Abstract
Motivation: Previous research in the biomedical text-mining domain has historically been limited to titles, abstracts and metadata available in MEDLINE records. Recent research initiatives such as TREC Genomics and BioCreAtIvE strongly point to the merits of moving beyond abstracts and into the realm of full texts. Full texts are, however, more expensive to process not only in terms of resources needed but also in terms of accuracy. Since full texts contain embellishments that elaborate, contextualize, contrast, supplement, etc., there is greater risk for false positives. Motivated by this, we explore an approach that offers a compromise between the extremes of abstracts and full texts. Specifically, we create reduced versions of full text documents that contain only important portions. In the long-term, our goal is to explore the use of such summaries for functions such as document retrieval and information extraction. Here, we focus on designing summarization strategies. In particular, we explore the use of MeSH terms, manually assigned to documents by trained annotators, as clues to select important text segments from the full text documents.
Results: Our experiments confirm the ability of our approach to pick the important text portions. Using the ROUGE measures for evaluation, we were able to achieve maximum ROUGE-1, ROUGE-2 and ROUGE-SU4 F-scores of 0.4150, 0.1435 and 0.1782, respectively, for our MeSH term-based method versus the maximum baseline scores of 0.3815, 0.1353 and 0.1428, respectively. Using a MeSH profile-based strategy, we were able to achieve maximum ROUGE F-scores of 0.4320, 0.1497 and 0.1887, respectively. Human evaluation of the baselines and our proposed strategies further corroborates the ability of our method to select important sentences from the full texts.
Contact: sanmitra-bhattacharya@uiowa.edu; padmini-srinivasan@uiowa.edu
1 INTRODUCTION
Information systems in the biomedical domain, serving goals such as information extraction and retrieval, have historically been designed in the context of MEDLINE records. These records, limited to titles and abstracts and some metadata, are very different from full text documents. Recent papers in BMC Bioinformatics elaborate on these differences (Cohen et al., 2010; Lin, 2009). Moreover, recent research initiatives in bioinformatics such as TREC Genomics (http://ir.ohsu.edu/genomics/) and BioCreAtIvE (http://www.biocreative.org/) strongly point to the merits of moving beyond abstracts and taking advantage of full texts. But full texts are more expensive to process not only in terms of their much larger sizes and their often varying structural characteristics but also in terms of what it takes to achieve high levels of accuracy. Since full texts contain embellishments that elaborate, contextualize, contrast, supplement, etc., the core ideas of a paper, there is greater opportunity for false positives. For example in a retrieval context, a full text may be retrieved because of query terms being present in (relatively) unimportant sections. In an information extraction context a gene, though correctly identified in a document, may be of low interest in terms of the document's core emphasis.
In this article, we explore document summarization as a compromise between the two extremes of abstracts versus full texts. Specifically, we create a reduced version of each full text document that contains only its important portions. This reduced version may be viewed as a ‘summary’ but our interest is not to generate a human readable summary, rather it is to have an intermediate representation that may later be used for algorithmic functions such as to serve text retrieval and information extraction.
Our approach for full text reduction is novel, particularly in the context of various document summarization methods that have been explored. We capitalize on the MeSH terms present in MEDLINE records. Our hypothesis is that MeSH terms may be used to identify the important portions of full text documents. In essence, we suggest that the MeSH indexing, accomplished by trained indexers, offers a useful window into full text. We treat MeSH terms as high-quality hooks used to select important text segments in the full document. This idea, if it works, will give us a general strategy for reducing biomedical documents to their core portions.
Our long-term motivation for building reduced versions of full texts is to use these as document representations in biomedical systems designed for retrieval, information extraction and other high-level functions. Several use cases may be identified in the bioinformatics arena. One obvious use case is document retrieval. Indeed our own efforts with GeneDocs (Sehgal and Srinivasan, 2006), a document retrieval system for gene queries, and our efforts in BioCreAtIvE III (Bhattacharya et al., 2010a; b), motivate this research. These also offer retrieval test beds for follow-up research. Another use case is in information extraction, an area that has attracted significant attention. A third use case is in text mining, particularly mining for new ideas and hypotheses. Again, our algorithms here offer test beds for future work (Srinivasan, 2004; Srinivasan and Libbus, 2004). Our document summarization methods are also general enough to be applied to any biomedical domain that employs MeSH indexing. So it may be tested and utilized by other researchers in the context of their text-based biomedical applications.
Given the successful efforts of archival organizations such as PubMed Central and the Open Access initiative, our goal is to explore a new basis for biomedical systems; one that utilizes reduced versions of full texts that contain only their core portions. We hope to show in future work that these reduced versions lead to more efficient and effective results than using either abstracts or full-text-based collections.
We first present our MeSH-based methods for extracting core portions of full text documents (Section 3). We then compare these extracts with the abstracts and with summaries generated at random and summaries generated by MEAD, a well-accepted text summarization system. Comparisons are done in multiple ways including by experts assessing the importance of selected sentences (Sections 5 and 6). We also perform error analysis (Section 7) that will inform our future research. Related research and conclusions are in Sections 2 and 8, respectively.
2 RELATED RESEARCH
Text summarization can be broadly divided into two main categories, namely, extractive summarization and abstractive summarization. Extractive summarization is where the most important sentences are selected based on the presence of important terms or concepts. On the other hand, abstractive summarization deals with the construction of novel sentences that represent the important concepts in a more concise way. Due to the underlying challenges of abstractive summarization, more effort has been directed toward the extractive summarization research as seen in TREC (http://trec.nist.gov/), DUC (http://duc.nist.gov/) and TAC (http://www.nist.gov/tac/) conferences. However, these summarization efforts focus mainly on newswire data, which is significantly different from scientific literature.
Most of the early work in automated text summarization was directed toward single document summaries, (Brandow et al., 1995; Johnson et al., 1997; Luhn, 1958) and typically used statistical measures for extracting sentences. Machine Learning-based techniques like Naïve-Bayes method (Aone et al., 1999); Kupiec et al., 1995) and Hidden Markov Models (Conroy and O'leary, 2001) were also used for sentence extraction from documents. A notable shift was seen with the arrival of knowledge-based domain-specific and domain-independent summarization systems. These could handle both single and multidocument summarization tasks (Radev and McKeown, 1998; Radev et al., 2004b).
One of the first applications of text summarization in the biomedical domain was for generating graphical abstractive summarization based on the semantic interpretation of the biomedical text (Fiszman et al., 2004). Chiang et al. (2006) developed a text-mining-based gene information summarization technique for a cluster of genes based on information extracted from PubMed abstracts followed by processing using a Finite State Automaton. Yoo et al. (2007) propose a graph-based document clustering and summarization technique, which employs biomedical ontology-enriched graphs to represent document content. Reeve et al. (2007) used concept chaining to link together semantically related concepts identified from the Unified Medical Language System (UMLS) Metathesaurus and used these chains to extract sentences for summarization. Ling et al. (2007) propose a technique for semi-structured gene summaries from the literature based on sentence extraction depending upon several semantic aspects of genes. Their proposed probabilistic language model approach outperforms other baseline methods. Jin et al. (2009) propose another method for generating gene summaries from MEDLINE abstracts based on selection of information-rich sentences followed by inter-sentential redundancy removal. Recently, Agarwal and Yu (2009) proposed an extractive summarization technique for figures in biomedical literature which employs a sentence classification system for selection of sentences from the full text. There are also efforts to identify relevant text passages from patient records. For example, Cohen (2008) explores a hotspot identification method using specific words of interest. However, patient records offer a significantly different ‘document context’ for summarization research.
None of these methods directly employ the MeSH index terms for sentence extraction and thereby summarization from full text. This aspect is a key novel contribution of our research.
3 MESH-BASED SUMMARIZATION
Merits of MeSH have been studied extensively in the context for which it was designed, i.e. for information retrieval and text classification. MeSH has also been studied in several other research contexts such as for automatic annotation of documents [e.g. Trieschnigg et al. (2009)], for MEDLINE record clustering [e.g. Zhu et al. (2009)], etc. Here, we see an opportunity to use MeSH terms assigned to MEDLINE records in a novel way; namely to reduce the corresponding full text documents into summaries. Given the intellectual effort invested in MeSH-based indexing, we see MeSH as a solid basis for this reduction process. In our approach, a text segment is considered important if it relates to the MeSH terms used to index the document. A text segment may be an individual sentence or a larger unit such as a paragraph, though we only explore the former in this article. We assess the relationship between a sentence and the MeSH terms using similarity functions. Segments that are most similar to the MeSH terms are selected for inclusion in the reduced version. Essentially, we treat the MeSH terms as high-quality hooks that control the selection of sentences from the full document. We explore two different approaches for using MeSH terms in this manner; we refer to these as MeSH Term and MeSH Profile approaches. We present these next.
Note too that the amount of reduction can be varied. We can aim for reduced versions that are of a specified length (e.g. 500 words or 15 sentences) or that represent a percentage of reduction (60% reduction of full text). Here, we explore optimal values in terms of number of sentences.
3.1 MeSH term approach
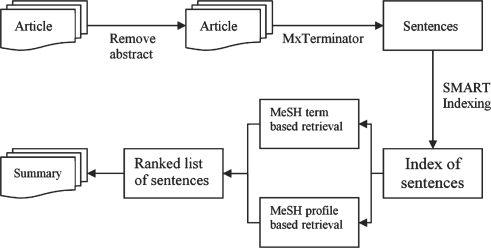
Starting with an individual full text document to reduce, we remove the abstract and break the rest apart into sentences using MxTerminator (Reynar and Ratnaparkhi, 1997), followed by some post-processing. We then index the collection of sentences using the SMART retrieval system (Salton, 1971). Note each sentence is regarded as an independent record. We then create a query consisting of the MeSH terms marked as major index terms/topics (marked with asterisks) in the corresponding MEDLINE record. We consider only the descriptor phrases of those terms. Finally, we run retrieval with this query, which returns a ranking of sentences. The top N (a parameter) sentences are selected for our document summary. Figure 1 shows the underlying algorithm. While indexing, term features are weighted using the standard atc approach; a for augmented term frequency weights, t for inverse document (in this case sentence) frequency weight and c for cosine normalization (Salton, 1971).
Fig. 1.
A simplified diagram of the summarization system.
3.2 MeSH profile approach
This approach addresses the vocabulary mismatch problem. MeSH terms come from a controlled vocabulary and so may not appear in the full text document in their exact form. There may be text segments that address the concepts underlying the MeSH terms without containing the terms directly. In this approach, we first find free-text terms that are related to a given MeSH term. These terms form the free-text profile of the MeSH term. The profile terms along with the starting MeSH term may be used to select sentences in the same way as described previously (Fig. 1). Essentially, this approach expands the MeSH terms with free-text terms.
There are two broad directions for finding related terms. The first is through an authoritative source such as the Unified Medical Language System, UMLS (http://www.nlm.nih.gov/research/umls/). The second direction, the one explored here, is to identify expansion terms from a relevant text corpus. We will explore the first direction in future work. Here, we build profiles from MEDLINE.
In essence, for each MeSH term, we retrieve up to 2500 MEDLINE records that are indexed by the term. We then create a term vector for each record and then calculate the average term vector for this document set; this we refer to as the free-text profile of the MeSH term. Terms are weighted statistically using the atc strategy described before. The resulting free-text profile is essentially a ranked set of free-text terms where the ranking is by calculated term weight. Since we are interested in using these profiles for sentence selection, we limit the MeSH term profiles to a few top ranking terms (parameter M). In preliminary experiments (with five full text documents), we explored summaries using profiles of top 5, 10, 15 and 20 terms. From these, we determined that (on average) MeSH profiles of top 5 and 10 free-text terms are the most interesting. Thus, we present results for M=5 and M=10 in our work here.
MeSH profile: varying length—Instead of limiting a MeSH term profile using an absolute threshold such as the top 5 or 10 free-text words, there may be a better way to determine profile length. For instance, it may be that some MeSH terms are better with a longer profile than others. We explore this notion via a retrieval experiment and it is the basis of our last strategy called Variable Length.
In essence, for each MeSH term, we identify two sets of MEDLINE documents. The first, a positive set (POS), includes up to 2500 records that have been indexed with this term. The second, a negative set (NEG), includes up to 2500 records that might have the individual words of the MeSH term but are not indexed by the MeSH term. As an example, for the MeSH term Adaptation, Physiological, a negative record may have the words adaptation and physiological in the title or/and abstract but the record itself is not indexed by the MeSH term.
Given a POS set and a NEG set of MEDLINE records as described above, we may test profiles of varying lengths on their ability to separate the two sets. In other words, if we use the profile terms as a query we should be able to rank the positive records above the negative records.1 In this manner, we may test profiles of different sizes and identify the best size for a given MeSH term. Note that in all these retrieval runs, the records used to build the profile in the first place were a different sample of POS documents. For each term, we explored profiles of length N=5, 10, 15 and 20 free-text terms. We then selected the best N for the term. Table 1 below shows the distribution of M (the number of expansion terms in the profile) over the terms in our dataset. It is interesting to observe that for 19% of the terms the best strategy is to not expand it with a free-text profile. Examples of such terms include Actins, Brain, DNA, Mutation, Oxygen, Prions, RNA Splicing, etc.
Table 1.
Distribution of MeSH terms for variable length profile cutoffs
| Percentage of MeSH terms | No. of expansion terms in best profile |
|---|---|
| 19% | 0 |
| 28% | 5 |
| 22% | 10 |
| 14% | 15 |
| 17% | 20 |
4 METHODS
4.1 Dataset
The document set used in our experiments comprises of 100 full text articles in HTML format selected from the full text article collection of the TREC 2006 Genomics Track (Hersh et al., 2006). MEDLINE records for these 100 documents were obtained from PubMed using application programming interfaces (API) provided by NCBI. For our experiments, we consider only the MeSH major terms, which represent the major topics covered in an article. For our document set, the number of MeSH major terms used to index varies between 1 and 10. The selected articles were verified for the presence of abstracts, which are later used to evaluate our experiments. We selected articles containing at least one passage which had been judged relevant to a topic used in TREC 2006 Genomics Track (Hersh et al., 2006). This allows us to run a small-scale retrieval experiment with the reduced set of documents.
4.2 Data preprocessing
The full text HTML documents were pre-processed to remove all HTML tags. These were then segmented into sentences using a maximum entropy-based sentence boundary detector, MxTerminator (Reynar and Ratnaparkhi, 1997). However, since MxTerminator was trained on a Wall Street Journal corpus, we had to follow additional steps to remove errors. These originated from incorrect sentence splitting based on words or phrases which are typical of scientific literature (et al., Fig., Ref., S.E., etc.).
4.3 Indexing and term profiles
Documents were reduced to generate summaries one at a time. For each document, the sentences obtained after the preprocessing step were indexed with SMART. Each sentence was treated as an individual record. The abstracts were not included. MeSH term profiles were built for the expanded strategy as described in Section 3.2.
4.4 MEAD baselines
Baselines were built using MEAD (Radev et al., 2004a), a state-of-the-art single and multidocument summarization system. MEAD is a centroid-based extractive summarizer which selects the most important sentences from a set of sentences based on the linear combination of three features, namely, the centroid score, the position score and the overlap-with-first score (Radev et al., 2001). We select MEAD as it has been used extensively both as a platform for developing summarization systems and also as a baseline system for testing novel summarizers. As a first baseline system, we use MEAD's random summarizer that randomly selects sentences to produce a summary of a given length. We repeat this randomization 100 times for each document and report the average of the evaluation scores (MEAD Random). As a second baseline system, we create true summaries of a fixed length using MEAD with its default settings (MEAD True).
4.5 Evaluation measures
We evaluate our system-generated summaries using the abstracts of the articles as the gold standard. Ideally, we would have gold standard summaries of varying lengths to assess system-generated summaries of varying lengths. However, since these are not available, we are constrained to using the readily available abstracts as the gold standard. It is important to note here that although this compromise is made for the evaluation we do not consider our summaries as surrogate abstracts. In fact, the more interesting summaries are likely to be those that are larger than the abstract and that augment the information content of the abstract. But given the practical limitations, we assess our system-generated summaries of varying lengths with the abstracts. We are also, therefore, limited to testing summaries of length close to that of the abstract. If these summaries turn out to be of high quality, then we are encouraged to explore larger sized summaries.
For our evaluations, we use ROUGE (Lin, 2004), an n-gram based measure. ROUGE compares the quality of a computer-generated summary versus a human-generated summary and is a widely accepted metric for automatic evaluation of summarization tasks (DUC 2004-2007 and TAC 2008-2010). In our experiments, we use the ROUGE-1, ROUGE-2 and ROUGE-SU4 F-scores. ROUGE-1 measures the overlap of unigrams between the two summaries while ROUGE-2 measures the overlap of bigrams. ROUGE-SU4 measures the overlap of skip-bigrams with unigram scoring for a maximum skip distance of four. Skip-bigram measures the overlap of any pair of words in their sentence order permitting arbitrary gaps. Unigram and bigram statistics have been shown to have the highest correlation with human assessments (Lin and Hovy, 2003). The ROUGE measures we compute are based on the parameters described for the DUC 2007 Summarization Task (http://duc.nist.gov/duc2007/tasks.html). However, we also evaluate under more strict conditions, namely using unstemmed words and after removing stop words. Overall, we found that the stricter settings resulted in a systematic drop of ROUGE scores although the relative order of performance was preserved among the various strategies tested. Thus, the scores we present in this article are limited to the F-scores of the ROUGE-1, ROUGE-2 and ROUGE-SU4 without stemming or stop word removal.
5 RESULTS
5.1 MeSH term-based summarization
Tables 2–5 present the ROUGE F-scores for MeSH term-based summaries and the MEAD summaries for various summary lengths. We can see that MeSH term-based summaries outperform the other summaries. As mentioned before, MEAD Random was run 100 times for each document and average scores are reported here. (Note that the SD of all the scores for varying number of sentences in these Random summaries ranged between 0.002 and 0.005.)
Table 2.
ROUGE F-Scores for baselines and MeSH term approach where number of sentences in summary = number of sentences in abstract
| MEAD Random | MEAD True | MeSH Term | |
|---|---|---|---|
| ROUGE-1 | 0.3781 | 0.3815 | 0.4150 |
| ROUGE-2 | 0.1062 | 0.1353 | 0.1435 |
| ROUGE-SU4 | 0.1428 | 0.1411 | 0.1782 |
Table 3.
ROUGE F-Scores for baselines and MeSH term approach where number of sentences in summary = 5
| MEAD Random | MEAD True | MeSH Term | |
|---|---|---|---|
| ROUGE-1 | 0.3074 | 0.3224 | 0.3385 |
| ROUGE-2 | 0.0774 | 0.0873 | 0.1126 |
| ROUGE-SU4 | 0.0884 | 0.1005 | 0.1094 |
Table 4.
ROUGE F-Scores for baselines and MeSH term approach where number of sentences in summary = 10
| MEAD Random | MEAD True | MeSH Term | |
|---|---|---|---|
| ROUGE-1 | 0.3731 | 0.3606 | 0.4083 |
| ROUGE-2 | 0.1031 | 0.1079 | 0.1401 |
| ROUGE-SU4 | 0.1317 | 0.1188 | 0.1665 |
Table 5.
ROUGE F-Scores for baselines and MeSH term approach where number of sentences in summary = 15
| MEAD Random | MEAD True | MeSH Term | |
|---|---|---|---|
| ROUGE-1 | 0.3662 | 0.3214 | 0.3878 |
| ROUGE-2 | 0.1127 | 0.1257 | 0.1441 |
| ROUGE-SU4 | 0.1217 | 0.0877 | 0.1424 |
MeSH term summaries are always the best, achieving the highest ROUGE-1, ROUGE-2 and ROUGE-SU4 scores of 0.4150, 0.1435 and 0.1782, respectively, when the summaries matched in number of sentences. ROUGE-1 and ROUGE-2 scores of MEAD True summary exceed the MEAD Random summary scores. Somewhat surprisingly, this is not true for the ROUGE-SU4 score.
When summary length is fixed to five sentences (Table 3), we see a drop in the overall ROUGE scores for all systems even though MeSH Term continues to outperform the other two. This drop can be attributed to the fact that the number of sentences was considerably smaller than average number of sentences in our gold standard abstracts (8.4 sentences).
For longer summaries containing 10 or 15 sentences, the ROUGE scores of the MeSH term-based strategy consistently surpass the scores of the other baseline strategies. While the ROUGE-2 scores of MEAD True summaries are better than the Random summaries, we found the MEAD Random summaries doing better than MEAD True summaries based on ROUGE-1 and ROUGE-SU4 scores. As noted by Ling et al. (2007) and Inouye, (2010), longer summaries tend to contain redundant information in which case evaluation methods might potentially favor a Random summary to a True summary downplaying the significance of the True summary.
For each of the different strategies, we also calculated the stopword removed and stopword removed with stemmed settings of ROUGE. The scores of both these stringent versions were found to be lower than the scores shown here but their relative orders were found to be identical with the stemmed version consistently scoring better than the other method.
Finally, the differences in ROUGE scores for the best-performing MEAD baseline summaries to the corresponding MeSH term-based summaries (Table 2) were determined to be statistically significant (using the Wilcoxon Signed Rank Test) with P<0.01.
5.2 MeSH Term versus MeSH Profile
Tables 6–9 show the results of ROUGE evaluations of the MeSH profile-based strategy compared with the MeSH term-based summarization. We observe that the ROUGE scores of MeSH profile-based summaries almost always exceed the scores for the MeSH term-based summaries. As with the results shown in Table 2, the best scores are achieved when the number of sentences of summaries is equal to the number of sentences of the abstract of the corresponding article.
Table 6.
ROUGE F-Scores for MeSH term and MeSH profile approaches where number of sentences in summary = number of sentences in abstract
| MeSH Term | MeSH Profiles |
|||
|---|---|---|---|---|
| 5-Terms | 10-Terms | VL | ||
| ROUGE-1 | 0.4150 | 0.4201 | 0.4220 | 0.4320 |
| ROUGE-2 | 0.1435 | 0.1375 | 0.1408 | 0.1497 |
| ROUGE-SU4 | 0.1782 | 0.1743 | 0.1753 | 0.1887 |
5-Terms, selecting top five terms from MeSH Profiles; 10-Terms, selecting top 10 terms from MeSH Profiles; VL, variable length; N is set specific to the MeSH Term.
Table 7.
ROUGE F-Scores for MeSH term and MeSH profile approaches where number of sentences in summary = 5
| MeSH Term | MeSH Profiles |
|||
|---|---|---|---|---|
| 5-Terms | 10-Terms | VL | ||
| ROUGE-1 | 0.3385 | 0.3538 | 0.3607 | 0.3661 |
| ROUGE-2 | 0.1126 | 0.1154 | 0.1169 | 0.1206 |
| ROUGE-SU4 | 0.1094 | 0.1165 | 0.1201 | 0.1246 |
5-Terms, selecting top five terms from MeSH Profiles; 10-Terms, selecting top 10 terms from MeSH Profiles; VL, variable length; N is set specific to the MeSH Term.
Table 8.
ROUGE F-Scores for MeSH term and MeSH profile approaches where number of sentences in summary = 10
| MeSH Term | MeSH Profiles |
|||
|---|---|---|---|---|
| 5-Terms | 10-Terms | VL | ||
| ROUGE-1 | 0.4083 | 0.4100 | 0.41293 | 0.4245 |
| ROUGE-2 | 0.1401 | 0.1342 | 0.13921 | 0.1485 |
| ROUGE-SU4 | 0.1665 | 0.1595 | 0.16265 | 0.1748 |
5-Terms, selecting top five terms from MeSH Profiles; 10-Terms, selecting top 10 terms from MeSH Profiles; VL, variable length; N is set specific to the MeSH Term.
Table 9.
ROUGE F-Scores for MeSH term and MeSH profile approaches where number of sentences in summary = 15
| MeSH Term | MeSH Profiles |
|||
|---|---|---|---|---|
| 5-Terms | 10-Terms | VL | ||
| ROUGE-1 | 0.3878 | 0.3893 | 0.3878 | 0.3954 |
| ROUGE-2 | 0.1441 | 0.1397 | 0.1431 | 0.1486 |
| ROUGE-SU4 | 0.1424 | 0.1375 | 0.1359 | 0.1428 |
5-Terms, selecting top five terms from MeSH Profiles; 10-Terms, selecting top 10 terms from MeSH Profiles; VL, variable length; N is set specific to the MeSH Term.
Comparing across the different profile lengths, top 5, top 10 and variable length (VL), we find that VL generates the best results. The ROUGE-1, ROUGE-2 and ROUGE-SU4 scores were 0.4320, 0.1497 and 0.1887, respectively, for this strategy.
We computed the statistical significance of the difference in ROUGE scores (Table 6) for the best-performing MeSH major term-based strategy against the best-performing profile-based strategy (variable length) and found the differences to be statistically significant with P<0.01 (using the Wilcoxon Signed Rank Test).
6 HUMAN EVALUATIONS
Besides the automatic evaluations with ROUGE, we ran a small-scale experiment with human judgments. Three assessors (2 PhDs and one MS in the biomedical sciences area) were asked to rate a set of sentences for each document. In essence, they were asked to rate the importance of a sentence in terms of selecting it if they were to write a summary of the document. They could also rate based on the presence of words or concepts that are important to the article. The rating scale was 1 to 5 with 5 indicating very important and 1 not at all important. Assessors were given the document (with the abstract removed) to read before rating the sentences. The aim of this human evaluation was to get a feel for the merits of our methods rather than to get quantitative assessment of the strategies tested.
For each document, we pooled the sentences present in the two baseline summaries (MEAD Random2 and MEAD True), and the sentences present in the best MeSH Term and MeSH Profile summaries. More specifically, these summaries correspond to the summaries evaluated using ROUGE in the MEAD Random, MEAD True, MeSH Term and MeSH Profiles (VL) columns of Tables 2 and 6. The summaries in each case were restricted to the length of the abstract in number of sentences. These summaries were chosen as they represent the best cases in terms of ROUGE evaluations. The sentences of the abstract (our gold standard) were also added to this pool. Sentences in a pool were given to the assessors in random order. The number of candidate sentences for each article was at most five times the number of sentences in its abstract. The number of sentences in the pool is influenced by the extent to which the methods identify overlapping sentences. We found that the overlap is usually small. The highest overlap was between the MeSH Term and MeSH Profile (VL) strategies (29%) with the second highest between MeSH Profile (VL) and MEAD True (only 7%) strategies. If we ignore MEAD Random, the least overlap was between MeSH Term and MEAD True (6%).
We compare summaries by calculating Accuracy for each summary as in Equation (1). Since the maximum score (rating) for each sentence is five, the maximum sum of sentence scores in a summary, i.e. denominator in Equation (1), is five times the length of the summary.
| (1) |
In this equation, s denotes a sentence from a summary. A total of eight full text articles were selected for human evaluations. Each assessor was required to evaluate sentence sets for four articles. Two of the evaluators were given four non-overlapping articles and one of the evaluators was given four articles such that half of the articles match the first evaluator and the other half matches the second evaluator. This resulted in unique ratings for four articles and overlapped ratings for the other four articles. Average scores were taken for the four overlapping articles (Article numbers 1, 2, 3 and 4).
Randolph's kappa statistics (Randolph, 2005) was used to calculate the overall agreement among the assessors for the four overlapping articles. This was ideal in our case as Randolph's kappa does not assume that assessors have a constraint on distribution of cases across different categories. The percentage of overall agreement was found to be 74% with a fixed-marginal kappa of 0.647 and free-marginal kappa of 0.675. A total number of 530 non-unique sentences were rated by the assessors.
The accuracy scores for abstract and system-generated summaries are presented in Table 10. As expected, the mean accuracy score of 0.82 for the abstracts was higher than any of the candidate summaries of the same size. The MeSH term-based and MeSH profile-based summaries perform better than the MEAD True and MEAD Random summaries. Moreover, their averages are closer to that of the abstract than the MEAD averages. Our MeSH profile-based strategy performs better than our MeSH term-based strategy for all documents except for one where performance is the same. MEAD True summaries are rated higher than the random summaries except for two documents (Articles numbers 6 and 8). The SD of the MEAD Random summaries is also noticeably higher than any of the other systems.
Table 10.
Accuracy scores: human evaluations of abstract and summaries
| Article | Abs | MeSH Term | MeSH Profile | M-True | M-Random |
|---|---|---|---|---|---|
| 1 | 0.74 | 0.68 | 0.70 | 0.54 | 0.46 |
| 2 | 0.76 | 0.69 | 0.71 | 0.60 | 0.38 |
| 3 | 0.70 | 0.63 | 0.67 | 0.47 | 0.45 |
| 4 | 0.80 | 0.64 | 0.71 | 0.51 | 0.44 |
| 5 | 0.85 | 0.73 | 0.75 | 0.65 | 0.45 |
| 6 | 0.90 | 0.80 | 0.80 | 0.65 | 0.70 |
| 7 | 0.84 | 0.76 | 0.78 | 0.67 | 0.58 |
| 8 | 0.97 | 0.71 | 0.83 | 0.66 | 0.76 |
| Mean | 0.82 | 0.71 | 0.74 | 0.59 | 0.53 |
| SD | 0.09 | 0.06 | 0.06 | 0.08 | 0.14 |
Abs, abstract; M-True, MEAD True Summary; M-Random, MEAD Random Summary.
As a preliminary test of the value of our summaries in a retrieval application context, we conducted a small-scale retrieval study. We indexed our collection of 100 documents that had been reduced in this experiment. Since these were TREC 2006 Genomics documents and were selected such that each contained at least one passage relevant to a topic, we were able to run a retrieval experiment, albeit on a small scale. There were 26 topics (2 topics had no relevant passages) that had at least one relevant document in the dataset. We generated a basic disjunctive query for each of these topics and ran them against different indexed versions of the document collection. These versions were the collection represented by (i) abstracts alone, (ii) full texts (including abstracts), (iii) abstract-length summaries generated by MeSH-Profile (VL) strategy, (iv) 10-sentence long summaries generated by MeSH-Profile (VL) strategy, and (v) a hybrid strategy combining sentences from strategies (i) and (iv). The results are in Table 11. We find that retrieval using full text gives the best mean average precision (MAP) score (0.1585) followed by the MAP score (0.1521) using the hybrid version. As expected the MAP score from version (iii) performs worse than the abstract following the same pattern as observed in the human evaluations (Table 10). However, it is interesting to note that version (v) performs significantly better than the versions (i) and (iv) alone. These results point to the potential of our system-generated summary to go beyond abstracts and select important sentences that augment the information in the abstract. Moreover, just by adding 10 sentences to the abstract the performance is close to that achieved using full text. Although the scale of this experiment is small, overall the retrieval results are encouraging and offer preliminary support to our ideas. In future work, we will explore these retrieval experiments more rigorously.
Table 11.
Document-level MAP scores for retrieval experiments
| Abs | Full | MeSHProf-1 | MeSHProf-2 | Hybrid | |
|---|---|---|---|---|---|
| Doc-MAP | 0.1368 | 0.1585 | 0.1307 | 0.1412 | 0.1521 |
Abs, abstract; Full, full text; MeSHProf-1, abstract-length MeSH Profile summary; MeSHProf-2, 10-sentence long MeSH Profile summary; Hybrid, combined summary from Abs and MeSHProf-2; Doc-MAP, document-level MAP.
7 ERROR ANALYSIS
We conducted error analysis on summaries for eight documents. These were the same set of documents which we used for the human evaluations (Section 6). In each case, we looked at errors made by MeSH profile-based strategy (variable length) and those made by MEAD True strategies. In this analysis, the abstracts are treated as the gold standard. Our aim is to get ideas for improving upon our methods.
Errors of omission: First we identified the ‘important’ words in the abstract that are not in our generated summary but were somewhere in the full text of the document (set A). We regard as important a word that refers to a key biomedical entity such as a gene, protein, disease, drug or enzyme. General words such as ‘common’, ‘find’, ‘additional’, ‘attachment’, ‘cell’, etc. were ignored. We then manually looked for reasons why our summaries may have missed these words.
Across the 8 documents, there were a total of 26 missed terms for the MeSH profile-based strategy and 36 missed terms for the MEAD True summary. Reasons we were able to identify are given below in Table 12.
Table 12.
Types of omission errors made by the summarization strategies
| Error type | No. of errors | Example |
|---|---|---|
| Acronym/synonym | 24 | SCA1, spinocerebellar ataxia type 1; HPE, holoprosencephaly; mAb1C2, 1C2 antibody, etc. |
| Missed non-major MeSH term | 9 | Phosphoinositide phospholipase C, glycosylphosphatidylinositols, hedgehog proteins, etc. |
| Singular/plural | 6 | Ataxia, ataxias, Ca2+-channel, Ca2+-channels; β-subunit, β-subunits, etc. |
| Condensed terms/concepts | 5 | Orexin-A and B, Orexin A and Orexin B; SCA1/SCA2: SCA1 and SCA2; SCA3/MJD, SCA3 or MJD, etc. |
| Missed major MeSH term | 5 | Central nervous system, prions, Machado-Joseph disease, etc. |
| Other | 13 | Creutzfeldt–Jakob syndrome, cerebral cortex, autocatalytic cleavage, cholesterol modification, etc. |
Results shown here are from a set of eight sample documents.
The most common type of error identified was the presence of acronyms/synonyms in place of its expansion/official name and vice versa. It is often observed in biomedical literature that authors introduce a new term along with its acronym in the abstract and from then on use the acronym to identify that term. In such a case, the ROUGE measures will penalize our system as no term that matches the expanded term (from the abstract) can be found in our summary which is derived from the rest of the article.
The second most frequent error was due to missed non-major MeSH terms. In this article, we considered only the major MeSH terms for our experiments and this observation leads to our future directions. Of the 163 non-unique MeSH terms used to index the 8 documents we studied here, we used only 35 non-unique MeSH major terms for our purpose. This shows the scope of expansion of our research to incorporate more MeSH terms and thereby MeSH term-based profiles to further improve our summarization system.
The next most frequent error type was the mismatch between the singular and plural nouns. However, this error can be handled easily if we are less stringent with our evaluation and allow stemming for the ROUGE evaluations. This change will also likely increase the overall performance of our system.
The error with condensed terms and concepts is a challenging problem in our opinion. This type of error cannot be handled without actually modifying the text of the article and needs further research in evaluation methods that can implicitly handle these conditions.
We found that errors arising due to missed major MeSH terms are always unique to the MEAD True summary. We found this error in five of the eight MEAD True summaries. This observation again reinforced our confidence in the direction of MeSH term-based summarization.
We group all the other omissions in this category. Misspellings (e.g. Creutzfeld–Jakob Syndrome instead of Creutzfeldt–Jakob Syndrome) and omission of crucial information which are fundamental to the article would fall into this category (e.g. location of a disease or effects of a disease).
Errors of commission: Using a similar strategy, we identified ‘important’ words in the summaries that were not in the abstract (set B). We then manually looked for reasons why our summaries included these words.
There are fewer varieties of error which are unique to this category. All the errors originating from acronyms/synonyms, missed major and non-major MeSH terms, singular/plural and condensed terms/concepts in the Errors of Omission also appear in this error type.
However, there is one additional dominant type of error in this category. We observed that the sentences in the MeSH profile-based summaries and MEAD True summaries are not as concise as the sentences in the abstract. Sentences selected from the Introduction section are often superfluous and contain extra information not associated to the major focus of the sentence. For example, a sentence about a disease and its effect in humans might inadvertently discuss the effects of the disease in other animals. Sentences extracted from the Methods and Materials and Results section often contain raw data accompanying justification of results. Sentences from tables and figure legends also seem to contribute to the redundant information. These are especially true in case of the MEAD True summary. Overall, there were many more errors of commission in MEAD True summaries than in MeSH Profile summaries. For our set of eight articles, we found that the average word-count of the MEAD True summaries were 58% more than the corresponding MeSH profile-based summaries.
8 CONCLUSION
Our research goal was to test a MeSH-based approach for reducing full text documents and constructing summaries. We explore two approaches: MeSH Term and MeSH Profile approaches. In the latter, the MeSH terms are expanded with free-text terms. In both cases, the MeSH terms are used to select portions of the full text and thereby create summaries. We compare these with the abstract using several ROUGE scores. MEAD, a state-of-the-art summarizer, was used as our baseline.
We find that our MeSH-based strategies work well. For ROUGE-based automatic evaluation, we found that our MeSH term-based strategy always outperforms both the MEAD Random and MEAD True baseline strategies. Our expanded approach using MeSH Profiles provides an added boost to the summarization performance compared with our MeSH term-based strategies. The differences in their ROUGE evaluation were also found to be statistically significant.
Manual judgments by three assessors indicate that our ROUGE scores are in line with human evaluations. For obvious reasons, Abstract sentences were found to be most important followed by MeSH profile- and MeSH term-based sentences. The MEAD baseline scores were found to be significantly less than our proposed methods. Percentage of overall agreement among assessors was found to be 74% using Randolph's kappa.
Our results confirm the ability of our MeSH-based methods to produce summaries that contain important sentences. Based on these results, we are hopeful that we may be able to achieve a middle ground such that we do not have to rely on full text and at the same time are not restricted to the information content of the abstract. Our small-scale experiments also provide encouraging, though preliminary, results supporting the benefits of using MeSH term-based summaries for information retrieval.
Our MeSH term-based summarization strategies likely benefit from the human expertise invested while assigning relevant MeSH terms to MEDLINE documents. Inaccuracies in annotation will likely impact the quality of the summarization in a trickle-down effect. Another limitation of our strategy is that it cannot be used for summarizing recently published literature that have not yet been annotated with MeSH terms. However, also note that our system can produce pseudo-abstracts for biomedical documents that do not have an explicit Abstract (several such instances exist in MEDLINE in the form of editorials, reviews, comments, responses, etc.).
The error analysis yields several potential directions for exploring improvements. Also we had limited the MeSH terms to only the major terms. That is, we ignored the MeSH non-major terms for these experiments. Note that we also ignored the qualifier phrases for a MeSH major term descriptor (e.g. genetics, metabolism, chemistry). The observations from the error analysis point to the necessity of including the non-major MeSH terms in our methods. These should give us a fine-grained extraction strategy which will help reduce the obvious errors and thus improve the performance of the system. A second direction to explore is to build profiles for the MeSH terms using vocabulary resources such as the UMLS or at least using the MeSH hierarchy itself. Each term could be expanded to include its synonyms, near-synonyms and closely related concepts. Given the intellectual efforts underlying these vocabularies, it is quite possible that these provide additional advantages over the methods used here. A third direction for future research is that instead of considering sentences as individual entities in our methods, we could consider larger groups of sentences as the base units. It could be that larger text segments are better at indicating importance. Furthermore, we might explore heuristics such as sentence location and the presence of cue words (e.g. ‘our goal’, ‘we find’, etc.) for improved summarization. It would also be interesting to apply our strategies to application-specific summaries as studied by Ling et al. (2007) and Jin et al. (2009). In our current approach, we have not considered inter-sentential redundancy between selected summary sentences. We would like to incorporate novelty detection in our future research and address aspect-level diversity as explored in TREC 2006 and 2007 Genomics Tracks (Hersh et al., 2006, 2007).
ACKNOWLEDGEMENT
We would like to thank the assessors for their contribution with the manual evaluations of the baseline and our systems.
Funding: National Science Foundation, USA (grant number 0960984 in part).
Conflict of Interest: none declared.
Footnotes
1We note that retrieval results are not the same as summarization results (via document reduction). However, these retrieval experiments do offer some idea of the relative merits of profiles of different lengths.
2For MEAD Random, we selected one of the random summaries.
REFERENCES
- Agarwal S., Yu H. FigSum: automatically generating structured text summaries for figures in biomedical literature. AMIA Annu. Symp. Proc. 2009;2009:6–10. [PMC free article] [PubMed] [Google Scholar]
- Aone C., et al. A trainable summarizer with knowledge acquired from robust NLP techniques. In: Mani I., Maybury M.T., editors. Advances in Automatic Text Summarization. 1999. pp. 71–80. [Google Scholar]
- Bhattacharya S., et al. In Proceedings of the BioCreative III workshop. Bethesda, MD, USA: 2010a. Cross-species gene normalization at the University of Iowa; pp. 55–59. [Google Scholar]
- Bhattacharya S., et al. Proceedings of the BioCreative III workshop. Bethesda, MD, USA: 2010b. Online gene indexing and retrieval for BioCreative III at the University of Iowa; pp. 52–54. [Google Scholar]
- Brandow R., et al. Automatic condensation of electronic publications by sentence selection. Inf. Process. Manage. 1995;31:675–685. [Google Scholar]
- Chiang J.-H., et al. GeneLibrarian: an effective gene-information summarization and visualization system. BMC Bioinformatics. 2006;7:392. doi: 10.1186/1471-2105-7-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.M. Five-way smoking status classification using text hot-spot identification and error-correcting output codes. J. Am. Med. Inform. Assoc. 2008;15:32–35. doi: 10.1197/jamia.M2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen K.B., et al. The structural and content aspects of abstracts versus bodies of full text journal articles are different. BMC Bioinformatics. 2010;11:492. doi: 10.1186/1471-2105-11-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy J.M., O'leary D.P. Proceedings of the 24th Annual Iternational ACM SIGIR Conference on Research and Development in Information Retrieval, SIGIR '01. New York, NY, USA: ACM; 2001. Text summarization via hidden Markov models; pp. 406–407. [Google Scholar]
- Fiszman M., et al. Proceedings of the HLT-NAACL Workshop on Computational Lexical Semantics, CLS '04. Morristown, NJ, USA: Association for Computational Linguistics; 2004. Abstraction summarization for managing the biomedical research literature; pp. 76–83. [Google Scholar]
- Hersh W., et al. TREC 2007 genomics track overview. Gaithersburg, MD: 2007. TREC 2007 genomics track overview. [Google Scholar]
- Hersh W., et al. TREC 2006 genomics track overview. Gaithersburg, MD: 2006. TREC 2006 genomics track overview. [Google Scholar]
- Inouye D. Research Final Report. Colorado Springs, GA: University of Colorado at Colorado Springs; 2010. Multiple post microblog summarization. [Google Scholar]
- Jin F., et al. Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing, BioNLP '09. Stroudsburg, PA, USA: Association for Computational Linguistics; 2009. Towards automatic generation of gene summary; pp. 97–105. [Google Scholar]
- Johnson F.C., et al. The Application of Linguistic Processing to Automatic Abstract Generation. San Francisco, CA, USA: Morgan Kaufmann Publishers Inc.; 1997. [Google Scholar]
- Kupiec J., et al. Proceedings of the 18th annual international ACM SIGIR conference on Research and development in information retrieval, SIGIR '95. New York, NY, USA: ACM; 1995. A trainable document summarizer; pp. 68–73. [Google Scholar]
- Lin C.-Y. ROUGE: a package for automatic evaluation of summaries. In: Moens M.-F., Szpakowicz S., editors. Text Summarization Branches Out: Proceedings of the ACL-04 Workshop. Barcelona, Spain: Association for Computational Linguistics; 2004. pp. 74–81. [Google Scholar]
- Lin J. Is searching full text more effective than searching abstracts? BMC Bioinformatics. 2009;10:46. doi: 10.1186/1471-2105-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-Y., Hovy E. Proceedings of the 2003 Conference of the North American Chapter of the Association for Computational Linguistics on Human Language Technology - Volume 1, NAACL '03. Morristown, NJ, USA: Association for Computational Linguistics; 2003. Automatic evaluation of summaries using N-gram co-occurrence statistics; pp. 71–78. [Google Scholar]
- Ling X., et al. Generating gene summaries from biomedical literature: a study of semi-structured summarization. Inf. Process. Manage. 2007;43:1777–1791. [Google Scholar]
- Luhn H.P. The automatic creation of literature abstracts. IBM J. Res. Dev. 1958;2:159–165. [Google Scholar]
- Radev D.R., McKeown K.R. Generating natural language summaries from multiple on-line sources. Comput. Linguist. 1998;24:470–500. [Google Scholar]
- Radev D., et al. Proceedings of the 4th International Conference on Language Resources and Evaluation (LREC 2004) Lisbon, Portugal: 2004a. MEAD - a platform for multidocument multilingual text summarization. [Google Scholar]
- Radev D.R., et al. Centroid-based summarization of multiple documents. Inf. Process. Manage. 2004b;40:919–938. [Google Scholar]
- Radev D., et al. Proceedings of the Document Understanding Conference. New Orleans, LA: 2001. Experiments in single and multidocument summarization using MEAD. [Google Scholar]
- Randolph J.J. Joensuu University Learning and Instruction Symposium 2005. Joensuu, Finland: 2005. Free-marginal multirater kappa: an alternative to Fleiss' fixed-marginal multirater kappa. [Google Scholar]
- Reeve L.H., et al. Biomedical text summarisation using concept chains. Int. J. Data Min. Bioinform. 2007;1:389–407. doi: 10.1504/ijdmb.2007.012967. [DOI] [PubMed] [Google Scholar]
- Reynar J.C., Ratnaparkhi A. Proceedings of the Fifth Conference on Applied Natural Language Processing, ANLC '97. Washington, DC: Association for Computational Linguistics; 1997. A maximum entropy approach to identifying sentence boundaries; pp. 16–19. [Google Scholar]
- Salton G. The SMART Retrieval System – Experiments in Automatic Document Processing. Upper Saddle River, NJ, USA: Prentice-Hall, Inc.; 1971. [Google Scholar]
- Sehgal A.K., Srinivasan P. Retrieval with gene queries. BMC Bioinformatics. 2006;7:220. doi: 10.1186/1471-2105-7-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan P. Text mining: generating hypotheses from MEDLINE. J. Am. Soc. Inf. Sci. Technol. 2004;55:396–413. [Google Scholar]
- Srinivasan P., Libbus B. Mining MEDLINE for implicit links between dietary substances and diseases. Bioinformatics. 2004;20(l. 1):i290–i296. doi: 10.1093/bioinformatics/bth914. [DOI] [PubMed] [Google Scholar]
- Trieschnigg D., et al. MeSH Up: effective MeSH text classification for improved document retrieval. Bioinformatics. 2009;25:1412–1418. doi: 10.1093/bioinformatics/btp249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo I., et al. A coherent graph-based semantic clustering and summarization approach for biomedical literature and a new summarization evaluation method. BMC Bioinformatics. 2007;8(Suppl. 9):S4. doi: 10.1186/1471-2105-8-S9-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., et al. Enhancing MEDLINE document clustering by incorporating MeSH semantic similarity. Bioinformatics. 2009;25:1944–1951. doi: 10.1093/bioinformatics/btp338. [DOI] [PubMed] [Google Scholar]