Abstract
Summary: The large number of genomes that will be sequenced will need to be annotated with genes and other functional features. Aligning gene sequences from a related species to the target genome is an economical and highly reliable method to identify genes; unfortunately, existing tools have been lacking in sensitivity and speed. A program we reported, sim4cc, was shown to be highly accurate but is limited to comparing one cDNA with one genomic sequence. We present here an optimization of the tool, implemented in the packages sim4db and leaff. The new tool performs batch alignments of cDNA and genomic sequences in a fraction of the time required by its predecessor, and thus is very well suited for genome-wide analyses.
Availability: Sim4db and leaff are written in C, C++ and Perl for Linux and other Unix platforms. Source code is distributed free of charge from http://sourceforge.net/projects/kmer/.
Contact: florea@umiacs.umd.edu
Supplementary information: Supplementary data are available at Bioinformatics Online.
1 INTRODUCTION
The number and variety of sequenced genomes will continue to grow spectacularly. The 10 000 Genomes Project (G10KCOS, 2009) alone will catalog and sequence more than 10 000 species from across the spectrum of verterbate evolution, and there are numerous ongoing projects to sequence plants and animals of agricultural importance, or of more specialized scientific interest (Plant and Animal Genome Conference; http://www.intl-pag.org/). As sequencing becomes increasingly accessible, one can expect many more groups and even individual investigators to sequence the genome of their studied organism. These genomes will need to be annotated with genes and other functional features. A key resource of gene information are the cDNA (mRNA, EST) sequences already in the databases, which can be readily aligned to a target genome to produce gene models. To further facilitate this comparative annotation approach, an increasing number of projects are producing mixed collections of resources from several related species, which are then used to analyze each of those genomes (The Fagaceae Genomics Project, http://www.fagaceae.org; The Genome Database for Rosaceae, http://www.rosaceae.org).
Most spliced alignment tools were designed for comparing highly similar sequences and perform poorly on cross-species comparisons, where sequence similarity drops. Few programs have been adapted for aligning sequences cross-species, most notably BLAT (Kent, 2002) and GMAP (Wu and Watanabe, 2005). These, however, produce output that is often less accurate than required, more so as the distance between species increases. Other tools, reviewed in Zhou et al. (2009), employ probabilistic or exact dynamic programming methods and are capable of aligning sequences cross-species, but lack the speed required for whole-genome annotation and are limited to comparisons between close species. The main difficulty in aligning cross-species is detecting weakly similar regions, which leads to incomplete gene models and incorrect exon boundaries. Differences in the gene models of orthologs caused by evolutionary block insertion and deletion events are a further challenge. We recently developed a program, sim4cc (Zhou et al., 2009), which produces more accurate alignments and for a wider range of evolutionary distances than previously possible. Sim4cc followed the ‘seed-and-extend’ approach of its predecessor sim4 (Florea et al., 1998), one of the earliest spliced alignment tools, but incorporated specialized features, such as mathematically optimized spaced seeds, sophisticated splice site models and evolution-sensitive alignment algorithms. Sim4cc's basic use is a ‘one-to-one’ alignment of one cDNA with one genomic region, intended for the analysis of a small number of samples, which makes it less friendly for pipeline applications.
We developed an optimized framework for running batch sim4cc tasks, called sim4db, which speeds up processing by two orders of magnitude for a typical application. The main improvements are a fast sequence indexing and compression mechanism, implemented in the package leaff and medium-grained multi-threading. Source code and online documentation for the program are available free of charge and under an open-source model from http://sourceforge.net/projects/kmer/.
2 APPROACH
Sim4db takes as input two multifasta files containing the genomic and the cDNA sequences, respectively, and optionally a script specifying a set of pairings among the sequences. For instance, the script line ‘-e 5 -D 0 14 500 60 000 -r’ indicates that the reverse complement of the sixth sequence in the cDNA file should be aligned to the region 14 500–60 000 in the first genomic sequence. If no script is given, the default operation is an all-against-all comparison. This new streamlined format makes incorporation into an automated analysis pipeline considerably easier. Sim4db then determines a sim4cc alignment for each sequence pair specified in the script. The process is multithreaded, with several concurrent threads handling the reading and writing of sequences and alignments and the alignment calculation. Sim4db returns matches that pass user-defined cutoffs for coverage, percent sequence identity or alignment length, or the best result for each run. One important feature of sim4db not available with sim4cc is the ability to detect multiple cDNA matches in a genomic region, which is essential when searching for cDNAs from a gene family. For broader utility, sim4db can be used to align sequences from either different species (option ‘-interspecies’), using the sim4cc algorithm, or the same species, using the sim4 algorithm.
A key feature of sim4db is the ability to perform fast random access to sequences. The leaff package is used to generate a random access index into the supplied multifasta file, which then allows one or several sequences, or intervals within these sequences, to be retrieved by index number or keyword. Leaff is incorporated into the sim4db code as a library, but can also be used from the command line. In addition to sequence retrieval, the package contains utilities for diverse sequence manipulations, including simulating errors in sequences for testing alignment programs, partitioning a large sequence into equally sized pieces and generating random subsets of sequences from a multifasta file.
3 RESULTS
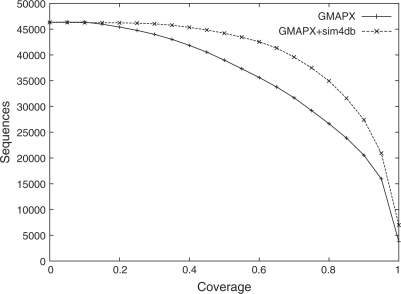
Sim4db can be used to align a small or a very large number of pairs of sequences, sequentially (option ‘-pairwise’) or as prescribed by a script, or to perform an all-against-all comparison. While the genomic sequences can be chromosomes, a time-saving strategy for genome-wide applications is to use sim4db to refine alignments in regions detected by a less accurate but fast high-throughput mapper, such as GMAP or BLAT. Figure 1 shows a typical user scenario where 92 142 zebrafinch GenBank ESTs were aligned to the recently assembled 1 GB turkey genome (Turkey Genome Sequencing Consortium, 2010). First, sequences were mapped to the genome using GMAP in cross-species mode (′−X′ option); then, sim4db realigned each cDNA to its matching region extended on both sides by 50 KB (Supplementary Material S1). As Figure 1 illustrates, adding sim4db produces longer alignments, and leads to more mapped sequences for any coverage cutoff than using GMAP alone. Overall, sim4db extends GMAP's alignments by more than 10% on average, a significant gain in sensitivity. (A more detailed analysis of accuracy, and comparison to other tools, is included in Supplementary Material S2).
Fig. 1.
Mapping rates of zebrafinch ESTs to the turkey genome with varying coverage cutoffs (horizontal axis), using GMAP only versus combining GMAP and sim4db.
We also compared the run time of sim4db versus a standard pipelined invocation of multiple sim4cc's. For this purpose, pairings in the script were sorted by genomic sequence index to minimize computationally expensive accesses to the genome file. Thus, each genomic sequence was read the first time its index was encountered, and then reused on subsequent calls involving that index. Two tests were performed. In the first test, a Perl script read pairs of sequences directly from the multi-fasta files, invoking sim4cc for each pair. In the second test, the leaff utility was used to retrieve the sequences. Sim4db had a 275-fold speedup (from 5 h 33 min to 1 min 12.5 s on a 3.0 GHz quad-core Intel Xeon processor) over the simple scan, and 145-fold when the leaff utility was used. These improvements are truly significant, and are critical for mapping very large datasets efficiently.
4 CONCLUSION
We described a utility, sim4db, for performing batch spliced alignment of cDNA and genomic sequences from the same or related species. The tool has the high accuracy of its predecessors, sim4 and sim4cc, but is two orders of magnitude faster, owing to its multithreaded design and fast sequence indexing. Sim4db can be invoked from the command line or can be combined with a fast sequence search engine for automated use. Compact, fast and accurate, and delivering provenly more accurate alignments than existing tools, we believe sim4db is a much needed addition to every genome annotator's repertoire of tools.
Funding: National Institutes of Health (R01-LM006845 to Steven L. Salzberg).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Florea L., et al. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genome 10K Community of Scientists. Genome 10K: a proposal to obtain whole-genome sequence for 10 000 vertebrate species. J. Hered. 2009;100:659–674. doi: 10.1093/jhered/esp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT – the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkey Genome Sequencing Consortium. Changing the landscape of genome sequencing: the domestic turkey (Meleagris gallopavo) genome assembly and analysis. PLoS Biol. 2010;8:e1000475. doi: 10.1371/journal.pbio.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.D., Watanabe C.K. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- Zhou L., et al. Sim4cc: a cross-species spliced alignment program. Nucleic Acids Res. 2009;37:e80. doi: 10.1093/nar/gkp319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.