Abstract
Genetic engineering is the process of modifying an organism’s genetic composition by adding foreign genes to produce desired traits or evaluate function. Dr. Jon W. Gordon and Sterling Professor Emeritus at Yale Dr. Frank H. Ruddle were pioneers in mammalian gene transfer research. Their research resulted in production of the first transgenic animals, which contained foreign DNA that was passed on to offspring. Transgenic mice have revolutionized biology, medicine, and biotechnology in the 21st century. In brief, this review revisits their creation of transgenic mice and discusses a few evolving applications of their transgenic technology used in biomedical research.
Keywords: transgenic, transgenic technology, Frank Ruddle, Jon Gordon, somatic cell genetics, gene mapping, overexpression, knockout, genetic modification
Introduction
After service in the U.S. Air Force following World War II, Dr. Frank H. Ruddle completed his undergraduate education at Wayne State University in Detroit, Michigan, in 1953 and two years later received a master’s degree in science from the same institution. In 1960, Dr. Ruddle earned his doctorate in biology from the University of California at Berkeley, where he studied chromosome patterns in established cell cultures. Dr. Morgan Harris, a prominent cell biologist, mentored Dr. Ruddle during his time at Berkeley. Immediately after graduate school, Dr. Ruddle went to the University of Glasgow to pursue his interests in somatic recombination. As a postdoctoral associate, he worked with Drs. John Paul and Guido Pontecorvo, leaders in modern genetics at the time. Dr. Ruddle was greatly interested in Pontecorvo’s work on fungal parasexuality, or processes other than sexual reproduction that lead to gene recombination.
After brief postdoctoral training at Glasgow, Dr. Ruddle became an assistant professor in the Yale Biology Department in 1961. Once at Yale, Dr. Ruddle continued to pursue his interests in somatic cell genetics. Transfer of genetic material from a human cell to another type of cell in culture was state of the art for somatic cell genetics in the 1960s, since this “somatic cell hybrid” system allowed recombination and segregation of genes. Development of techniques to stain chromosomes made it possible to distinguish between mouse and human chromosomes. In a mouse-human mixture of cells, many human chromosomes were lost from the hybrid cell lines. Furthermore, with the absence or presence of certain chromosomes, one could assign genes to particular chromosomes, as determined by the correlation of known gene products, such as enzymes. Once gene location was established, genetic linkage could be determined by analyzing other proteins to determine if they were also lost or gained. This system simulated genetic segregation in meiosis, whereby genes that are physically close to each other on the same chromosome usually do not separate. These predictions could be confirmed with in situ hybridization to probe specific genes of metaphase chromosomes. Dr. Ruddle’s laboratory adopted this somatic cell hybrid approach to determine the physical location of genes on chromosomes, which proved a difficult task at the time. Dr. Ruddle’s lab made valuable contributions to the exploration of the genome, including mapping of the human interferon, pro-collagen, and β-globin genes [1-3]. With enthusiasm for the growing field of gene mapping, Dr. Ruddle initiated a Human Gene Mapping Workshop, which began in 1973. Located at Yale, this workshop offered hundreds of geneticists the opportunity to discuss recent literature in order to assign particular genes to precise locations on chromosomes. Almost 100 genes were cited in the report of the first Human Gene Mapping Workshop, and this number grew to more than 2,000 entries by the Tenth Human Gene Mapping Workshop in 1989 [4]. Furthermore, Dr. Ruddle created a database at Yale, called the Human Gene Map (HGM) library, which was the main source of human gene mapping information for those in the gene mapping community [5]. In many aspects, Dr. Ruddle was a pioneer of the Human Genome Project before its inception in 1989.
Application of somatic cell genetics to produce transgenic mice
In the late 1970s, Dr. Ruddle began the first of two appointments as chairman of the Biology Department, now Molecular, Cellular, and Developmental Biology, at Yale University. He also held a joint appointment in Yale School of Medicine’s new Department of Human Genetics. As his professional duties expanded, his laboratory interests did, too, to include applications of somatic cell genetic techniques to mammalian gene transfer, with the hopes of creating a mouse model to study developmental gene regulation and human disease. Previously, Rudolf Jaenisch and Beatrice Mintz from Fox Chase Cancer Center in Philadelphia showed that when mouse embryos were infected with Simian virus (SV40), this viral DNA could be integrated into the germ line [6,7]. However, it was unclear whether specific genes, previously manipulated in vitro, could be stably introduced into mice if one used a similar approach as Jaenisch and Mintz. Also in the late 1970s, Dr. Jon W. Gordon had completed his PhD at Yale University. In Dr. Clement L. Markert’s laboratory, Dr. Gordon’s graduate training focused on mouse developmental biology as he used mouse embryos to study how single genes can affect determination of cell phenotype. Dr. Gordon decided to stay at Yale as a postdoctoral fellow in Dr. Ruddle’s lab to pursue a novel project. In collaboration, Dr. Gordon and Dr. Ruddle produced the first “transgenic” animal. They generated a mouse in which specified foreign genes were inserted into its genome and transmitted to offspring, indicating stable inheritance. Furthermore, injection of this cloned DNA into freshly fertilized mice embryos reduced the occurrence of mosaicism, or somatic cells with more than one genotype, as seen in previous attempts [6,8,9]. This work was published in seminal papers in Proceedings of the National Academy of Sciences and Science in 1980 and 1981, respectively [10,11].
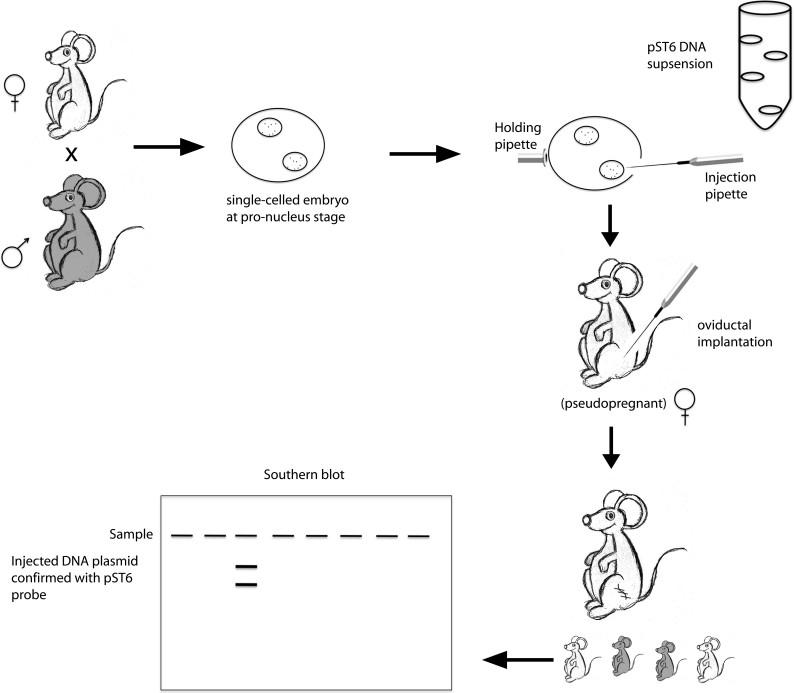
To summarize their work, several recombinant plasmids were created and modified for the purpose of tracking specific genes after introduction into mammalian embryos. The first plasmid, named pST6, was a derivative of pBR322, a widely used bacterial plasmid for cloning at the time. The authors inserted a portion of the SV40 virus and the herpes simplex virus thymidine kinase (TK) gene. These sequences would allow for DNA replication and confirmation of gene transfer, respectively. For negative controls, they created variations of the pST6 plasmid, which included reversal of the SV40 sequence and formation of dimerized pST6 plasmids. These were termed pST9 and pST12, respectively. A second type of plasmid, pRH 1.3Mm 1, also was derived from the pBR322 plasmid. It contained a random and interspersed sequence. If DNA integration was mediated by homologous recombination, this sequence would increase integration frequency, as it was shown that repetitive sequences caused a high frequency of recombination and formation of recombinant genomes in yeast [12]. Next, Dr. Ruddle and Dr. Gordon aimed to inject their purified DNA into single-celled embryos to ensure passage to all daughter cells. However, this would be technically challenging because of the small size of the embryos. To solve this problem, very fine microneedles were created from capillary tubes. In addition, holding pipettes were constructed from capillary tubes in order to stabilize the embryo. By capillary action, DNA was collected into the microneedle for injection. Their recombinant plasmids were then “microinjected” into single-celled mouse embryos at the pronuclear stage of development. Surviving eggs were implanted into psuedopregnant females. Offspring were born 3 weeks later, and they were subjected to DNA analysis by Southern blot (Figure 1). Of the 78 mice injected with the original pST6 plasmid, only two of 78 animals were positive for transformation, as determined by Southern blot hybridization to the pST6 probe. Furthermore, only one animal contained the DNA in integrated form as probed with TK, albeit the gene was modified. The other animal appeared to keep portions of the plasmid DNA without integration of the TK gene. Animals that were uninjected, or injected with the other plasmids mentioned, were not positive for DNA transformation. Despite a low success rate, this study served as proof of principle that cloned DNA could be directly inserted into the mouse genome. Furthermore, incomplete inter-species sequence homology between donor and recipient was not an obstacle to prevent transformation, as human TK was expressed in mice, suggesting that it might be possible to transfer any gene between distinct species. Lastly, failure to see any transformed mice with the pRH 1.3Mm 1 plasmid suggested that this phenomenon appeared to be predominantly random and not mediated by homologous recombination. One year later, Dr. Ruddle and Dr. Gordon reported in Science that these transgenic mice could pass the inserted genes to their offspring [11]. Hence, these sequences had become a stable part of the genome.
Figure 1.
Figure 1 (modified from [38]). Generation of the first transgenic mouse. To create transgenic animals (mice shown here), male and female mice were mated. The single-celled embryo was obtained from the pregnant female. DNA was microinjected into the pronucleus of a fertilized ovum. Once injected, surviving embryos were re-implanted into the oviduct of pseudopregnant female recipients. These females gave birth about 3 weeks after implantation. Transgene integration from litters was assessed by tissue analysis. This DNA injection into the pronucleus was the first and still most commonly used technique to make transgenic animals.
After the initial finding, several groups rapidly published similar findings that also showed integration and germ line transmission of foreign DNA. While the TK gene that Dr. Gordon and Dr. Ruddle injected did not induce an obvious phenotypic change, studies were published soon after that phenotypically demonstrated overexpression of exogenous genes. Proof of principle was dramatically manifested in mice transgenic for the rat and human growth hormone gene, which stimulated significant growth in the transgenic mice compared to their littermates, described by Palmiter and Brinster [13,14]. Taken together, these exciting findings set the stage for a revolution in the use of genetically modified organisms as a standard investigative tool in research.
Advances in transgenic technology
Compared to the 2 percent success rate of the original studies by Dr. Gordon and Dr. Ruddle, rapid refinement in the methodology led to 15 percent of offspring carrying the target gene [15,16], which today remains virtually unchanged, but depends on the size of DNA injected, species, and other variables. In addition, new ways to make transgenics were subsequently discovered and include embryonic stem cell transfer into embryos and recombinant virus infection of embryos [17,18]. Today, transgenic overexpression and targeted deletion are the two most common approaches to genetic manipulation. Gene null (knockout and knockin) mice are expansions of the fundamental principle of transgenesis. Similar to transgenic mice, knockout mice also are created using cloned DNA in embryonic stem cells (which in this case has similarities to the gene of interest) that allows for homologous DNA recombination to achieve site-specific disruption.
In conventional transgenic mice, the gene is introduced in the first cell of the embryo and is passed to all cells of the adult animal, presumably allowing constitutive expression of the protein. However, innovative approaches have led to the ability to make a transgenic mouse capable of expressing protein in a cell/organ specific manner by using tissue specific promoters. Examples include placing a gene under the control of the K14 or Vav promoter, which are specific for keratinocytes and hematopoietic cells, respectively [19,20]. Therefore, genes of interest will be uniquely overexpressed in the desired cell type. In addition to spatial specificity, temporal specificity is important since genes can play many roles dependant on the developmental stage of an organism. To overcome this, systems of inducible gene expression systems have been designed in which the expression of the desired gene is placed under the control of a cis acting element, which will respond to specific molecules that are introduced into the system. Examples include steroid hormone-based, antibiotic, heavy metal ion, and heat shock inducible systems [21]. In addition to overexpressing foreign genes into organisms, transgenes also have the potential to silence desired genes by introduction of DNA encoding short hairpin RNAs, which lead to cleavage of specific messenger of RNA. Similarly, transgenic mice can be used to create a conditional knockout mouse. Mice transgenic for Cre, a DNA recombinase, can be crossed with transgenic mice in which loxP sites have been introduced that flank a gene of interest (DNA is then said to be “floxed”). Since loxP sites contain a binding region for Cre, the gene of interest will be deleted as a result of the Cre-lox recombination. As discussed earlier, the inducible systems of gene expression can be used to turn on Cre, which then lead to deletion of the floxed gene. These mice are known as conditional knockout mice. This system is very useful when normal gene disruption results in embryonic lethality. Importantly, these mice also can be under spatial control, again by using using a tissue specific promoter to overexpress Cre. In addition, the Cre-lox system can be used to induce genes in spatio-temporal fashion, by mechanisms not described here.
Dr. Ruddle’s lab has continued to use transgenic technology while focusing on the Hox family genes and their roles in development and evolution. His lab has realized that variations in regulatory regions of these developmental control genes can influence their gene transcription and may contribute toward evolution [22]. Similarly, it was realized that production of some transgenic mice fail because the relatively small DNA construct injected does not contain critical regulatory regions of the gene, such as a section of the promoter or enhancers that may be necessary for transgene expression. Therefore, expression of transgene is subjected to being silenced by surrounding host sequences. This phenomena is known as positional effect [23]. Such obstacles have been overcome through bacterial artificial chromosomes (BACS) and yeast artificial chromosomes (YACS) [24,25]. These DNA “vehicles” allow transfer of large fragments of cloned genomic DNA, which contain necessary regulatory elements for expression. Due to the large size of YACS, the location of the desired gene will not interfere with expression. However, YACS lead to high rates of chimerism, difficulty with DNA isolation and cloning, and insert instability [26]. BACS have emerged as a solution to this problem due to their relative ease of handling and insert stability. However, the maximum insert size that BACS can accommodate is much smaller than that of YACS. With further contribution to transgenic technology, Dr. Ruddle’s lab has engineered a system that uses a yeast-bacteria shuttle vector (pClasper), which combines the two strengths of the systems without the disadvantages [27]. Using this vector, large regions from a YAC or BAC can be cloned and stably maintained in both yeast and bacteria. Further DNA manipulation can be made relatively easily in yeast and finally transferred to bacteria for simple isolation. A recent example of this technology has been used to isolate large stretches of DNA from a BAC clone that includes upstream and downstream noncoding sequences of the Chst4 (Hec-6st) gene, which is specific for high endothelial venules (HEVs). Additionally, reporter genes, β-galactosidase or green fluorescent protein (GFP), were inserted into an exon of the Chst4 gene, allowing simultaneous expression of the endogenous gene and transgene and real time visualization of HEVs in vivo [28,29].
Applications of transgenic technology
As more information is discovered concerning the genetic etiology of diseases, transgenic and null mice have been developed in a concerted fashion to allow researchers an opportunity to study and understand the function of gene products. One such example is an animal model used to study hepatitis B virus (HBV) infection, developed shortly after Dr. Gordon and Dr. Ruddle’s transgenic mouse [30]. To mimic the situation found in HBV carriers, mice transgenic for a hepatitis B surface antigen (HBsAg) were produced. Similar to the pathology in humans, increased expression of this antigen leads to severe liver cell damage and an inflammatory response. Many details on the mechanism of HBV virus infection and pathological consequences have been gained through this model. Likewise, many other other transgenic mice created have become valuable models to study mechanistic aspects of human disease.
In addition to using animal models for mechanistic studies, the pioneering transgenic technology created by Dr. Gordon and Dr. Ruddle has applications in medicine. For example, donor animals suitable for xenotransplantation, such as pigs, have emerged as a solution to the shortage of allogeneic organs [31]. To prevent human rejection of pig grafts, pigs transgenic for human leukocyte antigen and other corrections for immunological incompatibilities have been generated in order to attenuate the risk of acute rejection [32]. Transgenic animals also are used for large-scale production of therapeutic proteins, such as blood clotting factors (i.e., Factor VIII), which is otherwise dependant on isolation from scarce sources of human plasma [33]. In addition, human monoclonal antibodies used to treat cancer and autoimmune diseases are produced in transgenic mice. Antibodies produced in transgenic animals are typically less expensive and provide a greater yield than mammalian cell culture. Lastly, the idea of gene therapy seeks to use genetic engineering to benefit humans by replacing defective genes with normal copies. A classic example occurs in sickle cell anemia, in which a point mutation in the gene for beta globin, a protein component of hemoglobin, leads to sickle cell pathology. Replacing this mutated gene with a normal beta globin gene is a promising strategy, although this approach has been limited by technical challenges such as the immunogenicity of viral vectors used to deliver the genes and delivery to the appropriate cell type.
In agriculture, crops traditionally have been altered by crossing plants with individual favorable traits to produce new variety that contain both traits. This method relies on homozygous recombination that by chance may generate genetic diversity. Alternatively, transgenic plants provide an efficient solution to select desired traits. This can, in turn, reduce crop losses and improve plant growth, ultimately providing a stable supply of food for the increasing global population. For example, plants transgenic for toxin genes from the bacteria, Bacillus thuringiensis, are insect resistant [34]. Furthermore, genes that are important for herbicide resistance also can be introduced into plants [34]. Meanwhile, depletion of fossil fuel reserves raises interest in the use of renewable energy sources such as biofuels. Many biofuels are made from plant mass. Similar to plants for nutritional consumption by humans, transgenic plants engineered for biofuel production have important traits such as pest and herbicide resistance, resistance to disease, and other modifications that will allow it to thrive in sub-optimal conditions.
Concluding Remarks
Prior to the transgenic mouse, knowledge of gene function in mice only came about through forward genetics in extensively inbred mice, which used radiation or a different mutagen to affect the genotype, with the hopes of observing a resulting phenotype. As mentioned earlier, favorable traits in animals and plants were selected by outbreeding. Molecular biology techniques and genetically modified organisms have, in parallel, allowed rapid acceleration of science. Dr. Ruddle and Dr. Gordon’s first demonstration of foreign gene transfer began a revolution in the use of animal models for biomedical research. Importantly, their contributions did not end there. Dr. Jon Gordon has mentored the next generation of scientists since he joined the faculty of Mount Sinai School of Medicine in 1982. As an expert in recombinant DNA technology, he has shared his insights in the form of books, reviews, and primary research articles [35,36]. As mentioned earlier, Dr. Ruddle has continued to improve transgenic technology, while applying it to study fundamental biological questions. Additionally, he has helped advance the field of genomics, a word that he helped coin [37]. As a Sterling Professor Emeritus at Yale, he continues to publish primary research articles and is active in the scientific community. He has received many well-deserved awards, including induction into the National Academy of Science.
In summary, the pioneering work of Dr. Gordon and Dr. Ruddle has opened new avenues for research. Transgenic technology continues to hold great promise for the future but must be met with sound ethical reasoning by scientists, the biotechnology industry, government and the public.
Acknowledgments
The author would like to thank Dr. Nancy Ruddle, Laura DeMare, and Elenoe Smith for helpful comments in editing this article.
Glossary
- HGM
Human Gene Map
- TK
thymidine kinase
- BACS
bacterial artificial chromosomes
- YACS
yeast artificial chromosomes
- HEVs
high endothelial venules
- GFP
green fluorescent protein
- HBV
hepatitis B virus
- HBsAg
hepatitis B surface antigen
References
- Tan YH, Creagan RP, Ruddle FH. The somatic cell genetics of human interferon: assignment of human interferon loci to chromosomes 2 and 5. Proc Natl Acad Sci USA. 1974;71(6):2251–2255. doi: 10.1073/pnas.71.6.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundar Raj CV, Church RL, Klobutcher LA, Ruddle FH. Genetics of the connective tissue proteins: assignment of the gene for human type I procollagen to chromosome 17 by analysis of cell hybrids and microcell hybrids. Proc Natl Acad Sci USA. 1977;74(10):4444–4448. doi: 10.1073/pnas.74.10.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth A, Nienhuis A, Lawrence J, Giles R, Turner P, Ruddle FH. Chromosomal localization of human beta globin gene on human chromosome 11 in somatic cell hybrids. Proc Natl Acad Sci USA. 1978;75(3):1456–1460. doi: 10.1073/pnas.75.3.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddle F. Hundred-year search for the human genome. Annu Rev Genomics Hum Genet. 2001;2:1–8. doi: 10.1146/annurev.genom.2.1.1. [DOI] [PubMed] [Google Scholar]
- Pearson PL. Genome mapping databases: data acquisition, storage and access. Curr Opin Genet Dev. 1991;1(1):119–123. doi: 10.1016/0959-437x(91)80052-n. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Mintz B. Simian virus 40 DNA sequences in DNA of healthy adult mice derived from preimplantation blastocysts injected with viral DNA. Proc Natl Acad Sci USA. 1974;71(4):1250–1254. doi: 10.1073/pnas.71.4.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R. Germ line integration and Mendelian transmission of the exogenous Moloney leukemia virus. Proc Natl Acad Sci USA. 1976;73(4):1260–1264. doi: 10.1073/pnas.73.4.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz B. Formation of Genetically Mosaic Mouse Embryos, and Early Development of "Lethal (T12/T12)-Normal" Mosaics. J Exp Zool. 1964;157:273–292. doi: 10.1002/jez.1401570210. [DOI] [PubMed] [Google Scholar]
- Papaioannou VE, McBurney MW, Gardner RL, Evans MJ. Fate of teratocarcinoma cells injected into early mouse embryos. Nature. 1975;258(5530):70–73. doi: 10.1038/258070a0. [DOI] [PubMed] [Google Scholar]
- Gordon JW, Scangos GA, Plotkin DJ, Barbosa JA, Ruddle FH. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc Natl Acad Sci USA. 1980;77(12):7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JW, Ruddle FH. Integration and stable germ line transmission of genes injected into mouse pronuclei. Science. 1981;214(4526):1244–1246. doi: 10.1126/science.6272397. [DOI] [PubMed] [Google Scholar]
- Fonty G, Goursot R, Wilkie D, Bernardi G. The mitochondrial genome of wild-type yeast cells. VII. Recombination in crosses. J Mol Biol. 1978;119(2):213–235. doi: 10.1016/0022-2836(78)90435-7. [DOI] [PubMed] [Google Scholar]
- Palmiter RD, Brinster RL, Hammer RE, Trumbauer ME, Rosenfeld MG, Birnberg NC. et al. Dramatic growth of mice that develop from eggs microinjected with metallothionein-growth hormone fusion genes. Nature. 1982;300(5893):611–615. doi: 10.1038/300611a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter RD, Norstedt G, Gelinas RE, Hammer RE, Brinster RL. Metallothionein-human GH fusion genes stimulate growth of mice. Science. 1983;222(4625):809–814. doi: 10.1126/science.6356363. [DOI] [PubMed] [Google Scholar]
- Brinster RL, Chen HY, Trumbauer M, Senear AW, Warren R, Palmiter RD. Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell. 1981;27(1 Pt 2):223–231. doi: 10.1016/0092-8674(81)90376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter RD, Chen HY, Brinster RL. Differential regulation of metallothionein-thymidine kinase fusion genes in transgenic mice and their offspring. Cell. 1982;29(2):701–710. doi: 10.1016/0092-8674(82)90186-6. [DOI] [PubMed] [Google Scholar]
- Gossler A, Doetschman T, Korn R, Serfling E, Kemler R. Transgenesis by means of blastocyst-derived embryonic stem cell lines. Proc Natl Acad Sci USA. 1986;83(23):9065–9069. doi: 10.1073/pnas.83.23.9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Putten H, Botteri FM, Miller AD, Rosenfeld MG, Fan H, Evans RM. et al. Efficient insertion of genes into the mouse germ line via retroviral vectors. Proc Natl Acad Sci USA. 1985;82(18):6148–6152. doi: 10.1073/pnas.82.18.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Rosenberg M, Ross S, Tyner A, Fuchs E. Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice. Proc Natl Acad Sci USA. 1989;86(5):1563–1567. doi: 10.1073/pnas.86.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvy S, Metcalf D, Gibson L, Bath ML, Harris AW, Adams JM. Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood. 1999;94(6):1855–1863. [PubMed] [Google Scholar]
- Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2(10):743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- Shashikant CS, Kim CB, Borbely MA, Wang WC, Ruddle FH. Comparative studies on mammalian Hoxc8 early enhancer sequence reveal a baleen whale-specific deletion of a cis-acting element. Proc Natl Acad Sci USA. 1998;95(26):15446–15451. doi: 10.1073/pnas.95.26.15446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B. Molecular biology. Enhancers, chromosome position effects, and transgenic mice. Nature. 1983;306(5941):313–314. doi: 10.1038/306313a0. [DOI] [PubMed] [Google Scholar]
- Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotechnol. 1997;15(9):859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- Jakobovits A, Moore AL, Green LL, Vergara GJ, Maynard-Currie CE, Austin HA. et al. Germ-line transmission and expression of a human-derived yeast artificial chromosome. Nature. 1993;362(6417):255–258. doi: 10.1038/362255a0. [DOI] [PubMed] [Google Scholar]
- Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 2001;10(2):83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- Bradshaw MS, Bollekens JA, Ruddle FH. A new vector for recombination-based cloning of large DNA fragments from yeast artificial chromosomes. Nucleic Acids Res. 1995;23(23):4850–4856. doi: 10.1093/nar/23.23.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Bentley K, Lebrun M, Lesslauer W, Ruddle FH, Ruddle NH. Transgenic LacZ under control of Hec-6st regulatory sequences recapitulates endogenous gene expression on high endothelial venules. Proc Natl Acad Sci USA. 2007;104(11):4577–4582. doi: 10.1073/pnas.0700334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley KL, Stranford S, Liao S, Mounzer RM, Ruddle FH, Ruddle NH. High endothelial venule reporter mice to probe regulation of lymph node vasculature. Adv Exp Med Biol. 2011;691:35–44. doi: 10.1007/978-1-4419-6612-4_4. [DOI] [PubMed] [Google Scholar]
- Chisari FV, Pinkert CA, Milich DR, Filippi P, McLachlan A, Palmiter RD. et al. A transgenic mouse model of the chronic hepatitis B surface antigen carrier state. Science. 1985;230(4730):1157–1160. doi: 10.1126/science.3865369. [DOI] [PubMed] [Google Scholar]
- Weiss RA. Transgenic pigs and virus adaptation. Nature. 1998;391(6665):327–328. doi: 10.1038/34772. [DOI] [PubMed] [Google Scholar]
- Weiss EH, Lilienfeld BG, Muller S, Muller E, Herbach N, Kessler B. et al. HLA-E/human beta2-microglobulin transgenic pigs: protection against xenogeneic human anti-pig natural killer cell cytotoxicity. Transplantation. 2009;87(1):35–43. doi: 10.1097/TP.0b013e318191c784. [DOI] [PubMed] [Google Scholar]
- Paleyanda RK, Velander WH, Lee TK, Scandella DH, Gwazdauskas FC, Knight JW. et al. Transgenic pigs produce functional human factor VIII in milk. Nat Biotechnol. 1997;15(10):971–975. doi: 10.1038/nbt1097-971. [DOI] [PubMed] [Google Scholar]
- Prieto-Samsonov DL, Vazquez-Padron RI, Ayra-Pardo C, Gonzalez-Cabrera J, de la Riva GA. Bacillus thuringiensis: from biodiversity to biotechnology. J Ind Microbiol Biotechnol. 1997;19(3):202–219. doi: 10.1038/sj.jim.2900460. [DOI] [PubMed] [Google Scholar]
- Gordon JW. The science and ethics of engineering the human germ line: Mendel’s maze. Hoboken, NJ: Wiley-Liss; 2003. [Google Scholar]
- Gordon JW. Genetic enhancement in humans. Science. 1999;283(5410):2023–2024. doi: 10.1126/science.283.5410.2023. [DOI] [PubMed] [Google Scholar]
- Yadav SP. The wholeness in suffix -omics, -omes, and the word om. J Biomol Tech. 2007;18(5):277. [PMC free article] [PubMed] [Google Scholar]
- Gordon JW, Ruddle FH. DNA-mediated genetic transformation of mouse embryos and bone marrow―a review. Gene. 1985;33(2):121–136. doi: 10.1016/0378-1119(85)90087-3. [DOI] [PubMed] [Google Scholar]