Abstract
Summary: A program package to enable genome-wide association of copy number variants (CNVs) with quantitative phenotypes in families of arbitrary size and complexity. Intensity signals that act as proxies for the number of copies are modeled in a variance component framework and association with traits is assessed through formal likelihood testing.
Availability and implementation: The Java package is made available at www.imperial.ac.uk/medicine/people/m.falchi/.
Contact: m.falchi@imperial.ac.uk
Copy number variants (CNVs) have recently attracted increasing interest as a source of phenotypic variation in humans, with both rare and common CNVs being associated with different complex diseases (e.g. Diskin et al., 2009; Walters et al., 2010) and gene expression levels (Henrichsen et al., 2009). The availability of high-coverage SNP arrays and the development of methods for making CNV calls from SNP data are now being exploited to evaluate genome-wide association with CNVs (e.g. Glessner et al., 2009). At present, FBAT (Ionita-Laza et al., 2008) is the only other available algorithm that performs association test of normalized signal intensity measurement variations, reflecting CNVs in family-based datasets. The family-based association test implemented in FBAT uses only the within-family variation. Although that is one of the possible approaches to control for population stratification, disregarding the between-family variation might lead to decreased statistical power (Aulchenko et al., 2007). famCNV uses information from both within- and between-family variations, while population stratification issues can be detected and corrected using standard methods. Although qualitative CNV calls from existing algorithms can be incorporated as multiallele loci in a family-based association test, direct testing of the underlying quantitative CNV measurement has been shown to be more robust in identifying genuine associations in many cases (McCarroll and Altshuler, 2007; Stranger et al., 2007). Moreover, quantitative trait designs are less likely to be affected by differential errors that can arise in case-control studies. We have implemented a Java program, famCNV, that uses intensity signals such as the log2 ratio from array comparative genomic hybridization (aCGH) or the log ratio of observed to expected signal intensity (LRR) from Illumina genotyping arrays. The quantitative trait of interest is analysed in a variance component framework to model the resemblance among relatives and to remove possible bias from familiality (Abecasis et al., 2000). A linear mixed model is fitted to data to partition the total phenotypic variance into a polygenic component σa2 determined by additive genetic effects and a residual variance σe2 determined by the unique individual environment, including measurement errors. The variance covariance matrix Ω for the subjects in each family is Ω=2Φσa2+lσe2, where 2Φ is the matrix of the expected proportion of alleles that are identical-by-descent among the family members used to correct for the sample structure and l is an identity matrix.
Variance component analysis allows simultaneous modeling of the trait mean as μ=γ+Aβ, where γ is the trait mean, A is a matrix of covariate effects and β the corresponding regression coefficients. The test is evaluated by comparing the likelihood of the full model, where signal intensities are included in the mean model and the likelihood of the null model where the effect of the signal intensities is constrained to zero. Twice the difference between the logarithms of the full and the null likelihoods asymptotically follows a chi-square distribution with one degree of freedom and can be used to evaluate the difference in goodness-of-fit between the two models (Amos, 1994).
When using data from genotype arrays, the B Allele frequency (BAF—representing the proportion of the total signal deriving from the B allele) can also be included in both the full and null models, to control for misleading differences in an LRR distribution between genotypes that may have arisen due to errors during the array normalization process (Peiffer et al., 2006).
The identified loci, either via the reported P-value or via meeting a threshold for the false discovery rate (FDR; Storey and Tibshirani, 2003), are expected to reflect probes that are both genuinely copy number-variable and whose copy number is associated with the quantitative trait under examination. To evaluate this approach, we used famCNV to identify CNVs that were associated in cis with variation in gene expression. We applied our approach to a cohort of 154 nuclear families, including 732 subjects for which both genotyping and gene expression data were available (Carlsson et al., 2009). For all siblings, gene expression levels were measured in subcutaneous adipose tissue using Affymetrix U133Plus2.0 microarrays and DNA from peripheral blood was genotyped using the Illumina 610K-Quad array. As quantitative traits, we selected 28 580 transcripts with known position in the genome and 275 348 SNPs for which intensity data were available. For each of these transcripts, our preliminary analysis focussed on SNP markers lying within the transcript plus 10 kb upstream and downstream. The median (first–third quartile) number of markers tested per transcript was 10 (6–21).
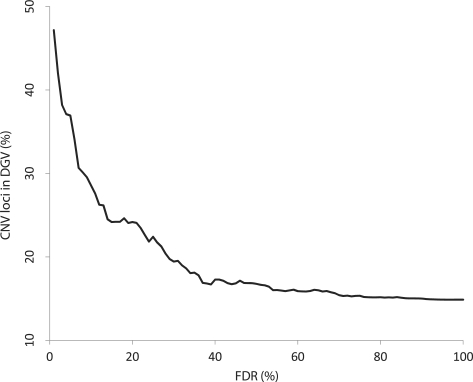
Using this dataset, 189 probes were identified as putative CNV loci associated with transcript level of the neighbouring gene (161 different transcripts), at FDR of 10%. We performed an in-silico experiment to determine whether these regions have been independently identified as copy number-variable in other studies. The probe locations were checked against CNVs and indels catalogued in the Database of Genomic Variants (DGV, version 9). We selected from the database CNVs and indels detected through aCGH and sequencing studies, and determined the number of our association signals that were included in the DGV-reported boundaries. Figure 1 shows the results for CNVs identified by famCNV at different FDR levels, demonstrating that the proportion lying within reported CNVs rises with an increasingly stringent significance threshold.
Fig. 1.
Overlap of CNVs identified by famCNV with CNV loci catalogued in the Database of Genomic Variants at different FDR levels. At an FDR of 1%, 47% of QT-associated probes lay within reported CNVs. In contrast, only 15% of non-associated probes (FDR > 80%) lay within DGV CNVs.
As a second level of validation of the presence of common CNVs at the predicted loci, we examined the overlap between the CNV regions identified by famCNV in the family study and the calls made by an alternative CNV prediction algorithm, cnvHap (Coin et al., 2010), in an independent dataset of Caucasian ancestry. For this, we used a cohort of 902 adult French subjects genotyped using the Illumina Human CNV370-duo array (Meyre et al., 2009). No phenotypic data were considered by cnvHap, as this analysis was aimed solely at confirmation of the presence of common structural variants.
For this comparison, we first limited the CNV calls of cnvHap to those that spanned at least two SNPs and were present in at least five samples. We found that the regions identified by famCNV at 5% FDR were enriched 6.7-fold (P < 0.01) for probes that intersect with cnvHap CNV calls, when compared to the frequency observed in 100 random datasets of probes not found to show association with transcript levels by famCNV (FDR > 80%). Similarly significant results were obtained after applying a wide range of alternative CNV-calling criteria to cnvHap.
The package is entirely written in the Java language and is therefore executable on all supported operating systems. The variance component algorithm follows the QTDT implementation (Abecasis et al., 2000). Files can be imported into famCNV in the PED and MAP file formats as used by the PLINK package (Purcell et al., 2007). For analysis of gene expression data, transcript locations can be provided to allow the restriction of the analyses within a user-defined window around each transcript. To help distinguish between putative rare and common CNVs, famCNV also calculates the Gini coefficient (Gini, 1921) to determine whether the positive contribution to the final chi-square statistics has been determined by a uniformly distributed effect or by a small number of strongly associated families.
The program, documentation and example files are available at http://www.imperial.ac.uk/medicine/people/m.falchi/.
ACKNOWLEDGEMENTS
The authors thank Leonardo Bottolo, Adam J. De Smith and the staff of the Imperial College High-Performance Computing Service for their advice and support.
Funding: This study was funded by Grant no. 079534/z/06/z from the Wellcome Trust. The SOS Sibpair study is supported by the Swedish Research Council (K2010-55X-11285-13), the Swedish Foundation for Strategic Research to Sahlgrenska Center for Cardiovascular and Metabolic Research, the Swedish Diabetes foundation and the Swedish federal government under the LUA/ALF agreement.
Conflict of Interest: none declared.
REFERENCES
- Abecasis G.R., et al. A general test of association for quantitative traits in nuclear families. Am. J. Hum. Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos C.I. Robust variance-components approach for assessing genetic linkage in pedigrees. Am. J. Hum. Genet. 1994;54:535–543. [PMC free article] [PubMed] [Google Scholar]
- Aulchenko Y.S., et al. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics. 2007;177:577–578. doi: 10.1534/genetics.107.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson L.M., et al. ALK7 expression is specific for adipose tissue, reduced in obesity and correlates to factors implicated in metabolic disease. Biochem. Biophys. Res. Commun. 2009;382:309–314. doi: 10.1016/j.bbrc.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coin L.J., et al. cnvHap: an integrative population and haplotype-based multiplatform model of SNPs and CNVs. Nat. Methods. 2010;7:541–546. doi: 10.1038/nmeth.1466. [DOI] [PubMed] [Google Scholar]
- Diskin S.J., et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gini C. Measurement of inequality of incomes. Econ. J. 1921;31:124–126. [Google Scholar]
- Glessner J.T., et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichsen C.N., et al. Segmental copy number variation shapes tissue transcriptomes. Nat. Genet. 2009;41:424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- Ionita-Laza I., et al. On the analysis of copy-number variations in genome-wide association studies: a translation of the family-based association test. Genet. Epidemiol. 2008;32:273–284. doi: 10.1002/gepi.20302. [DOI] [PubMed] [Google Scholar]
- McCarroll S.A., Altshuler D.M. Copy-number variation and association studies of human disease. Nat. Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- Meyre D., et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat. Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- Peiffer D.A., et al. High-resolution genomic profiling of chromosomal aberrations using infinium whole-genome genotyping. Genome Res. 2006;16:1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger B.E., et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters R.G., et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]