Abstract
Mutations in Interferon Regulatory Factor 6 (IRF6) have been identified in two human allelic syndromes with cleft lip and/or palate: Van der Woude (VWS) and Popliteal Pterygia (PPS) Syndromes. Furthermore, common IRF6 haplotypes and single nucleotide polymorphisms (SNP) alleles are strongly associated with non-syndromic clefting defects in multiple ethnic populations. Mutations in the mouse often provide good models for the study of human diseases and developmental processes. We identified the cleft palate 1 (clft1) mouse mutant in a forward genetic screen for phenotypes modeling human congenital disease. In the clft1 mutant, we have identified a novel missense point mutation in the mouse Irf6 gene, which confers an amino acid alteration that has been found in a VWS family. Phenotypic comparison of clft1 mutants to previously reported Irf6 mutant alleles demonstrates the Irf6clft1 allele is a hypomorphic allele. The cleft palate seen in these mutants appears to be due to abnormal adhesion between the palate and tongue. The Irf6clft1 allele provides the first mouse model for the study of an etiologic IRF6 missense mutation observed in a human VWS family.
INTRODUCTION
Non-syndromic orofacial clefts are among the most common human congenital disorders, occurring in 1-2 births per thousand (Mossey and Little, 2002). Orofacial clefting occurs as both isolated non-syndromic cleft lip with or without cleft palate (NSCL/P) or as part of a clefting syndrome, in which other anomalies are associated with the clefting phenotype. A large number of studies, including genome scans for linkage and association studies with candidate genes, have been carried out with the intent of identifying the genes responsible for cleft palate (CP) and cleft lip and palate (CL/P) (Vieira et al., 2008).
One of the few mutant loci proven to be a monogenic cause of orofacial clefting in humans is IRF6, for which mutations have been found in Van der Woude syndrome (VWS; MIM 119300) and Popliteal Pterygium syndrome (PPS; MIM 119500). VWS is the most common syndromic form of clefting, but its phenotypic similarity to NSCL/P, which differs only by the presence of bilateral lower lip pits, led to subsequent studies that uncovered a common haplotype associated with IRF6 that accounts for 12% of the genetic contribution of all common forms of CL/P across populations (Kondo et al., 2002; Schutte et al., 2000; Zucchero et al., 2004). More recently, a common SNP that disrupts a TFAP2A transcription factor binding site in highly evolutionarily conserved region upstream of the IRF6 transcriptional start site was shown to be associated with cleft lip only (Rahimov et al., 2008). PPS cases exhibit orofacial anomalies similar to VWS cases, but may present with a mixture of oral adhesions, eyelid adhesions (ankyloblepharon), pterygia, syndactyly and genital anomalies as well. Null and knock-in mutant alleles of Irf6 in the mouse exhibit CP that likely occurs secondary to abnormal oral adhesions (Ingraham et al., 2006; Richardson et al., 2006).
The initial report of etiologic IRF6 mutations in human VWS and PPS families by Kondo et al., suggested a genotype-phenotype correlation (Kondo et al., 2002). Nonsense or frameshift mutations that result in the premature termination of IRF6 were significantly more common in VWS. VWS missense mutations were evenly distributed throughout IRF6 and the DNA-binding and protein-binding domains, whereas PPS missense mutations were almost exclusively located within the DNA-binding domain and, more specifically, altered amino acids predicted to directly contact DNA.
The development of robust tools for genomic analysis in the mouse has facilitated the use of forward genetics as a method to isolate new mutations in an unbiased manner. N-ethyl-N-nitrosourea (ENU) is an efficient chemical mutagen that induces single nucleotide substitutions, making it ideal for use in such genetic screens. These sequence changes can create a wide spectrum of etiologic mutations of the types commonly identified in human disease. As craniofacial phenotypes are readily ascertained by examination, this is an especially fruitful means to generate mouse models of human craniofacial disease.
Here we report the cloning and characterization of clft1, an ENU-induced mutant allele of Irf6. We show that the clft1 mutant phenotype is caused by a missense mutation that alters the Proline-39 amino acid residue, which has also been found to be mutated in a human VWS family (Kondo et al., 2002). Phenotypic characterization of the cleft palate, skeletal and skin phenotypes suggests that clft1 is a hypomorphic allele of Irf6 and a potential model of VWS.
RESULTS
Clft1 mutants have cleft palate defects
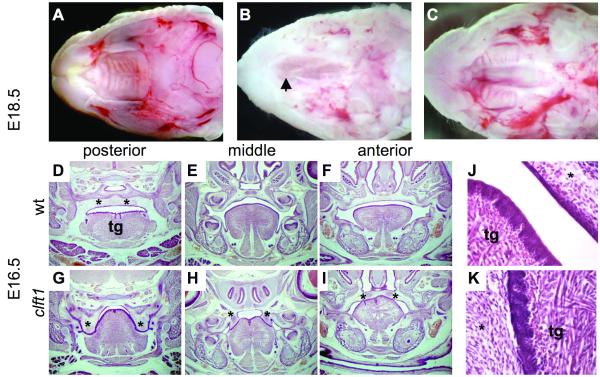
We recovered the cleft palate 1 (clft1) mutation in an ENU mutagenesis screen in the mouse designed to identify recessive mutations that model human congenital defects. We have previously described this strategy in which we examine potentially mutant embryos at embryonic day (E) 18.5 (Herron et al., 2002). This is one day before birth, facilitating the recovery of mutants with defects such as craniofacial clefting, which might be perinatal lethal and consequently not readily ascertained at postnatal stages. Clft1 mutants were easily detected during our screening because we were unable to open their mouths to assess secondary palate fusion. Further analysis revealed that the tongue was abnormally adhered to the roof of the mouth. Upon removal of the mandible and tongue to examine the palate, we observed two classes of cleft palate in homozygous clft1 mutants. The first class of mutants has partial fusion of the palate shelves in the anterior (7/11 animals examined both histologically and in whole mount; Fig. 1B,I). Remaining mutants show a complete cleft of the secondary palate (4/11; Fig. 1C).
Figure 1. Clft1 palatal defects.
(A-C) E18.5 whole mount preparations with mandible and tongue removed. Clft1 mutants (B,C) show posterior cleft secondary palate, with normal elevation and fusion anteriorly (B), or complete cleft secondary palate (C). Arrowhead in B indicates closure of anterior palate. (D-K) Histological analysis at E16.5. Coronal sections from the posterior, middle and anterior portion of the oral cavity as indicated (*, palatal tissue; tg, tongue). (J,K) Higher magnification view of the oral adhesions between tongue and palate seen in clft1 mutants
We performed a histological analysis of the clft1 mutant. At E16.5, wild-type embryonic palatal shelves have undergone normal elevation and fusion above the tongue to create a distinct oral cavity and nasopharynx (Fig. 1D-F). Clft1 mutants, however, have severe defects with aberrant epithelial adhesions throughout the oral cavity. In the posterior palate, clft1 palatal shelves have failed to elevate and are fused to the lateral aspects of the tongue (Fig. 1G). In the mid-palate region, the palatal shelves are still fused to the tongue but show partial elevation (Fig. 1H). In the anterior secondary palate, the majority of mutants had normal palate shelf elevation and fusion, although the inferior aspects of the palate shelves remained fused to the dorsal tongue (Fig. 1I). Thus, we conclude our inability to open the mouths of clft1 mutants is the result of the abnormal adhesion between the tongue and oral epithelium (Fig. 1J,K).
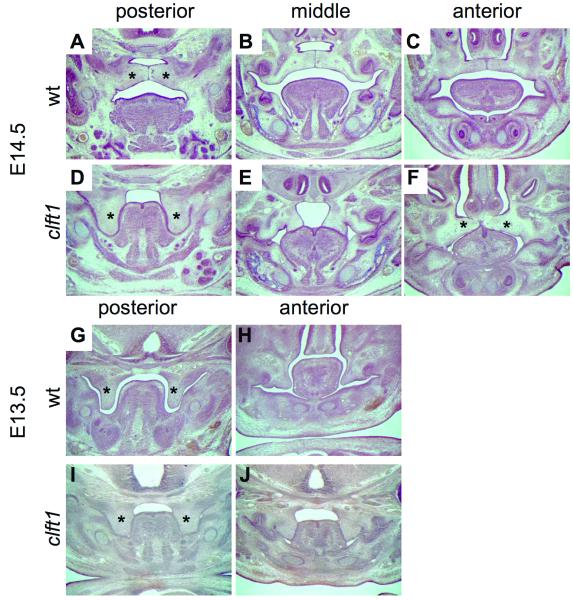
By E14.5, wild-type palatal shelves normally show complete elevation above the tongue and apposition with the presence of an epithelial seam (Fig. 2A-C). In contrast, at E14.5 mutant palate shelves have not elevated and were adhered to the sides of the tongue posteriorly (Fig. 2D). In the mid-palate region, the palate shelves have elevated but are tethered by epithelial fusion with the dorsolateral tongue (Fig. 2E). As noted above, some embryos show palate fusion in the anterior region (Fig. 2F). Palate shelf elevation cannot be assessed prior to E14.5, but the presence of oral adhesions between the palate and tongue are evident in E12.5 and E13.5 clft1 mutants (Fig. 2G-J; 4/4 mutants, data not shown). We have comprehensively examined six heterozygotes histologically at these stages and have not observed any abnormal phenotypes. We have examined over 25 heterozygous animals with no incidence of clefting.
Figure 2. Developmental analysis of Clft1 cleft palate.
Histological analysis at E14.5 (A-F) and E13.5 (G-J). Sections are from the posterior, middle and anterior portion of the oral cavity as indicated (*, palatal tissue; tg, tongue).
Cloning of the clf1 mutation
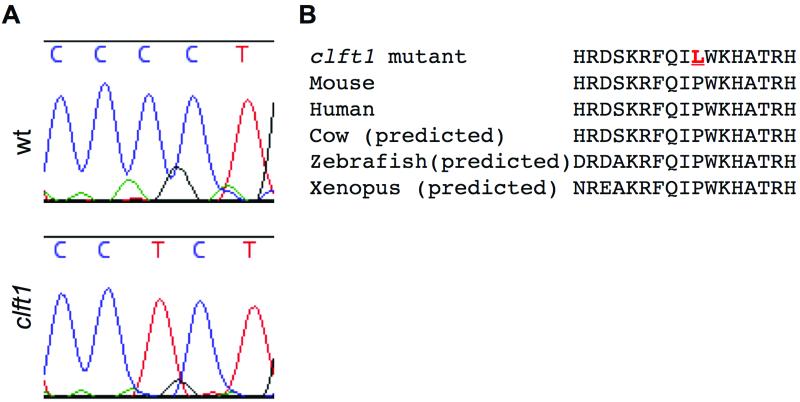
Our initial genome scan to map the clft1 mutation localized the mutation to a 14.7 Mb interval on distal chromosome 1. Phenotypic similarity between clft1 mutants and existing mouse mutants in Interferon regulatory factor 6 (Irf6) suggested Irf6 as a candidate gene. Genomic sequence analysis of the Irf6 locus in clft1 mutants revealed a missense mutation altering the thirty-ninth amino acid of the protein, a highly conserved proline, to a leucine (Fig. 3). Quantative RT-PCR analysis did not reveal differences in Irf6 transcript levels between wild-type and clft1 embryos, consistent with a missense mutation. Of note, this same residue is mutated in a family with Van der Woude syndrome (Kondo et al., 2002).
Figure 3. Clft1 mutants carry a mutation in the Irf6 gene.
Exon-directed sequencing of Irf6 in clft1 mutants revealed a sequence change creating a missense mutation in a well-conserved residue in the Irf6 gene.
The clft1 mutant is a hypomorphic allele of Irf6
Two mouse mutants have been generated to study Irf6 loss of function in the mouse (Ingraham et al., 2006; Richardson et al., 2006). Both of these mutants have cleft secondary palate and epidermal adhesions in the oral cavity, as well as skeletal defects. The oral epidermal adhesions observed in clft1 mutants appear to be less severe than those observed in the previously reported Irf6 mutants (Ingraham et al., 2006; Richardson et al., 2006). The palate shelves in these alleles show complete adherence to the tongue and oral epithelium and never elevate (B. C. Schutte, personal communication). We commonly see partial palate shelf elevation, although they always remain adhered to the tongue.
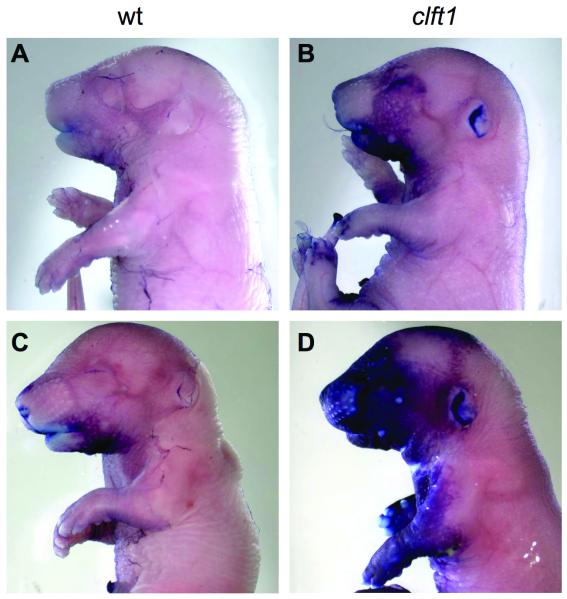
Both of the previously described mutant lines have defects in epidermis development: specifically, failure of terminal keratinocyte differentiation. To further examine the similarity of our Irf6clft1allele to these other Irf6 mutants, we performed the toluidine blue dye exclusion assay to assess the integrity of the skin permeability barrier. The barrier forms around E16 in the mouse in a pattern where the dorsal side is the first to become permeable with skin differentiation proceeding ventrally (Hardman et al., 1998). At E16.5 none of the embryos of any genotype we analyzed had formed the permeability barrier (n=8; data not shown). At E17.5, we found a range of dye exclusion patterns. Wild-type embryos were largely impermeable but some had slight uptake of the dye in the craniofacial region, indicating incomplete barrier formation (Fig. 4A,B). Clft1 mutants (n=4) showed complete barrier formation dorsally but exhibited some variability in the progression of the barrier ventrally (Fig. 4B,D). At E18.5 all mutant embryos had no observable difference in dye exclusion as compared to wild-type controls (n=5; Fig. 5C and data not shown). We further examined the skin histologically and saw no differences in skin differentiation between mutants and wild-type, and the cornified layer appears present in all sections examined (data not shown).
Figure 4. Clft1 mutants form a skin permeability barrier.
Wild-type (A,B) and mutant (C,D) embryos are E16.5 were treated with toluidine blue to asses skin differentiation. Wild-type embryos have almost complete exclusion of the dye while mutants are slightly delayed in forming the barrier.
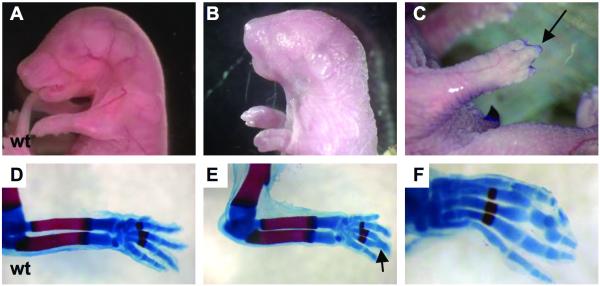
Figure 5. Clft1 skeletal defects.
A clft1 mutant at P0.5 (B) with apparently shortened forelimbs as compared to E18.5 wild-type (A). (C) An E18.5 clft1 embryo with syndactyly. (D-F) Skeletal preparations to highlight bone and cartilage. Note the fusion of phalanges on an otherwise normal clft1 limb (E,F).
We did observe a low incidence of skeletal abnormalities in Irf6clft1 mutants. We observed 31 mutants between E16.5 and E18.5 and detected skeletal abnormalities in four embryos. One mutant had very short forelimbs (Fig. 5B) and a curly tail; a second had hindlimbs that appeared fused to the body. Two more mutants showed fusion of middle digits (syndactyly; Fig 5C). To determine if these were due to underlying skeletal abnormalities, we performed skeletal preps to highlight embryonic bones and cartilaginous elements. All long bones appeared normal: the embryo with apparent hindlimb fusion had all appropriate skeletal elements. The syndactyly phenotype, however, was not just an epithelial abnormality. Rather, we observed three mutants with fused elements of the third and fourth proximal and intermediate phalanges (Fig. 5E and F). The mild skeletal defects and the lack of a skin permeability defect suggest that the Irf6clft1 allele is a hypomorphic mutation in Irf6.
DISCUSSION
Human IRF6 mutations cause two allelic autosomal dominant mixed clefting type syndromes: VWS and PPS, in which clefts of both the primary and secondary palates may be observed even within families. Recently, two Irf6 mutant alleles were reported that partially model the VWS and PPS phenotypes and identified a role for Irf6 as a key determinant of the keratinocyte proliferation-differentiation switch. Embryos homozygous for a gene trap null allele (Ingraham et al., 2006) have defects in stratified epidermis formation, skeletal defects secondary to the skin defect, cleft secondary palate and epidermal adhesions in the oral cavity. These oral adhesions are found at much reduced frequency in heterozygous embryos. Interestingly, the cleft palate in the homozygous embryos seems to be a defect in elevation of the palatal shelves. A second knock-in allele has been made with a mutation identical to that found in PPS patients (Richardson et al., 2006). These have a highly penetrant heterozygous phenotype of mild intraoral adhesions between the ventral surface of the tongue and the mandible, but no cleft lip or palate. In the homozygous state, the mutation causes similar defects in skin development and cleft secondary palate due to abnormal adhesion between the palate and the tongue.
The Irf6clft1mutant allele appears to be a milder allele than either previously reported for Irf6. We observe no gross defects in skin development (as determined by the permeability assay), and only rare expression of more severe phenotypes, including a loop tail and forelimb defects (Fig. 2B). We observe no defects in heterozygous mice, and a significant fraction of clft1 mutants with cleft palate show anterior secondary palate shelf elevation and fusion, which is never seen in more severe Irf6 mutant alleles. This finding is contradictory to our expectation that palate shelf fusion would not occur if palate shelf elevation could be uncoupled from the oral adhesions, based upon the fact that human VWS and PPS cases exhibit palate and lip fusion defects in the absence of severe oral adhesions. This observation is similar to that found for the PPS-like Irf6R84C heterozygous mice, in which multiple intraoral adhesions occur but palate fusion is complete.
The difference in allelic expressivity may be due to differences in genetic backgrounds. Previously published Irf6 alleles were on a mixed 129, C57BL/6 background (Ingraham et al., 2006; Richardson et al., 2006), and our allele is on a mixed A/J, FVB background. As such, we cannot formally exclude strain-specific modifier effects contributing to these phenotypic differences.
The coding mutation in the Irf6clft1 allele is in the Proline-39 amino acid that is mutated in a family with VWS. Biochemical studies have shown that the majority of VWS and PPS mutations that alter amino acids within the IRF6 DNA-binding domain abrogate binding to the IRF6 target DNA binding sequence motif, regardless of whether they make contact with DNA, (Little et al., 2009). The Proline-39 residue is not predicted to make direct contact with DNA (Kondo et al., 2002), but alteration of a proline within the IRF6 DNA-binding domain, which contains three α-helices, is likely to perturb its conformation. However, while the Irf6clft1 carries the same mutation as found in an affected human family, it does not appear to have cleft palate as a primary defect. Nonetheless, the Irf6clft1 hypomorphic allele is the first to contain a specific human VWS-like point mutation and will facilitate the molecular and biochemical study of its etiology in mice.
METHODS
Mouse husbandry
Animals were maintained as a mixed A/J, FVB stock. The allele was isolated in an ENU mutagenesis experiment in which A/J males were mutagenized and outcrossed to FVB females. Upon isolation of the clft1 mutation, the colony was maintained by a combination of intercross and outcross to FVB. Matings were monitored and noon of the day of copulation plug was determined to be E0.5. Routine genotyping was performed with an RFLP marker at ch1:193.1Mb (F primer: CCTTGGAGAGGTTCCTCCTATT; R primer TTACAGCGGGGCTACATTTAAG) which amplifies a polymorphic SNP identified with the use of the SNP2RFLP internet utility (http://genetics.bwh.harvard.edu/snp2rflp; Beckstead et al., 2008). Digestion of the resulting PCR product with RsaI enzyme cuts the AJ allele but not the FVB allele. All animals were maintained in accordance with HMS IACUC guidelines.
Genetic mapping
An initial genome scan was done with multiple mutant embryos using a 768 marker whole genome SNP panel to identify a region of shared A/J homozygosity among mutants, similar to method described previously (Moran et al., 2006). Irf6 was a candidate gene in the genetic interval found to carry the clft1 allele and subsequent exon-directed sequencing of tissues from clft1 mutants identified the mutation.
Histology and other molecular analyses
Samples for histological anlaysis were fixed in Bouins fixative, prepared using a Leica TP1020 automated tissue processor, sectioned at 14 μm and stained using established protocols. Skeletal preparations and the toluidine permeability assay were as described (Hogan et al., 1994); (Hardman et al., 1998). For qPCR analysis, total RNA from embryos was prepared with TRIZOL reagent, cDNA was prepared with the qScript cDNA supermix (Quanta). PCR analysis was with Pefecta SYBR Green Supermix (Quanta) and performed on a BioRad iCycler. Primers were: Irf6-F (CATGCCATTTATGCCATCAG ), Irf6-R (AAAAGGCGGCTGCTTCTCTA), Gapdh-F (ACTCCACTCACGGCAAATTC), and Gapdh-R (TCTCCATGGTGGTGAAGACA).
ACKNOWLEDGEMENTS
We thank Mingyue Lun and Mary Prysak for assistance with mouse husbandry and Brian Schutte for discussions of this data before publication. This work was supported by the N.I.H. with grants HD36404 to D.B. and HD531982 to R.S.
REFERENCES
- Beckstead WA, Bjork BC, Stottmann RW, Sunyaev S, Beier DR. SNP2RFLP: a computational tool to facilitate genetic mapping using benchtop analysis of SNPs. Mamm Genome. 2008;19:687–690. doi: 10.1007/s00335-008-9149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman MJ, Sisi P, Banbury DN, Byrne C. Patterned acquisition of skin barrier function during development. Development. 1998;125:1541–1552. doi: 10.1242/dev.125.8.1541. [DOI] [PubMed] [Google Scholar]
- Herron BJ, Lu W, Rao C, Liu S, Peters H, Bronson RT, Justice MJ, McDonald JD, Beier DR. Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nat Genet. 2002;30:185–189. doi: 10.1038/ng812. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo. Second ed Cold Spring Harbor Press; New York: 1994. [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, Murray JC, Schutte BC. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6) Nat Genet. 2006;38:1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, McDonald-McGinn DM, Zackai EH, Lammer EJ, Aylsworth AS, Ardinger HH, Lidral AC, Pober BR, Moreno L, Arcos-Burgos M, Valencia C, Houdayer C, Bahuau M, Moretti-Ferreira D, Richieri-Costa A, Dixon MJ, Murray JC. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little HJ, Rorick NK, Su LI, Baldock C, Malhotra S, Jowitt T, Gakhar L, Subramanian R, Schutte BC, Dixon MJ, Shore P. Missense mutations that cause Van der Woude syndrome and popliteal pterygium syndrome affect the DNA-binding and transcriptional activation functions of IRF6. Hum Mol Genet. 2009;18:535–545. doi: 10.1093/hmg/ddn381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JL, Bolton AD, Tran PV, Brown A, Dwyer ND, Manning DK, Bjork BC, Li C, Montgomery K, Siepka SM, Vitaterna MH, Takahashi JS, Wiltshire T, Kwiatkowski DJ, Kucherlapati R, Beier DR. Utilization of a whole genome SNP panel for efficient genetic mapping in the mouse. Genome Res. 2006;16:436–440. doi: 10.1101/gr.4563306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossey PA, Little J. Epidemiology of oral clefts: an international perspective. In: Wyszynski DF, editor. Cleft lip & palate. From origin to treatment. Oxford University Press; New York: 2002. pp. 127–158. [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C, Melbye M, Jugessur A, Lie RT, Wilcox AJ, Fitzpatrick DR, Green ED, Mossey PA, Little J, Steegers-Theunissen RP, Pennacchio LA, Schutte BC, Murray JC. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40:1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Malhotra S, Hardman MJ, Knowles L, Boot-Handford RP, Shore P, Whitmarsh A, Dixon MJ. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat Genet. 2006;38:1329–1334. doi: 10.1038/ng1894. [DOI] [PubMed] [Google Scholar]
- Schutte BC, Bjork BC, Coppage KB, Malik MI, Gregory SG, Scott DJ, Brentzell LM, Watanabe Y, Dixon MJ, Murray JC. A preliminary gene map for the Van der Woude syndrome critical region derived from 900 kb of genomic sequence at 1q32-q41. Genome Res. 2000;10:81–94. [PMC free article] [PubMed] [Google Scholar]
- Vieira AR, McHenry TG, Daack-Hirsch S, Murray JC, Marazita ML. Candidate gene/loci studies in cleft lip/palate and dental anomalies finds novel susceptibility genes for clefts. Genet Med. 2008;10:668–674. doi: 10.1097/GIM.0b013e3181833793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, Caprau D, Christensen K, Suzuki Y, Machida J, Natsume N, Yoshiura K, Vieira AR, Orioli IM, Castilla EE, Moreno L, Arcos-Burgos M, Lidral AC, Field LL, Liu YE, Ray A, Goldstein TH, Schultz RE, Shi M, Johnson MK, Kondo S, Schutte BC, Marazita ML, Murray JC. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]