic Graphical Models: Principles and Techniques. Cambridge, MA: The MIT Press; 2009. [Google Scholar]
- Koranda M., et al. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature. 2000;406:94–98. doi: 10.1038/35017589. [DOI] [PubMed] [Google Scholar]
- Kundaje A., et al. A predictive model of the oxygen and heme regulatory network in yeast. PLoS Comput. Biol. 2008;4:e1000224. doi: 10.1371/journal.pcbi.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Lee S-I., et al. Identifying regulatory mechanisms using individual variation reveals key role for chromatin modification. Proc. Natl Acad. Sci. USA. 2006;103:14062–14067. doi: 10.1073/pnas.0601852103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIsaac K., et al. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC. Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov S., Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- Mizuno T., et al. Role of the N-terminal region of Rap1p in the transcriptional activation of glycolytic genes in Saccharomyces cerevisiae. Yeast. 2004;21:851–866. doi: 10.1002/yea.1123. [DOI] [PubMed] [Google Scholar]
- Mnaimneh S., et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Nariai N., et al. Using protein–protein interactions for refining gene networks estimated from microarray data by bayesian networks. Pac. Symp. Biocomput. 2004:336–347. doi: 10.1142/9789812704856_0032. [DOI] [PubMed] [Google Scholar]
- Olson K., et al. Two regulators of Ste12p inhibit pheromone-responsive transcription by separate mechanisms. Mol. Cell Biol. 2000;20:4199–4209. doi: 10.1128/mcb.20.12.4199-4209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ourfali O., et al. SPINE: a framework for signaling-regulatory pathway inference from cause-effect experiments. Bioinformatics. 2007;23:i359–i366. doi: 10.1093/bioinformatics/btm170. [DOI] [PubMed] [Google Scholar]
- Peleg T., et al. Network-free inference of knockout effects in yeast. PLoS Comput. Biol. 2010;6:e1000635. doi: 10.1371/journal.pcbi.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramila T., et al. The forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006;20:2266–2278. doi: 10.1101/gad.1450606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey S.A., et al. Uncovering a macrophage transcriptional program by integrating evidence from motif scanning and expression dynamics. PLoS Comput. Biol. 2008;4:e1000021. doi: 10.1371/journal.pcbi.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E., et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- Segal E., et al. Learning module networks. J. Mach. Learn. Res. 2005;6:557–588. [Google Scholar]
- Shapira S., et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber C., Esposito R. UME6 is a central component of a developmental regulatory switch controlling meiosis-specific gene expression. Proc. Natl Acad. Sci. USA. 1995;92:12490–12494. doi: 10.1073/pnas.92.26.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. How to reconstruct a large genetic network from n gene perturbations in fewer than n2 easy steps. Bioinformatics. 2001;17:1183–1197. doi: 10.1093/bioinformatics/17.12.1183. [DOI] [PubMed] [Google Scholar]
- Yeang C., et al. Physical network models. J. Comput. Biol. 2004;11:243–262. doi: 10.1089/1066527041410382. [DOI] [PubMed] [Google Scholar]
- Yeger-Lotem E., et al. Bridging high-throughput genetic and transcriptional data reveals cellular responses to alpha-synuclein toxicity. Nat. Genet. 2009;41:316–323. doi: 10.1038/ng.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N., et al. Toward accurate reconstruction of functional protein networks. Mol. Syst. Biol. 2009;5:248. doi: 10.1038/msb.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou M., Conzen S.D. A new dynamic bayesian network (DBN) approach for identifying gene regulatory networks from time course microarray data. Bioinformatics. 2005;21:71–79. doi: 10.1093/bioinformatics/bth463. [DOI] [PubMed] [Google Scholar]