Abstract
Motivation: Next-generation sequencing techniques allow us to generate reads from a microbial environment in order to analyze the microbial community. However, assembling of a set of mixed reads from different species to form contigs is a bottleneck of metagenomic research. Although there are many assemblers for assembling reads from a single genome, there are no assemblers for assembling reads in metagenomic data without reference genome sequences. Moreover, the performances of these assemblers on metagenomic data are far from satisfactory, because of the existence of common regions in the genomes of subspecies and species, which make the assembly problem much more complicated.
Results: We introduce the Meta-IDBA algorithm for assembling reads in metagenomic data, which contain multiple genomes from different species. There are two core steps in Meta-IDBA. It first tries to partition the de Bruijn graph into isolated components of different species based on an important observation. Then, for each component, it captures the slight variants of the genomes of subspecies from the same species by multiple alignments and represents the genome of one species, using a consensus sequence. Comparison of the performances of Meta-IDBA and existing assemblers, such as Velvet and Abyss for different metagenomic datasets shows that Meta-IDBA can reconstruct longer contigs with similar accuracy.
Availability: Meta-IDBA toolkit is available at our website http://www.cs.hku.hk/~alse/metaidba.
Contact: chin@cs.hku.hk
1 INTRODUCTION
Metagenomic research studies the genetic information in an entire microbial community. It plays an important role in microbiology because over 99% of microbes can neither be isolated nor cultured (Wooley et al., 2010). Recent advances in next-generation sequencing technology allow us to generate reads from genomes of multiple species in these samples in an effective manner. The set of reads obtained is very complicated which makes the assembling of genomes of species that exist in the sample extremely difficult. There are two main approaches to study reads from these samples. One is to group (called binning) the reads according to some biological markers or structural features (Huson et al., 2007; Krause et al., 2008; Yang et al., 2010). Then, reads belonging to different species are studied. The other is to deduce the potential biological functions of the whole community by studying the reads directly using gene prediction or function annotation (Mavromatis et al., 2007; Qin et al., 2010; Wooley et al., 2010). Since the reads from the next-generation sequencing technologies are still relatively short, it is more effective if longer contigs can be constructed from the reads before conducting the study even though we are not able to assemble the genome of every species. High quality assembly results of the contigs are desirable in both approaches. If the assembled contigs are short and erroneous, the accuracy of binning, gene prediction, function annotation, etc. will be impaired. Thus, assemblers that can generate longer and more accurate contigs will definitely facilitate the study of metagenomic data.
If similar reference genomes exist in the database, one can assemble reads by first aligning them to the reference genomes (Gnerre et al., 2009). However, as over 90% of microbes in metagenomic data are unknown (Wooley et al., 2010), de novo assemblers are needed to assemble reads without any reference genomes. To our knowledge, currently there are no de novo metagenome-specific assemblers available. Assemblers such as EULER (Chaisson et al., 2009; Chaisson and Pevzner, 2008; Pevzner et al., 2001), Velvet (Zerbino and Birney, 2008; Zerbino et al., 2009), Abyss (Simpson et al., 2009), SOAPdenovo (Li et al., 2010) are for single genome, but are used in metagenomic study (Wooley et al., 2010). All these assemblers are based on the de Bruijn graph (Pevzner et al., 2001), which is a common approach to perform de novo assembly. In the de Bruijn graph, a vertex represents a length-k substring called k-mer and an edge connecting vertices u and v represents u and v appearing consecutively in a read. All these assemblers generate the de Bruijn graph from reads, and then apply some error removal methods (Simpson et al., 2009; Zerbino and Birney, 2008), e.g. removing tips and merging bubbles, to modify the graph based on its topological structure. Simple paths in the graph are outputted as contigs and paired-end information might be applied to further merge the contigs.
These existing assemblers do not work well for metagenomic datasets, except for some very small datasets containing specific species (Pop, 2009). Two main properties of a reasonably complicated metagenomic dataset make these assemblers fail to produce long contigs: (i) polymorphisms among similar subspecies and common genomic regions shared by different species; and (ii) uneven abundance ratios of species in a sample. The polymorphism of similar subspecies, especially subspecies of the same species, consists of very similar sequences with few variations (single nucleotide variation, short insertion or deletion or genomic rearrangements, etc.) and each variation introduces a branch in the de Bruijn graph (we call these branches sp-branches). Another source of branches is due to the common or similar genomic regions, say housekeeping genes, shared by different species (we call these branches cr-branches). These two types of branches, which do not exist when assembling single genomes, would make the de Bruijn graph for metagenomic data very complicated. Since existing assemblers output simple paths in the graph as contigs, these extra branches caused by common regions in different species prevent the construction of long contigs.
Some assemblers resolve branches by merging similar sequences as bubbles into one sequence. A bubble is defined as several similar paths with the same start vertex and the same end vertex (Zerbino and Birney, 2008) in the de Bruijn graph. Bubble merging helps to merge similar regions and reduce complexity of the de Bruijn graph. An important assumption used by assemblers to remove bubbles for single genome assembly is that the bubble is caused by a few single nucleotide polymorphisms (SNP) or errors in reads; thus, the simple paths inside a bubble are very similar, except for a few nucleotides. However, the ‘bubbles’ found in the graph for metagenomic dataset do not follow this assumption. Different bubbles mix together to make the start vertex and the end vertex very difficult to be identified. Some of these bubbles are formed by a mixture of sp- and cr-branches. Figure 1 shows an example of this phenomenon in which every simple path is contracted into a vertex for visualization. All branches at a vertex in this graph normally lead to some other vertex in the same component, but it is uncertain that these are bubbles for merging. If we look closer at these bubbles, even if the bubble is formed only by sp-branches (because of variations in subspecies), the multiple paths inside the bubble may differ a lot (maybe with larger insertion/deletion). Existing approaches for merging bubbles for single genome assembly do not work for this case; thus, they will fail to resolve these ‘bubbles’ and are unable to construct long contigs. Even if all bubbles can be identified, it is not easy to merge them together to form a consensus.
Fig. 1.
A component in de Bruijn graph of five E.coli subspecies.
To resolve branches in a de Bruijn graph, existing assemblers also try to use paired-end information to help find paths with paired-end reads support so as to eliminate those branches caused by erroneous reads and to construct longer contigs. In the case of multiple subspecies, each path will have a lot of support since they are not caused by erroneous reads, but variations in subspecies and the assemblers are not able to resolve these branches easily. Moreover, since the contigs are short, applying paired-end information becomes difficult, because usually paired-end information can only be applied to connect long contigs.
To show the complexity of a de Bruijn graph for metagenomic dataset, Table 1 compares graphs and assembly results of simulated reads sampled from a single genome (Escherichia coli 536) and from five different E.coli subspecies genomes. Table 1 shows that the five E.coli subspecies contain about twice the number of k-mers as that of a single E.coli subspecies, but 150 times more branches as that of single subspecies. This makes the graph complicated and genome assembly difficult. In fact, the performance of all the assemblers is poor when there are a lot of subspecies. The N50 values of Velvet , Abyss and SOAPdenovo drop from 178 914, 32 440 and 125 404 bp for single genome data to 875, 849 and 713 bp, respectively, for metagenomic data with five subspecies. Uneven abundance ratios in metagenomic data introduce another problem in assembly, because existing assemblers cannot distinguish erroroneous reads sampled from genomes with high abundance ratios and reads from genomes with low abundance ratios. Thus, it is difficult to identify species with the low abundance ratios.
Table 1.
Assembly results of E.coli 563 and five E.coli subspecies
| Assembler | E.coli 536 | Five E.coli subspecies |
|---|---|---|
| Perfect de Bruijn graph Assembler | ||
| k | 50 | 50 |
| No. of k-mers | 4 859 649 | 11 533 119 |
| No. of branches | 810 | 130 045 |
| Velvet | ||
| No. of contigs | 226 | 13 516 |
| N50 | 178 914 | 875 |
| Coverage (%) | 99.33 | 90.24 |
| Abyss | ||
| No. of contigs | 337 | 27 428 |
| N50 | 32 440 | 849 |
| Coverage (%) | 99.77 | 94.15 |
| SOAPdenovo | ||
| No. of contigs | 247 | 21 589 |
| N50 | 125 404 | 713 |
| Coverage (%) | 99.81 | 94.20 |
| Meta-IDBA | ||
| No. of contigs | 256 | 9292 |
| N50 | 122 317 | 5781 |
| Coverage (%) | 99.64 | 88.37 |
Simulated length-75 reads are sampled randomly from references with 1% error and 250 insert distance with a depth of 30. The value of k is set to 50 for all assemblers.
Resolving the branches (both sp- and cr-branches) in the graph is one of the key issues for solving the problem of metagenomic assembly. In this article, we focus on this issue. We remark that the issue of uneven abundance ratios of subspecies is also very important and difficult to solve and demands dedicated research effort.
Conceptually, we look at the problem from the following perspective. The cr-branches (caused by common regions in different species) should be removed to isolate one species from another. For sp-branches, since they are caused by variations of subspecies, instead of producing separate contigs for individual subspecies, it is preferable to represent possible variations (e.g. insertion) of similar sequences. The core of our proposed metagenomic assembler, Meta-IDBA, has two steps. The first step is to identify and remove cr-branches in the de Bruijn graph leaving a set of connected components, each of which hopefully corresponds to a set of subspecies of the same species. The second step is to transform each component into a multiple alignment with consensus so as to represent the contigs of different subspecies of the same species.
To distinguish cr-branches from sp-branches, the main idea is based on the fact that the genome sequences of subspecies in the same species are more similar than those from different species. Thus, more common k-mers are shared by the genome sequences of subspecies of the same species. Table 2 summarizes the k-mer similarity between subspecies within different taxonomic levels.
Table 2.
k-mer Similarity (k=50) in different taxonomic level
| Species | Genus | Family | Order | Class | Phylum | |
|---|---|---|---|---|---|---|
| Similarity (%) | 63.2 | 7.3 | 2.3 | 0.06 | 0.02 | 0.01 |
For each level, 1000 pairs of subspecies with lowest common ancestor of that level are generated randomly for k-mer similarity calculation.
The similarity inside the same species is much higher than that of the genus. Sixty percent similarity means that on average two k-mers of a subspecies will share at least one with another subspecies. Moreover, although the average similarity at the genus level is 7.3%, the common k-mers concentrate at some common regions, shared by different species, and is <1% at other regions. Nevertheless, reads from species further apart in higher taxonomic levels will share fewer k-mers.
Similar observations are also made in Fofanov et al. (2004) that a k-mer, with k≥20, tends to occur uniquely in the genome of a single species. Therefore in the de Bruijn graph, sp-branches started from a common k-mer in similar subspecies usually have (converge to) another common k-mer within a short distance, while cr-branches started from a common k-mer in multiple species seldom have another common k-mer within a short distance. By removing those branches that cannot converge to another k-mer within a short distance, the genome of a species with several subspecies can be represented by connected components in the de Bruijn graph, where each component represents similar contigs in the genomes from similar subspecies. As the contigs for each subspecies in the same component are with slight differences, a consensus contig can be found by multiple alignment (to capture the variations of the genome of the subspecies) to represent the genome of the species. Thus, instead of outputting simple paths in the de Bruijn graph to represent contigs, connected components representing consensus contigs of species will be output. As shown in Table 1, our assembler called Meta-IDBA is more effective in assembling reads in metagenomic data, where the N50 of Meta-IDBA is 5781 bp, about seven times longer than those of Velvet, Abyss and SOAPdenovo. We agree that the improvement in contig lengths mainly stems from the multiple alignment with consensus representation of the components, which seems to be a more appropriate output for contigs belonging to different subspecies due to variations. Moreover, if the similar regions cannot be separated, it will be too expensive to do multiple alignment among resulting contigs. To summarize, our work has two major contributions: to isolate components that derive from similar subspecies of the same species in the complicated de Bruijn graph of a metagenomic dataset; and to report the contigs of the subspecies using multiple alignment to highlight possible variants.
2 METHODS
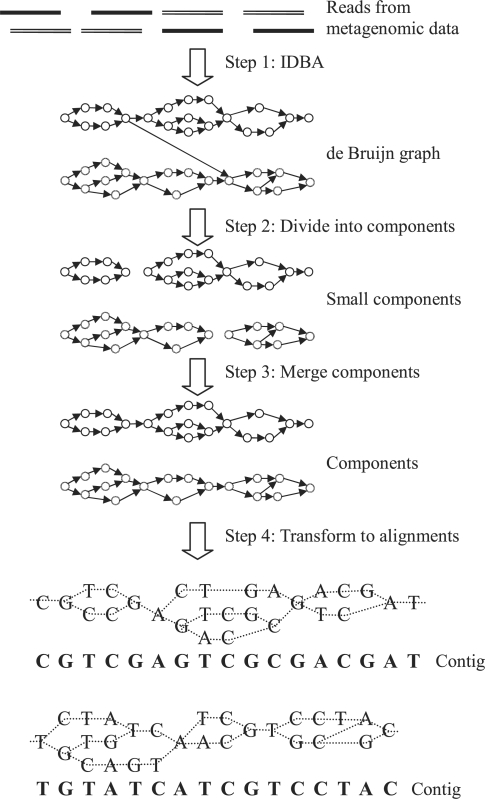
In this section, we will describe our algorithm, Meta-IDBA, for assembling reads from multiple genomes of subspecies in different species. There are two main steps in Meta-IDBA as shown in Figure 2. Initially (Step 1) sequencing reads are used to construct a de Bruijn graph using any de Bruijn graph-based assembler (Chaisson and Pevzner, 2008; Peng et al., 2010; Simpson et al., 2009; Zerbino and Birney, 2008). Each simple path in the de Bruijn graph might represent a contig of the genome of some species or subspecies. As there are some sequences appearing in multiple species, the de Bruijn graph of reads from different species are interconnected by cr-branches. In the second step (Step 2), based on the assumption that the genomes of subspecies from the same species share more similar regions than the genomes of subspecies from different species, Meta-IDBA divides the de Bruijn graph into many small connected components by removing cr-branches. As the genome sequences of subspecies even from the same species are not exactly the same, the consensus contig may not be represented by a simple path in the de Bruijn graph, but may be represented by a component with multiple paths (due to sp-branches) from a source vertex to a single sink vertex, so as to confine the variations of the similar regions in each genome. These small components are then merged into bigger components, which represent longer consensus contigs using paired-end reads (Step 3). In the last step of Meta-IDBA (Step 4), each component is transformed to a multiple alignment of similar contigs of different subspecies from the same species. The consensus contig, which represents the similar contigs of the subspecies in a species, is found. The steps of Meta-IDBA will be described in detail in the following sections.
Fig. 2.
Workflow of Meta-IDBA algorithm.
2.1 Construct de Bruijn graph
Any genome assembler based on the de Bruijn graph approach (Chaisson et al., 2009; Peng et al., 2010; Pevzner et al., 2001; Simpson et al., 2009; Zerbino and Birney, 2008) can be used to assemble reads to form contigs as if the reads were from a single genome. The de Bruijn graph-based algorithm first divides each reads into length-k substrings called k-mers. Each k-mer is represented by a vertex in the de Bruijn graph. There is an edge from u to v if the length k −1 suffix of k-mer u is the same as the length k −1 prefix of k-mer v and k-mers u and v occur adjacently in at least one read. In the single genome assembly problem, each simple path in the de Bruijn graph represents a contig of the genome and each branch represents a repeat region in the genome or an error in the read. The IDBA genome assembler (Peng et al., 2010) is used in our implementation as IDBA iteratively increases the value of k and removes k-mers with few supports from reads, more branches can be resolved and longer contigs of the genome can be obtained. However, metagenomic data contains read from genomes of multiple species, and the sp- and cr-branches in the de Bruijn graph represent common regions occurring in genomes of different species or subspecies. These kinds of branches cannot be removed by the single genome assembler and thus very short contigs will be produced especially for samples with many subspecies of the same species. Thus, additional steps are required for resolving these branches.
2.2 Divide graph into connected components
Based on the well-accepted idea (Pop, 2009) that genomes of subspecies in the same species are more similar than genomes of subspecies from different species, Meta-IDBA divides the de Bruijn graph into connected components such that each component represents a consensus contig of a species. With the assumption that genomes of subspecies from the same species are very similar, there are many similar regions along the genome, where common k-mers (e.g. k=50) do not appear too distinct and at least one common k-mer exists within a relative short region of length w (e.g. w=300 bp). On the other hand, the genomes of subspecies from different species seldom contain a common k-mer so frequently. Based on this assumption, we divide the de Bruijn graph into components by solving the following graph partitioning algorithm.
Graph partition problem: given a directed graph G, the graph partition problem is to partition the graph into maximal connected components that satisfy the following property. For each vertex u in a component C, there is another vertex v in C such that: (i) starting from each out-going edge e of u, there is at least a path from u to v in C with length at most w; or (ii) for every in-coming edge e of u, there is at least a path from v to u in C with length at most w.
The idea behind the formulation of the graph partition problem is as follows. If a k-mer u can reach another k-mer v in a de Bruijn graph through a path of length at most w starting from each of its out-going edges (or vice versa), k-mers u and v are likely to occur in the genomes of subspecies from the same species, which represent high similarity among the genomes of the subspecies. Otherwise, k-mers u and v may occur in genomes of different species and they should be separated.
The graph partition problem can be solved by a greedy algorithm, which repeatedly checks all out-going (in-coming) edges of each vertex. If there are no paths of length at most w starting from (ending with) the out-going (in-coming) edges of a vertex u that ends with (start from) another vertex v, all the out-going (in-coming) edges of vertex u are removed from the de Bruijn graph. The removing process will be repeated until all components satisfy our requirements. The correctness of this greedy algorithm is proved in Theorem 1. The process will remove the cr-branches as well as some of the sp-branches, resulting in many small connected components with close common regions (k-mers) representing consensus contigs of a single species.
Theorem 1. —
The greedy algorithm solves the graph partition problem.
Proof. —
By induction of the number of removing edges. When the first edge e1=(u1, u1′) is removed by the greedy algorithm, edge e1 should not be in any component, because either there is no vertex v such that all out-going edges of u1 can access v in a path in G with length at most w or there is no vertex u such that u can access u1′ by paths in G with length at most w that end with each in-coming edges of u1′. Consider the k-th edge ek=(uk,uk′) removed by the greedy algorithm. Either there is no vertex v such that all out-going edges of uk can access v in a path in G/{e1,…,ek−1} with length at most w or there is no vertex u such that u can access u1′ by paths in G/{e1,…,ek−1} with length at most w that end with each in-coming edges of uk′. Since the edges e1,…,ek−1 are not in any component, edge ek should not be in any component. ▪
2.3 Connect components by paired-end reads
After solving the graph partition problem, larger components may be broken down into smaller components, because of the erroneous reads and common regions among different species and each component will represent a consensus contig of subspecies from a single species. Meta-IDBA merges the components into larger components using paired-end reads. A paired-end read represents two reads appearing in the same genome with known order and distance (insert distance). One end of a paired-end read is considered to appear in a component if a k-mer of the read appears in the component. If the two ends of a paired-end read appear in different components, the paired-end read is considered as a support that the two components should be merged into a larger component. Meta-IDBA merges the components if there are at least α (e.g. α=10) supports from paired-end reads and the two components are connected by the shortest path, which matches with the insert distance of the pair-end reads only if the merging is unambiguous (i.e. the merging will not be performed if a component has enough supports to connect to more than one component). Moreover, only the components with consensus contig longer than insert distance are considered in this procedure, because otherwise a component caused by a repeat region shorter than insert distance will be connected to multiple components with enough supports.
2.4 Construct multiple alignment and consensus contigs
Since the genomes of the subspecies from the same species are similar with small variants, it is very difficult to determine long contigs (simple path) from each subspecies, because these contigs are interconnected in the de Bruijn graph. Instead of representing a contig of a species by a simple path, a consensus contig of the species (which has many subspecies) is represented by a component, which is usually a direct acyclic graph (with exception when there is repeat region in the contig) to capture the small variants among the genomes of different subspecies in the same species. Each simple path in the component represents a short contig of a subspecies and the relative positions of these short contigs can be obtained from the structure of the directed acyclic graph. In order to obtain a contig of a single species from the component, multiple alignment of the short contigs represented by simple paths is performed. Meta-IDBA starts with an alignment of some short simple subpaths at the source vertex of the component and progressively constructs the alignments of longer contigs with reference to the longest subpath, because it provides more information of the genomes of the subspecies than the short subpaths with deletions. As the relative position of each simple path can be determined, the alignment between each simple subpath can be found locally. Finally, each component is represented by a consensus contig obtained through multiple alignment.
3 RESULTS
3.1 Simulated data
To compare the performance of Meta-IDBA with the other assemblers, we constructed three datasets with different complexities. The NCBI RefSeq (Pruitt et al., 2009) database was used for generating the simulated sequencing reads. Table 3 summarizes the composition and experiment result of each dataset.
Table 3.
The compositions and experiment results of simulated datasets
| Features | Low-complexity |
Medium-complexity |
High-complexity |
|||
|---|---|---|---|---|---|---|
| Taxonomic level | ≤ Genus | ≤ Family | ≤ Class | |||
| No. of species | 2 | 5 | 10 | |||
| No. of cases | 10 | 10 | 5 | |||
| Expression level | Uniform | Log normal | Uniform | Log normal | Uniform | Log normal |
| Meta-IDBA | ||||||
| Component accuracy (%) | 98.82 | 98.80 | 98.32 | 98.16 | 99.35 | 99.1 |
| N50 | 18 729 | 20 689 | 11 111 | 14 610 | 8246 | 9553 |
| Coverage (%) | 91.76 | 89.20 | 87.39 | 81.62 | 91.47 | 84.16 |
| No. of contigs | 2674 | 2180 | 13 627 | 7716 | 55 249 | 38 500 |
| No. of bases | 5 418 695 | 5 223 474 | 20 615 422 | 17 644 012 | 68 465 188 | 58 088 054 |
| No. of error contigs | 9 | 9 | 26 | 21 | 90 | 223 |
| No. of error bases | 57 645 | 44 427 | 115 050 | 82 159 | 336 429 | 371 334 |
| Time (min) | 12.5 | 12.1 | 54.1 | 55.2 | 120.1 | 115.7 |
| Velvet | ||||||
| N50 | 11 437 | 9771 | 3356 | 3433 | 1983 | 1997 |
| Coverage (%) | 87.29 | 83.70 | 86.83 | 84.88 | 92.39 | 75.77 |
| No. of contigs | 2309 | 2230 | 12 839 | 11 208 | 106 917 | 37 567 |
| No. of bases | 4 876 110 | 4 674 621 | 15 824 798 | 16 017 085 | 70 072 915 | 44 948 173 |
| No. of error contigs | 9 | 7 | 16 | 14 | 196 | 59 |
| No. of error bases | 54 364 | 34 796 | 62 470 | 70 148 | 401 434 | 230 525 |
| Time (min) | 16.8 | 15.2 | 44.7 | 45.0 | 96.4 | 95.8 |
| Abyss | ||||||
| N50 | 2395 | 3608 | 1188 | 1570 | 1484 | 2511 |
| Coverage (%) | 95.06 | 93.40 | 93.80 | 88.97 | 94.56 | 86.53 |
| No. of contigs | 10 724 | 8448 | 48 409 | 35 796 | 123 583 | 85 837 |
| No. of bases | 8 199 665 | 8 151 930 | 31 072 592 | 26 252 905 | 94 857 363 | 81 759 539 |
| No. of error contigs | 15 | 20 | 45 | 42 | 426 | 389 |
| No. of error bases | 24 035 | 22 733 | 43 112 | 39 118 | 173 856 | 163 246 |
| Time (min) | 38.3 | 37.5 | 147.7 | 145.7 | 319.0 | 323.1 |
| SOAPdenovo | ||||||
| N50 | 7457 | 8233 | 2502 | 2171 | 1806 | 1351 |
| Coverage (%) | 93.57 | 93.91 | 94.97 | 93.77 | 97.27 | 87.37 |
| No. of contigs | 7742 | 7566 | 42 158 | 41 446 | 124 756 | 116 723 |
| No. of bases | 6 253 699 | 6 210 923 | 25 421 067 | 24 160 482 | 80 261 153 | 63 438 459 |
| No. of error contigs | 6 | 7 | 20 | 26 | 94 | 159 |
| No. of error bases | 16 337 | 28 987 | 57 283 | 60 601 | 185 439 | 160 779 |
| Time (min) | 11.3 | 10.2 | 39.8 | 37.6 | 84.7 | 85.1 |
For low-complexity datasets, two species, each having at least two subspecies within a genus, were selected. Length-75 sequencing reads were sampled from the selected reference genome at 30× depth. In all datasets, the error rate and insert distance were set to 1% and 250, respectively. Medium-complexity and high-complexity datasets were generated similarly according to the properties shown in Table 3. Two distributions of abundance ratios were used to generate simulated sequencing reads. The first one was uniform distribution, for a situation without uneven abundance ratios. The other one was log normal distribution, since some research on species richness estimation in metagenomic data shows that log normal distribution fits the data well (Hong et al., 2006; Youssef and Elshahed, 2008).
For comparison, Velvet, Abyss, SOAPdenovo and Meta-IDBA were executed on the above three datasets. In all experiments, the k-value of the de Bruijn graph was set to 50. Default values were used for all assemblers, except option ‘-M 3 -F’ is activated to merge similar regions for SOAPdenovo. The quality of assembler output was measured in three aspects on resultant contigs: N50 for contiguity, coverage for completeness and number of erroneous bases for accuracy. If the assembler generates scaffolds with wildcard nucleotide symbol ‘N’ inside, the ‘N’ will be removed to split the scaffolds into contigs. Correctness was checked by alignment, and a contig is considered as correct if it can be aligned to a reference with 95% similarity by BLAT (Kent, 2002). The 95% similarity means that the sum of mismatch bases and the in-del length should not be >5% of a contig. In the calculation of N50 and coverage, only correct contigs are considered. For Meta-IDBA, one more measure, is considered. The component accuracy indicates how accurate graph separation method works. A component is considered as correct only if 95% of the bases, inside one component, are actually from the same species. The experimental results for datasets of different complexities are shown in Figures 3–5, and the average values of all measurements are summarized in Table 3.
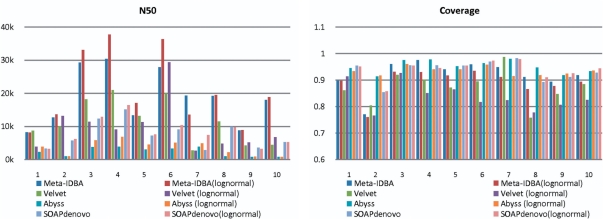
Fig. 3.
Experiment results of low-complexity datasets.
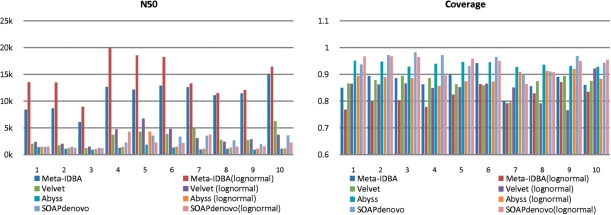
Fig. 4.
Experiment results of medium-complexity datasets.
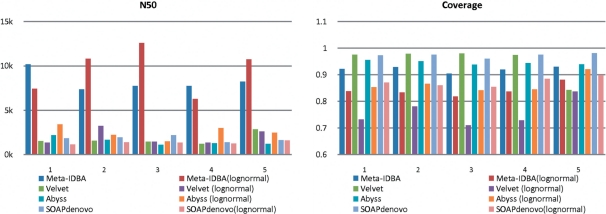
Fig. 5.
Experiment results of high-complexity datasets.
For the low-complexity dataset, Meta-IDBA had the longest N50 in most cases, i.e. about ≥1.5 times that of Velvet, Abyss and SOAPdenovo. In a few cases, Velvet had similar or better N50 than Meta-IDBA, because the graph separation algorithm in Meta-IDBA partitioned some parts of the graph into components which could be better handled by bubble merging for low-complexity datasets. When considering coverage, all the assemblers had similar performances. In general, Abyss and SOAPdenovo had slightly better coverage and produced more contigs. Velvet and Meta-IDBA had similar numbers of erroneous contigs, which are a little more than Abyss and SOAPdenovo generated. Component accuracy showed that Meta-IDBA can separate contigs from different species accurately. When considering log normal distribution, it is interesting to note that N50 of the assembly results increased in some cases. This is because some subspecies, having low coverage after introducing different abundance ratios, can be considered removed from the dataset, reducing the complexity of the dataset. Consequently, the resultant contigs from the other similar species did not form complicated components. Overall, the N50 of Meta-IDBA for the log normal distribution was always better than that of the uniform distribution, because we applied the de Bruijn graph based assembler IDBA (Peng et al., 2010), which does not rely on the read coverage of the genome very much.
For medium-complexity datasets, Meta-IDBA always gave the best N50 among all assemblers. This means Meta-IDBA is able to group similar regions from different subspecies of the same species together effectively when the complexity of graph increases, while the bubble merging method failed in this situation. The performances of all assemblers in coverage and accuracy were similar to those of low-complexity data.
The performance of the high-complexity datasets further confirmed that Meta-IDBA can handle complicated graphs. The N50 of the other assemblers is much shorter, while that of Meta-IDBA only decreased slightly. The other measures remained similar as before.
All the experiments were executed in an eight-core machine with 144 GB memory. The runtime of Meta-IDBA and Velvet shown in Table 3 is more or less the same, but shorter than that of Abyss.
3.2 Real data
A real metagenomic sequencing dataset (SRX024329) from NCBI (http://www.ncbi.nlm.nih.gov/) was used to evaluate the performance of all assemblers in practice. It is a human metagenome sample from the G_DNA_Tongue dorsum of a female participant in the dbGaP study ‘HMP Core Microbiome Sampling Protocol A (HMP-A)’. Meta-IDBA provided the longest contigs among the assemblers (Table 4). It is difficult to access the accuracy of these results, since there are no references for most of the species in the real dataset. However, based on the results of simulated experiments, we have high confidence that Meta-IDBA can produce correct contigs and components.
Table 4.
Experimental results of real data
| Assembler | No. of contigs | Total bases | N50 | Maximum |
|---|---|---|---|---|
| Meta-IDBA | 121 924 | 74 493 748 | 2380 | 371 462 |
| Velvet | 199 310 | 80 297 709 | 738 | 207 709 |
| Abyss | 203 983 | 102 106 241 | 956 | 121 166 |
| SOAPdenovo | 271 500 | 110 655 983 | 591 | 367 374 |
3.3 Multiple alignment of component
In the last step of Meta-IDBA, multiple alignments are performed among contigs in each component. Because of the restrictive properties of the components, all contigs in one component represent similar subsequences in some subspecies of the same species. Figure 6 presents part of the multiple alignment of a component from five E.coli subspecies, which shows similar contigs with a small number of variants. We can confirm confidently that the multiple alignment, as shown in Figure 6, represents contigs from similar subspecies.
Fig. 6.
Multiple alignment of a component in five E.coli subspecies. Consensus is shown in the first row. Contigs are separated by spaces. The conserved nucleotides are represented by dots. The difference between contigs and consensus is highlighted.
4 DISCUSSION AND CONCLUSION
We tackled the assembly difficulty caused by the polymorphism in similar species in the metagenomic environment. Similar regions between species make the de Bruijn graph more complicated. Based on the observation that the genomes of subspecies from the same species share much more common k-mers than the genomes of subspecies from different species, we define the component to present the similar regions among subspecies from the same species. The assembly problem can then be modeled as a graph partition problem. After that, we designed an algorithm to identify the components from de Bruijn graph. Finally, paired-end reads are used to further connect components together.
Experiments on datasets of different complexities showed that Meta-IDBA can usually produce the longest contigs with similar accuracy and coverage when compared with other assemblers, especially for high-complexity datasets which contain more branches since there are more genomes in the dataset. Besides reconstructing longer contig, Meta-IDBA provides a multiple alignment of similar contigs from different subspecies in the same species, which represents the variants among genomes of these subspecies. The multiple alignment may be used to study the structural variations of genomes of different subspecies or determine conserved regions, which have biological functions for the subspecies. As the multiple alignments are constructed by a greedy algorithm, further study on finding the optimal multiple alignment may improve the accuracy and the usage of Meta-IDBA in analyzing metagenomic data. At present, Meta-IDBA cannot reconstruct the contigs of each single subspecies, because their genomes have a lot of common regions. Further study on using pair-end information and read coverage for reconstructing these contigs will be carried out.
There are cases in which Meta-IDBA fails to separate reads from different species into components. One case is in low- complexity dataset, which consists of Streptococcus pyogenes and S.dysgalactiae. The k-mer similarity of these two species is 17.69%, which is relatively higher than the other pair of genomes (~1%). On the other hand, the component accuracy of this dataset is 93%, which means many reads in the components are shared by these two species and some components represent similar regions of these two species. Therefore, it might not be necessary to separate the reads in these components, since these components may represent regions with the significant biological functions necessary for both species.
For metagenomic dataset with uneven abundance ratios, because the IDBA genome assembler does not depend much on coverage to create the de Bruijn graph, the change in abundance ratios will not affect its performance too much. If each species is sampled with high enough coverage (the required coverage depends on the error rate and read length), they can be assembled by Meta-IDBA. However, uneven abundance ratios will affect the sampling rates of reads of different species, but the difference in sampling rates can also provide information to separate the reads sampled from species with low abundance ratios and those from species with high abundance ratios (Wu and Ye, 2010). More research should be performed for studying how to make use of this information to improve the accuracy of Meta-IDBA.
Since our graph partition algorithm cannot distinguish erroneous edges and correct edges inside the graph, it relies very much on the quality of the de Bruijn graph generated by the assembler. If there are many false positive edges, the graph may be partitioned into many small components. Similarly, if there are many species (in practice, there are many more species (>1000) than those used in our simulation), there would be more edges in the graph, which also leads to many small components. As shown in Table 1, there is a big gap between the assembly results of single species and metagenomic. Much can be done to improve the quality of metagenomic assembly.
Funding: This research was supported in parts by HKU Genomics SRT funding, GRF HKU 711611 and GRF HKU 719709E.
Conflict of Interest: none declared.
REFERENCES
- Chaisson M.J., et al. De novo fragment assembly with short mate-paired reads: does the read length matter? Genome Res. 2009;19:336–346. doi: 10.1101/gr.079053.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M.J., Pevzner P.A. Short read fragment assembly of bacterial genomes. Genome Res. 2008;18:324–330. doi: 10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fofanov Y., et al. How independent are the appearances of n-mers in different genomes? Bioinformatics. 2004;20:2421–2428. doi: 10.1093/bioinformatics/bth266. [DOI] [PubMed] [Google Scholar]
- Gnerre S., et al. Assisted assembly: how to improve a de novo genome assembly by using related species. Genome Biol. 2009;10:R88. doi: 10.1186/gb-2009-10-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.H., et al. Predicting microbial species richness. Proc. Natl Acad. Sci. USA. 2006;103:117–122. doi: 10.1073/pnas.0507245102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., et al. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause L., et al. Phylogenetic classification of short environmental DNA fragments. Nucleic Acids Res. 2008;36:2230–2239. doi: 10.1093/nar/gkn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavromatis K., et al. Use of simulated datasets to evaluate the fidelity of metagenomic processing methods. Nat. Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- Peng Y., et al. IDBA—A Practical Iterative de Bruijn Graph De Novo Assembler. Lisbon: RECOMB; 2010. [Google Scholar]
- Pevzner P.A., et al. An Eulerian path approach to DNA fragment assembly. Proc. Natl Acad. Sci. USA. 2001;98:9748–9753. doi: 10.1073/pnas.171285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop M. Genome assembly reborn: recent computational challenges. Brief. Bioinformatics. 2009;10:354–366. doi: 10.1093/bib/bbp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., et al. NCBI reference sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J.T., et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooley J.C., et al. A primer on metagenomics. PLoS Comput. Biol. 2010;6:e1000667. doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.-W., Ye Y. A Novel Abundance-Based Algorithm for Binning Metagenomic Sequences Using l-Tuples. Lisbon: RECOMB; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., et al. Proceedings of the First ACM International Conference on Bioinformatics and Computational Biology. Niagara Falls, New York: ACM; 2010. MetaCluster: unsupervised binning of environmental genomic fragments and taxonomic annotation; pp. 170–179. [Google Scholar]
- Youssef N.H., Elshahed M.S. Species richness in soil bacterial communities: a proposed approach to overcome sample size bias. J. Microbiol. Methods. 2008;75:86–91. doi: 10.1016/j.mimet.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D.R., et al. Pebble and rock band: heuristic resolution of repeats and scaffolding in the velvet short-read de novo assembler. PLoS One. 2009;4:e8407. doi: 10.1371/journal.pone.0008407. [DOI] [PMC free article] [PubMed] [Google Scholar]