Abstract
Embryonic development marks a period of peak tissue growth and morphogenesis in the mammalian lifecycle. Many of the pathways that underlie cell proliferation and movement are relatively quiescent in adult animals but become reactivated during carcinogenesis. This phenomenon has been particularly well documented in pancreatic cancer, where detailed genetic studies and a robust mouse model have permitted investigators to test the role of various developmental signals in cancer progression. In this chapter, we review current knowledge regarding the signaling pathways that act during pancreatic development and the evidence that the reactivation of developmentally important signals is critical for the pathogenesis of this treatment-refractory malignancy.
I. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the 10th most prevalent cancer in the United States, with an estimated incidence of 42,000 people in 2009,1 and it is the fourth most common cause of cancer-related death. The prognosis of PDAC is poor, with only approximately 5% of patients surviving 5 years after diagnosis. The vast majority of PDAC patients die from complications of meta-static disease. Even patients who undergo pancreas resection for limited disease with no clinical evidence of metastasis have poor prognosis, as approximately 80% of these patients will also succumb to metastatic disease.2 Thus, while cancers of all origins share many of the same hallmarks,3 PDAC is unique in its ability to form large tumors and metastasize, ultimately killing the host and circumventing treatments that are efficacious in other malignancies. In order to intervene in this particularly lethal cancer, it will be essential to understand the unique features of pancreatic biology and the accumulation of molecular events that are seen exclusively in pancreatic cancer. We will begin with a brief review of the major themes of pancreas development and adult exocrine pancreas biology. Then, we will review the molecular underpinnings of PDAC, highlighting the reemergence of developmental pathways during cancer progression (Table I).
TABLE I.
Reemergent developmental pathways in PDAC
| Signaling pathway |
Effects in development | Effects in carcinogenesis |
|---|---|---|
| Hedgehog | Suppression required for pancreas specification | Canonical: Desmoplasia Noncanonical: Unclear |
| FGF | Pancreas specification, proliferation, differentiation, cell fate | Epithelial-to-mesenchymal transition (EMT), proliferation |
| Notch | Differentiation, cell fate | EMT, proliferation |
| TGF-β | Pancreas specification, proliferation, differentiation, cell fate | EMT, proliferation, desmoplasia |
| Retinoic acid | Pancreas specification, differentiation, cell fate | Unclear; putative marker of cancer stem cells |
| EGF | Proliferation, differentiation | Proliferation, desmoplasia |
| Wnt/β-catenin | Pancreas specification, cell fate | PanIN formation |
II. Anatomy of the Exocrine Pancreas
The pancreas is an endoderm-derived organ located in the upper abdomen of vertebrates, adjacent to the stomach, duodenum, and spleen. It maintains an independent blood supply from other abdominal organs. It also features a connection to the intestines via the main pancreatic duct, which serves to empty secretions from the pancreas into the intestinal lumen.
The pancreas comprises two compartments with distinct functions. The endocrine pancreas functions as a central regulator of glucose and metabolic homeostasis. Cells that compose the endocrine pancreas are organized within structures called the Islets of Langerhans (Fig. 1). Islets of Langerhans are nestled within the pancreatic parenchyma, often intimately associated with blood vessels. While islets are scattered throughout the pancreas, they are densely concentrated in the tail. Within these islets, α-, β-, δ-, γ-, and PP-cells secrete the hormones glucagon, insulin, somatostatin, ghrelin, and pancreatic polypeptide, respectively, in response to metabolic and chemical cues from the local blood supply. While carcinomas of the endocrine pancreas occur, these are relatively rare, and are beyond the scope of this chapter.
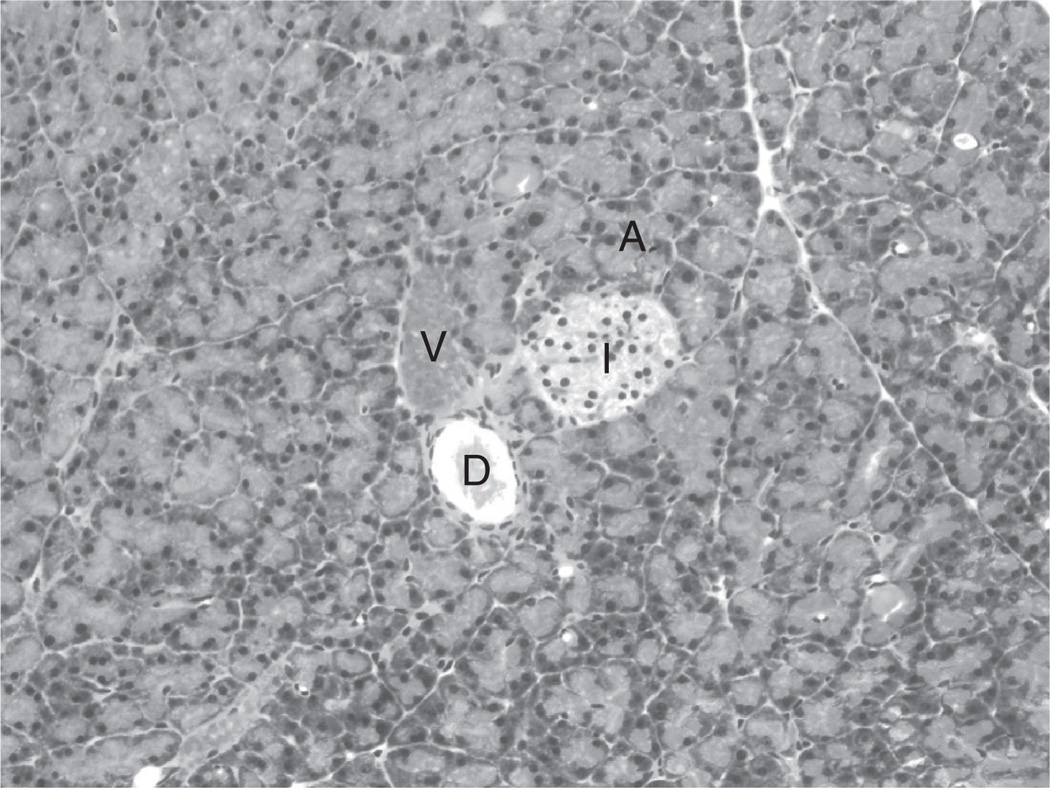
FIG. 1.
Histology of the pancreas. Hematoxylin and eosin staining of a normal pancreas, depicting an Islet of Langerhans (I), pancreatic duct (D), and blood vessel (V) surrounded by acinin(A). 10×.
The exocrine pancreas, from which PDAC arises, aids in the digestion of carbohydrates, fats, and proteins. Composing more than 90% of the organ, the exocrine compartment contains acinar and centroacinar cells interconnected by an elaborate epithelial-lined ductal network (Fig. 1). Acinar cells form discrete units, appropriately called acini, which, in cross-section, represent a circular cluster of polarized cells organized around a small concentric lumen. The apical portions of acinar cells face the middle of these acinar units, adjacent to the centrally positioned centroacinar cells. In response to the ingestion of food or drink, the upper intestine releases secretin and cholecystokinin, leading acinar cells to secrete inactive forms of digestive enzymes called zymogens. Zymogens undergo activation in an acidic environment; hence, concomitant release of bicarbonate by ductal cells maintains an alkaline microenvironment and prevents activation of digestive enzymes prior to exiting the pancreas. The lumens of the acinar units drain into a complex, interconnected network of epithelium-lined ducts which then anastomose to themain pancreatic duct, which in turn traverses the length of the pancreas. In most vertebrates, the main pancreatic duct represents the major conduit of drainage of pancreatic juice. The pancreatic duct then connects with the common bile duct, and pancreatic juice enters the intestine through the ampulla of Vater, located in the second portion of the duodenum.
Intercalated within the pancreatic parenchyma are mesenchyme-derived stromal cells, composed mostly of endothelial cells and quiescent fibroblasts. In non-diseased states, the stroma represents less than 1% of the pancreas. These cells are thought to primarily support the normal functioning of the epithelium by maintaining the intercellular structure of the organ. However, accumulating evidence has implicated pancreatic fibroblasts—especially the stellate cell, a specialized fibroblast—in additional tasks important for the normal functioning of the pancreas, including storage of Vitamin A, immune system regulation and surveillance, and maintenance of endothelial cell turnover.4,5 A complex, bidirectional interplay of signals between the stromal and epithelial compartments coordinates homeostasis within the organ. As will be discussed later, upon injury or carcinogenesis, signaling pathways active during pancreas development are reactivated, leading to the activation, accumulation, and diversification of the stromal compartment.
III. Development of the Exocrine Pancreas
Many cancers share common characteristics, as outlined previously3; however, each type of cancer is unique in its tissue of origin and developmental history. It is widely believed that the molecular events leading to carcinoma differ by tissue type and even by the specific cell of origin within a single organ. Consistent with these observations is the concept that key events leading to carcinogenesis involve the abnormal activation of developmental pathways specific to the ontogeny of the source or index cell.6 Indeed, recent work has supported the notion that this may be the case for PDAC. Thus, much insight into how PDAC forms has been attained by the assiduous study of normal exocrine pancreas development and mechanisms. Our understanding of early pancreas development comes from detailed studies using the mouse as a model system. By employing a variety of techniques, most importantly lineage labeling technology, investigators have been able to describe many of the morphogenic and genetic events involved in pancreas formation.
A. Early Steps Preceding Pancreatic Budding
Regional specification of the pancreas anlagen occurs as early as gastrulation. In studies of chicken development, fibroblast growth factor 4 (FGF4) released by the mesectoderm enables a region of the mesoderm to become responsive to propancreatic signals secreted by the mesoderm-derived notochord. Later in gastrulation, retinoic acid (RA) and bone morphogenic proteins (BMPs) are released, similarly allowing for the pancreatic anlagen to respond to pancreas-forming cues later in development.7,8 The pancreas derives from two primordia located in the dorsal and ventral aspects of the endoderm-derived primitive gut tube, forming distinct pancreatic buds that will eventually fuse. Upon completion of gastrulation, a patch of dorsal midgut endoderm finds itself in contact with the notochord through embryonic day 8 in the mouse (E8.0), eventually giving rise to the dorsal pancreas.
Precursors for the ventral pancreas are positioned adjacent to the lateral plate mesoderm. This geography leads to intimate interactions between the mesoderm and endoderm that are required for pancreas organogenesis.
B. Signals from the Notochord
The mesoderm-derived notochord is required for the eventual development of the dorsal pancreas.9,10 Excision of this structure at the 8–9-somite stage leads to abolition of subsequent expression of pancreatic marker genes, including pancreas and duodenal homeobox-1 (Pdx-1; Ref. 11). Several soluble notochord-derived factors have been implicated in pancreas genesis. The best characterized of these factors, activin-βB, a member of the transforming growth factor β (TGF-β) family, and FGF2, underscore the complexity of mesenchymal–endodermal interactions.12–14 Culture of the prepancreatic endoderm at the 9-somite stage with these factors was shown to be sufficient to induce the expression of pancreas-specific markers in the absence of notochord.15 In these experiments, physiologic levels of activin-βB and FGF2 both led to suppression of endodermal Sonic Hedgehog (Shh) ligand, a key event in pancreatic specification (see below). Indeed, in embryos at this stage cultured in vitro, dorsal Shh expression was repressed when the notochord remained intact, but this repression was lost upon excision of the notochord, resulting in dorsal pancreas agenesis. These observations suggest that patterning at the gastrulation stage leads to the geographic specification of the future dorsal pancreas via the silencing of Shh expression.
Activin and FGF signaling lead to a cascade of events in the pancreatic endoderm. Intracellular signaling through these pathways serves to inhibit Shh expression by these cells, thereby inhibiting Hedgehog signaling through the Gli family of transcription factors.16,17 These events are integral for the early expression of Pdx-1 at E8.5 (all developmental stages refer to the situation in mice unless otherwise noted). Pdx-1 is the first pancreatic transcription factor to be expressed, and it marks the dorsal and ventral pancreatic buds.18,19 Indeed, ectopic expression of Shh leads to loss of Pdx-1 expression in the pancreatic anlage. In a correlative experiment, Patched-null mice, in which Hedgehog signaling is permanently activated, die at E9.5 with no expression of Pdx-1 in the pancreatic anlage.15,17 Expression of Pdx-1 is critical for early events in pancreas formation, as Pdx-1 null mice have arrested pancreas development at the bud stage.18–20 FGF-receptor signaling in the endoderm also activates Notch signaling in the pancreatic endoderm at around E8–8.521,22 which may also play a role in the induction of Pdx-1 expression.
C. Early Signals from the Aortic Endothelium
Midline fusion of the dorsal aorta occurs at E8.5, leading to the interposition of the aorta between the notochord and the endoderm. Work by Melton and colleagues showed that endothelial cells from the aorta are required for dorsal budding as well as later endocrine differentiation.23 In fact, removal of the dorsal aortae in Xenopus embryos before fusion led to the lack of dorsal pancreas bud formation, presumably by preventing the induction of Pdx-1-expressing cells in the dorsal anlage.23,24 This observation led to the implication of vascular endothelial growth factor A (VEGF-A) as a factor involved in the specification of the developing pancreas. FGF10 is also released by the merged aortic endothelium and acts on the endoderm to promote the expression of Ptf1a, a key transcription factor necessary for normal pancreas development.25–27
D. Interactions with the Pancreatic Mesenchyme
After fusing of the aorta, the pancreatic mesenchyme becomes the primary signaling partner of the developing pancreatic endoderm. The early embryonic mesenchyme is thought to elaborate numerous signals that lead to the careful orchestration of important molecular events in the developing pancreas. As a direct result of midline aorta fusion, contact between the mesoderm-derived notochord and the underlying endoderm is disrupted, followed by condensation and brisk expansion of the surrounding mesenchyme around what will become the dorsal pancreas at approximately E9.0–9.5.9,28,29 Analogous events occur in the area of the ventral pancreas a short time thereafter. Subsequently, evagination of the pancreatic buds takes place, followed by elongation. It is at this point in time that mesenchymal proliferation rates are at their highest.30 The apical regions of the buds then undergo branching morphogenesis, forming outgrowths at acute angles, unique to pancreatic development. The sharp angles lead to the close approximation of the epithelium at branch points, with comparatively little mesenchyme between the basal ends of the branching epithelia, leading to the first signs of asymmetric distribution of mesenchyme within the developing pancreas. As will be discussed below, this redistribution of mesenchyme likely impacts the signals to the epithelia, affecting lineage decisions later in development. Fusion of the dorsal and ventral buds begins around E13 of mouse development, whereupon the cellular architecture of the pancreas starts to mature, and cells of the exocrine and endocrine lineage differentiate and organize accordingly. Throughout these stages of development, complex and diverse molecular interactions between the mesenchyme and the pancreatic epithelium take place. The careful geographic and time-dependent orchestration of epithelial–mesenchymal interactions has been well studied. However, given the complexity of these interactions, much is still unknown.
E. Key Signaling Pathways Involved in Exocrine Pancreas Development
Not surprisingly, a host of signal transduction pathways are utilized to form the pancreas. Because of space constraints, we will focus on pathways prominent in both exocrine pancreas development and in the progression of PDAC.
1. HEDGEHOG SIGNALING
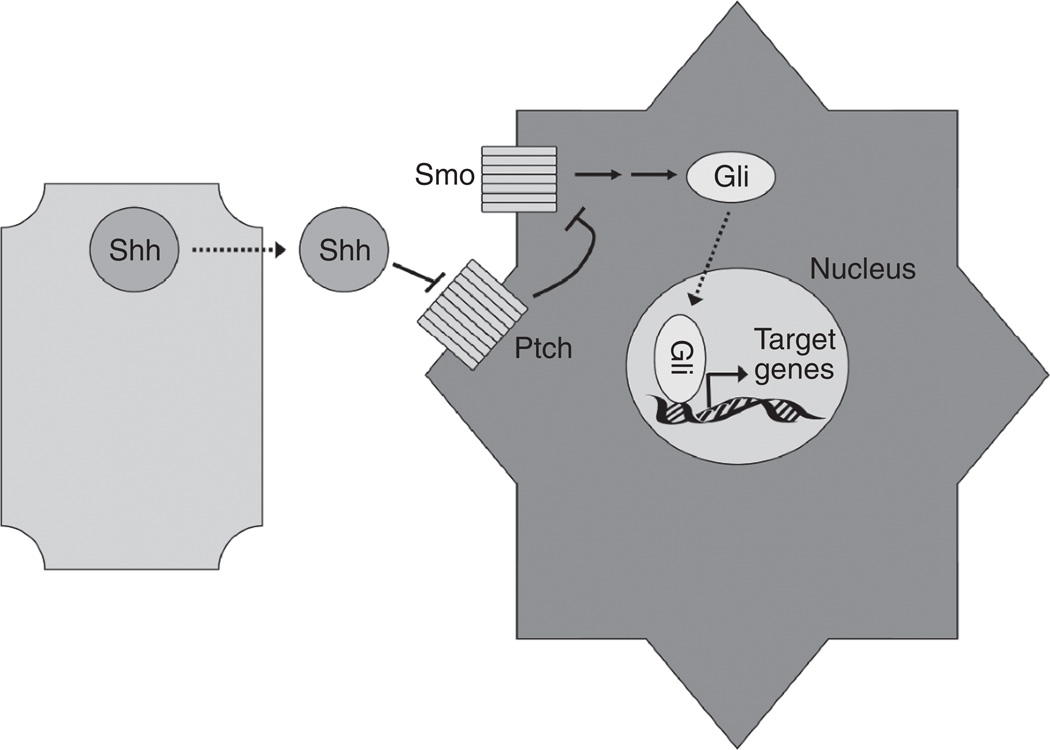
Hedgehog signaling occurs when a soluble ligand is secreted by a neighboring cell (Fig. 2). Three ligands are known to exist in mammals: Shh, Indian Hedgehog, and Desert Hedgehog. While each features different expression patterns and chemical structures, all elicit similar downstream effects as they bind to the same receptors.31 In the absence of ligand, the Patched receptor (Ptch) inhibits the activity of the G-coupled receptor protein Smoothened (Smo). Upon ligand binding, Smo dissociates from Ptch. Liberated Smo then initiates a cascade of events leading to the translocation of Gli transcription factors into the nucleus.
FIG. 2.
Hedgehog signaling. Paracrine Hedgehog signaling occurs when ligand (depicted here as Sonic Hedgehog, Shh) is secreted. Shh then binds to the Patched receptor (Ptch) on the target cell. When unbound by ligand, Ptch inhibits the transmembrane protein Smoothened (Smo). However, once ligand binds to Ptch, Smo is relieved of inhibition, leading to the activation of the Gli family of transcription factors and target gene expression.
As mentioned previously, repression of the Hedgehog signaling pathway is required for pancreas development. Forced expression of Shh in Pdx-1-expressing cells after E9.5 leads to loss of Pdx-1 expression in the dorsal pancreatic anlage and pancreatic agenesis, while ectopic expression of Shh later in pancreas development (E12.5) leads to the expected loss in pancreatic mass but also to expansion of the mesenchyme.32 In the intestine, Shh induces the adjacent mesenchyme to differentiate into smooth muscle16; hence, the absence of smooth muscle in the pancreas may be due to Shh repression at the earliest stages of development. The observation that epithelial-to-mesenchymal Hedgehog signaling, normally silenced during development, can induce stromal proliferation in the pancreas has provided clues about the mechanism of stromal expansion during pancreatic carcinogenesis. Interestingly, repression of Hedgehog signaling does not seem to be required for budding and development of the ventral pancreas.
2. FGF SIGNALING
The fibroblast growth factors comprise a group of heparin-binding proteins that share similar structures.33 These factors are involved in a variety of functions, including angiogenesis and mitogenesis. There are 14 FGFs known with varying expression domains and targets. FGFs mediate their actions after binding to one of four FGF receptors (FGFRs) on the target cell. FGFRs are subject extensive alternative splicing and glycosylation, resulting in a multitude of affinities and specificities for various FGF ligands. Binding of FGFR leads to the activation of a number of signal transduction pathways via tyrosine phosphorylation. 34 Multiple pathways are activated by FGFRs, including (1) the Ras-Raf1-MEK-MAPK pathway; (2) the phosphatidylinositol-3 kinase (PI3K) pathway; and (3) the phospholipase-C-gamma (PLC2γ)-phosphatidylinositol 4, 5-bisphosphate (PIP2) axis. These signaling cascades ultimately result in proliferative changes and effects on cellular differentiation.
Endoderm-to-mesenchymal FGF signaling plays an important role in pancreatic development. FGF4 elaborated by the pancreatic anlage, probably in concert with mesenchymal-derived FGF, serves to delineate the developing pancreatic mesenchyme early in development.35,36 Without this key molecular signal to the regional mesenchyme, the proper formation of the endoderm-derived pancreas does not take place.36 Later in development, once the pancreatic mesenchyme has undergone asymmetric distribution, FGF4 can cause the mesenchyme to persist, thereby enhancing exocrine differentiation of the adjacent epithelium.37
Several studies, including pioneering work from the Scharfman laboratory, have shown that mesenchyme-derived FGFs play a vital role in pancreas development. FGF1, 4, 7, 10 are all expressed by the pancreatic mesenchyme during development.38 These ligands bind to the dominant FGFR expressed in the pancreatic epithelium, FGFR2B.39 The first sign of mesenchymal-to-epithelial FGF signaling occurs at approximately E9.0, upon fusion of the aortic rings. FGF10 produced by the newly fused aorta leads to transcriptional changes in the dorsal pancreatic bud, resulting in the expression of pancreas-specific markers. FGFs elaborated from the cardiogenic mesenchyme similarly induce the expression of pancreatic markers in the ventral pancreas.40 FGF1, 7, and 10 signaling throughout the development of the pancreas promotes pancreatic epithelial proliferation, without eliciting cellular differentiation 38,41–43 before E16. Later in development, these mesenchymal factors are thought to guide pancreatic cells toward the exocrine lineage and inhibit endocrine differentiation by constraining the proliferation of endocrine progenitors. 37,44 Finally, it has been shown that FGF signaling can lead to Notch pathway activation in the pancreatic epithelium, which will be elaborated upon below.22,45 Later in development, signaling through FGFR3 inhibits endocrine differentiation and islet formation, possibly via the Notch pathway.46
3. NOTCH SIGNALING
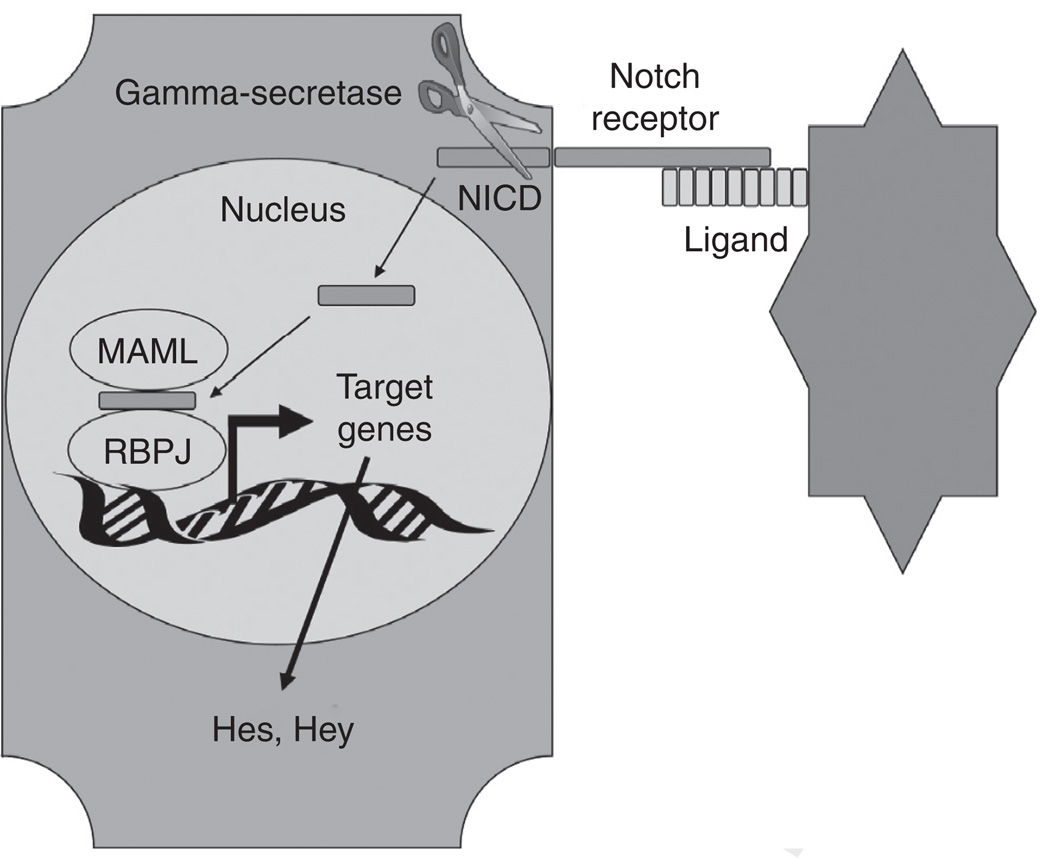
The Notch pathway has been implicated in many important basic events during mammalian organogenesis, including the delineation of tissue borders, the determination of cell fate, and differentiation within an organ.47–49 Signaling occurs when one of four receptors (Notch 1–4) binds to one of five known Notch ligands (Delta-like 1, 3, 4, and Jagged1, 2) expressed by an adjacent cell (Fig. 3). A series of proteolytic cleavages then occurs near the transmembrane domain of the Notch receptor, culminating in the release of Notch intracellular domain (NICD) by γ-secretase. NICD translocates to the nucleus and complexes with the transcription factor RBPJκ, switching it from a transcriptional repressor to activator. This leads to the transcription of select basic helix-loop-helix proteins, including hairy/enhancer of split 1 (Hes1), which mediate the downstream effects of Notch.
FIG. 3.
Notch signaling. Notch signaling is initiated when the extracellular portion of a Notch receptor contacts a Notch ligand from a signaling cell. This triggers the gamma-secretase-mediated cleavage of the intracellular domain of the engaged Notch receptor. This Notch intracellular domain (NICD) translocates to the nucleus where it complexes with Mastermind-like protein (MAML) and recombining binding protein suppressor of hairless (RBPJκ). This complex then mediates transcription of Notch target genes such as the Hes and Hey family of genes.
In specific reference to signals arising from mesenchymal–epithelial interactions, the Notch pathway is indirectly activated by paracrine FGF signaling. Mesenchymal FGF ligands bind to FGFR2B on the pancreatic epithelium during development. Through elegant transgenic experiments, Notch signaling was shown to be activated by FGFR2B signaling, culminating in an increase in the expression of the Notch effector molecule Hes1.22,36 Other downstream effects of Notch signaling include the downregulation of p57, a cyclin-dependent kinase inhibitor, and effects on the Ptf1a transcriptional complex.25,50–52 Later in development, mesenchymal-to-epithelial FGF signaling tilts the scales in favor of exocrine differentiation. After the appearance of mature pancreatic buds, there is a decline of FGF and Notch signaling in the developing exocrine pancreas,53 concurrent with the initiation of acinar differentiation.
It is thought that the bulk of Notch signaling in the pancreas occurs between epithelial cells, in a process commonly known as lateral inhibition. Several studies in mice have demonstrated that epithelial Notch signaling is critical for cell fate determination within the pancreas. But, as with most signaling pathways during development, the dynamics of lineage specification through Notch signaling is complex. Early in development, Hes1 expression serves to maintain cells in a progenitor state by inhibiting the expression of neurogenin3 (ngn3; Refs. 54,55;. Later in development, however, a shift in the glycosylation of the Notch receptor in a subpopulation of pancreatic progenitor cells leads to the induction of ngn3 expression, resulting in endocrine differentiation. 56 The timing of Ngn3 induction (secondary to repression of Notch signaling) may be the critical factor that determines endocrine cell fate.57 In addition, it was shown by conditional gene ablation that Jagged1, the dominant Notch ligand in the pancreas, can act as a competitive inhibitor of Notch signaling in the embryonic pancreas, while postnatally, it has its expected role as pathway activator.58 Notch signaling is thought to be persistently active in adult centroacinar cells, the terminal ductal epithelial cells found abutting the acinus.59 However, the precise lineage of these cells is unknown. Because Notch signaling is apparently quiescent in mature exocrine cells, the centroacinar cell has been theorized to be a rare progenitor cell present in the adult pancreas60 and a candidate cell of origin for PDAC.61
4. TGF-β SIGNALING
The TGF-β signaling family represents a group of diverse and versatile pathway constituents that play important roles in a number of biological processes, including embryonic development, extracellular matrix production, and apoptosis. For a more in-depth review of this pathway in pancreas development, the reader is referred to two comprehensive reviews in the subject. 62,63 Signaling is first initiated when a ligand from the TGF-β superfamily binds to a specific receptor. There are six types of ligands which, upon receptor engagement, result in unique downstream effects. Four of these play active roles in pancreas development and have been implicated in PDAC progression: (i) TGF-β family ligands, (ii) activins, (iii) BMPs, and (iv) growth differentiation factors (GDFs). Upon binding of ligand to a serine–threonine kinase receptor, the kinase domains undergo conformational change allowing for phosphorylation of receptor-regulated SMADs, intracellular proteins that shuttle from the plasma membrane to the nucleus.
The TGF-β family of ligands plays a key role during pancreas development, as well as mediating terminal differentiation events. All three TGF-β isoforms (TGF-β1, 2, and 3) are expressed in the embryonic pancreatic epithelium as early as E12.5.64,65 The mesenchyme does not express this ligand, and, during early exocrine differentiation, the major type II receptor for this ligand, TGFBRII, is exclusively located in the epithelium.64 As acinar cells are first specified, TGF-β ligand expression becomes sequestered in this compartment. 64,66 By E18.5, TGF-βRII expression is found only in the pancreatic ducts. These data suggest that in later development, acinar-to-ductal TGF-β signaling coordinates the precise formation of the ductal network. This notion was confirmed when mice expressing a dominant-negative form of TGF-βRII displayed aberrant redundant pancreatic ducts.67
The two activin ligands (Activin A and B) play important roles during pancreas development. One role of Activin B is to promote the specification of the dorsal pancreatic anlage by inhibition of Shh secretion (See Signals from the Notochord). Later in development, however, the expression of these ligands is restricted to cells of the endocrine lineage,68,69 and inhibition of Activin signaling is thought to be critical for the formation of the exocrine compartment through the secretion of follistatin, an inhibitor of Activin signaling. 70 Continued Activin signaling prevents branching morphogenesis.71 Thus, Activin signaling later in pancreas development provides a general proendocrine and antiexocrine cue. Indeed, exogenous activin supplementation has been used to promote beta cell differentiation in embryonic stem cells.72
BMP signaling is required for early pancreas development, cellular differentiation, and cell growth, though a thorough understanding of the role of this pathway later in development and in the adult pancreas is lacking. The most prominent BMPs expressed in the developing pancreas are BMP 4, 5, 6, and 7. BMPs 4, 5, and 6 are expressed in the pancreatic epithelium, whereas 4, 5, and 7 are expressed by the pancreatic mesenchyme.37,73,74 The receptors for these ligands, BMPR1A and B, and BMPR2 are expressed in both pancreatic epithelium and mesenchyme. As discussed earlier, BMP signaling from the mesoderm to the pancreatic anlage is one of the earliest steps in organogenesis. It has been shown recently that later in development, BMP signaling in the mesenchyme is required for proper pancreas size and branching.73 Interestingly, overexpression of BMP in Pdx-1-expressing cells led to pancreas agenesis, presumably due to stromal and smooth muscle hypertrophy in the overlying duodenal anlage.37
GDF11 is thought to be an important factor for acinar cell development. GDF11 is robustly expressed in the pancreas between E11.5 through E13.575 and is selectively expressed by acinar cells afterward. Two GDF11 null mice have been derived with slightly different phenotypes reported. Harmon and colleagues found that lack of this factor led to an expansion of Ngn3+ endocrine progenitor cells. They suggested that this might have been a result of HNF6 induction.76 Dichmann and colleagues found that knockout of GDF11 led to reduced pancreas size due to acinar cell loss, although the number of Ngn3+ cells was also elevated.77 Taken together, these experiments suggest that GDF11 expression promotes acinar cell survival and may influence endocrine cell fate.
5. RETINOID SIGNALING
Retinoids are a family of Vitamin A analogues that have a multitude of effects on various types of tissue both during development and in mature organs, including anterior/posterior patterning and cell fate decisions. Retinoids are produced by aldehyde dehydrogenase (ALDH). Multiple isoforms of ALDH exist, but the major isoform is ALDH2. Once synthesized, the RA analogues are secreted and bind in a mostly paracrine manner to heterodimers composed of a retinoic acid receptor (RAR) and a retinoic acid X receptor (RXR). This complex is then endocytosed into the cell where it mediates transcription via binding to retinoic acid response elements (RARE) in the regulatory domains of various genes.78
RA signaling has been shown to be critical to the development of both the endocrine and exocrine pancreas, though not as much is known regarding its role in the homeostasis of the adult pancreas. In zebra fish and mice, RALDH2 is expressed specifically by the mesenchyme; RA released by the mesenchyme serves to aid in the specification of the dorsal pancreatic bud.79,80 However, RA signaling was not evident in the ventral pancreatic anlage, even though the lateral plate mesoderm expressed RALDH2.81 Indeed, RALDH2-null mice fail to form a dorsal pancreas. Later in development, mesenchymal RA induces differentiation in insulin-positive cells in zebrafish,80 though this has not yet been fully confirmed in mice.79,80 With respect to the exocrine pancreas, the Gittes laboratory showed that RA elaborated by undifferentiated exocrine progenitor cells facilitates laminin upregulation in the surrounding mesenchyme, which then acts reciprocally on the epithelium to elicit ductal differentiation. 82 Thus, RA signaling occurs in a bidirectional manner in the developing pancreas, underscoring the complexity of epithelial–mesenchymal interactions.
6. EPITHELIAL GROWTH FACTOR SIGNALING
The epithelial growth factor (EGF) family comprises more than 30 ligands that can bind to multiple ErbB tyrosine kinase receptors. Several EGF ligands and receptors are expressed by the pancreatic epithelium and mesenchyme, which likely mediate reciprocal epithelial–mesenchymal interactions through the EGF receptor pathway.83 EGF ligands mediate a variety of biological effects, including proliferation and differentiation, depending upon the particular ligand–receptor pair. During development, the EGF receptors ErbB1 and ErbB4 are expressed throughout the pancreas starting at E12.5. After this time point, ErbB1 is expressed mainly in endocrine cells, whereas ErbB4 is relegated to the exocrine compartment. Exogenous EGF led to proliferation of progenitor cells in vitro and in vivo.84,85 Continued exogenous EGF treatment led to enhanced duct development,85 while withdrawal after treatment led to preferential endocrine differentiation.84
7. WNT SIGNALING
The Wnt signaling pathway controls a wide variety of cellular outputs depending on the identity and concentration of ligand and cell type studied. Signaling is initiated when soluble Wnt ligand binds to the extracellular portion of amember of the Frizzled family of receptors. In turn, a member of the Dishevelled family—in complex with a Frizzled receptor by way of lipoprotein-related peptide (LRP)—becomes activated. This event frees the protein Axin to inhibit a “β-catenin destruction complex” comprising GSK-3 and APC. This complex catalyzes the phosphorylation of cytoplasmic β-catenin, targeting this protein for proteolytic degradation, though the precise details of this interaction are still under intense scrutiny. When Wnt ligand is engaged with Frizzled, β-catenin can translocate to the nucleus to interact with the TCF/LEF family of transcription factors.
Like some of the aforementioned transduction pathways, Wnt signaling is required for normal pancreas development. Wnt8 has been implicated in early endodermal patterning, and Xenopus embryos lacking Wnt8 fail to develop pancreas or liver.86 Numerous Wnt ligands, Frizzled receptors and soluble inhibitors are expressed in the developing mouse pancreas,87 with ligand expression predominantly located in the mesenchyme and receptors and soluble inhibitors located in both compartments. Perhaps more than for any of the other pathway, the spatiotemporal expression of Wnt family members dictates the result of signaling. A multitude of different Cre lines have been used to perturb Wnt signaling in different tissue compartments at different time points during pancreatic development.
When Grapin-Botton and colleagues deleted β-catenin from pancreatic epithelial cells early in development, the resultant mice exhibited both endocrine and exocrine phenotypes.88 Later in development, β-catenin deletion led to almost complete absence of exocrine tissue, though the survival of Pdx-1-expressing progenitors was not affected.89,90 Constitutive activation of β-catenin at early time points led to pancreatic agenesis through inhibition of FGF10 and subsequent increase in Shh levels.44 Postnatally, β-catenin stabilization in Pdx-1-expressing progenitors resulted in a significant increase in the size of the pancreas, with equal effects on all lineages.91 Thus, the Wnt/β-catenin signaling pathway exerts diverse and complex effects during pancreas development depending on location and timing.
IV. Genetic Alterations in PDAC
While PDAC can occur in any individual, regardless of sex, race, or geography, there are certain underlying commonalities to all pancreatic tumors that not only aid in the diagnosis of the disease but also contribute to our evolving understanding of its pathogenesis. PDAC is a cancer of the exocrine pancreas which leads to an aggressive disease in which regions of duct-like epithelium are interspersed with less differentiated epithelial cells contained within a sea of proliferative stroma. Metastasis is extremely common. While most patients will succumb to their disease, the genetic events leading to metastatic spread differ from patient to patient. Nevertheless, a few genes are reproducibly mutated in pancreatic cancer, indicating that certain molecular changes are required for the development of this malignancy.
A. Kras
Kras was the first oncogene to be identified as playing a critical role in PDAC. A number of functions have been proposed for the Kras GTPase, including survival, proliferation, and differentiation.92 These diverse activities are driven by Kras-mediated activation of three important signaling cascades that also play prominent roles during pancreas development: the RAF/ERK/MAP kinase, PI3 kinase, and RalGDS pathways.93,94 Kras also activates the nuclear factor κB (NFκB) pathway, which is implicated in the generation of a rigorous inflammatory response.95 Pharmacologic suppression of each of these pathways separately effects pancreatic cancer cell growth in vitro, though none have resulted in a durable response in vivo. An activating mutation at codon 12 in Kras is detected in close to 100% of all PDACs and in many early neoplastic lesions.96–98 With the advent of conditional gene mutation in the mouse, the biology resulting from oncogenic Kras expression has been well documented.99 Targeted expression of activated Kras in the pancreatic epithelium promotes the formation of the precursor lesion “pancreatic intraepithelial neoplasia” (PanIN; further discussion below; Ref. 100), supporting the notion that Kras mutations are one of the earliest events leading to PDAC.101
Activating Kras mutations in PDAC exhibit the property of “oncogene addiction,”102 and three studies have shown that genetic silencing of Kras expression can singlehandedly arrest cancer cell growth.103–105 As a consequence, pharmacologic inhibitors of Kras have been rigorously pursued,106 but to date no clinical efficacy has been detected.107
Activating Kras mutations are not sufficient for PDAC formation. The observation that Kras mutations are common among patients with chronic pancreatitis supports this notion.108 While expression of oncogenic Kras in genetic mouse models is not sufficient to produce PDAC, the additional insult of chemical pancreatitis by cerulein elicits PDAC in most mice.109,110 This experimental model is consistent with the well-documented 16-fold increased risk for PDAC in patients with chronic pancreatitis over the general population. 111 However, confounding these experiments is the observation that cerulein and other CCK analogues increase Kras expression independently of any induced inflammatory response.112 Recent work from the Logsdon laboratory suggests that levels of Kras expression also drive the early stages of pancreatic carcinogenesis.113 In this elegant study, Ji and colleagues showed that forced expression of supraphysiologic levels of wild-type Kras was sufficient to induce inflammation reminiscent of human chronic pancreatitis and, later, produce PanIN lesions. When combined with loss of the tumor suppressor p53 (see below), these lesions progressed to PDAC. Taken together with the observation that many patients with chronic pancreatitis harbor mutations in Kras, without developing PDAC, these data suggest that increased Kras activity in itself is the key mechanism in PanIN initiation but is not sufficient to produce PDAC.
B. 9p21 Locus: Ink4a and ARF Tumor Suppressors
The second most common genetic events in PDAC are mutations occurring at the Ink4a/ARF tumor suppressor locus, occurring in up to 95% of all tumors.98,114 Promoter hypermethylation, mutation, and deletion have all been described as Ink4a-inactivating mechanisms. The sequence for Ink4a actually overlaps that of another tumor suppressor, ARF, in the 9p21 locus.115 While both genes have tumor-suppressing functions, they work independently of each other, and Ink4a loss is thought to be the primary event underlying PDAC.
Ink4a is involved in the initiation of cell senescence in times of environmental stress and aging, as well as in response to telomere erosion.116,117 Specifically, Ink4a halts entry into S-phase of the cell cycle by inhibiting CDK4/6-mediated phosphorylation of Rb. Germline mutations in Ink4a are associated with the Familial Atypical Mole-Malignant Melanoma syndrome (FAMMM). Patients with FAMMM display a 13-fold increased risk for developing PDAC.118,119 Interestingly, in addition to telomere shortening, high levels of Kras can induce Ink4a expression, leading to senescence.120 Thus, Ink4a loss may be required for cells to bypass the senescence phenotype promoted by activated Kras. Loss of Ink4a expression, in combination with the expression of activated oncogenic Kras, leads to rapid development of PDAC in mouse models.121 Thus, it is likely that in the setting of a mutation in Kras, deactivation of the senescence mechanisms is sufficient to lead to carcinogenesis in the pancreas.
The ARF tumor suppressor has divergent functions from Ink4a. The best studied function of ARF is the inhibition of MDM2-mediated proteolysis of the tumor suppressor p53, thus stabilizing this protein (see below). Indeed, the concentration of p53 protein in the cell is directly proportional to that of ARF.98,122 However, the entirety of ARFs functions has yet to be elucidated. Underscoring the presence of additional functions of the ARF tumor suppressor is the observation that mutations in both p53 and ARF are present in almost half of all PDAC.114,123 Indeed, tumor growth and mortality is greatly accelerated in genetic mouse models harboring oncogenic Kras and absence of ARF and p53, compared to mice with oncogenic Kras with either protein alone.124 Thus, ARF has tumor-suppressing functions independent of its effects of p53 stability.
C. The p53 Tumor Suppressor Protein
p53 serves as a tumor suppressor by mediating DNA checkpoint responses, usually in the setting of an accumulation of mutations from reactive oxygen species and telomere shortening, both of which take place during PanIN progression. Thus, it is not surprising that p53 mutations, usually in the form of missense mutations,98 are seen in later stage PanIN lesions.123,125 Following p53 loss, it is conceivable that the rate of genetic aberrancy increases drastically, possibly contributing to the immense heterogeneity of pancreatic tumors and their resistance to multiple chemotherapeutic agents.126 Even so, p53 is mutated in only about half of all PDAC cases,98 though p53 levels are suppressed in a much greater fraction.125
D. SMAD4/DPC Tumor Suppressor
SMAD4 (DPC) is another commonly mutated gene in PDAC, with mutations present in about 50% of all cases.127 Similar to p53, loss of SMAD4 usually occurs late in PanIN progression to PDAC.128 As touched upon earlier, SMAD4 is an integral member of the TGF-β signaling cascade. The effect of SMAD4 loss in the exocrine pancreas is further discussed below.
E. LKB1/STK11 Tumor Suppressor
The LKB1 protein product is a serine–threonine kinase involved in a multitude of epithelial cell processes, including the regulation of cell metabolism and establishment of cell polarity.129 Many of its diverse functions are mediated through the regulation of AMPK, thereby affecting signaling through the mTOR pathway.130–132 Germline mutations in LKB1/STK11 have been identified as the cause of Peutz-Jeghers syndrome, in which patients sustain benign intestinal polyposis as teenagers. These patients have a more than 40-fold increased risk of developing PDAC.133 LKB1 is also mutated in approximately 4% of all cases of spontaneous PDAC, though this rate is higher in intraductal papillary mucinous neoplasms.134
F. The BRCA2 Tumor Suppressor Protein
BRCA2, similar in function to BRCA1, is involved in the correction of double stranded breaks in DNA. In this way, BRCA2 is critical for the maintenance of genomic stability. Loss of BRCA2 leads to the rapid accumulation of chromosomal aberrations (Venkitaraman, 2002). Patients with BRCA2 mutations are known to be at elevated risk for developing PDAC, and mutations in BRCA2 account for up to one-fifth of all cases of familial PDAC (Murphy, 2002). BRCA2 mutations are also common in sporadic cases of PDAC.
Recently, the “partner and localizer” of BRCA2 (PALB2) was identified as a possible pancreatic cancer susceptibility gene.135–137 PALB2, involved in targeting of BRCA2 to the nucleus as well as in BRCA2-mediated repair of double-strand breaks, was initially identified as a susceptibility gene for breast cancer.138 Two years later, Jones and colleagues identified three separate mutations in PALB2 in families with familial pancreatic cancer, thus establishing PALB2 as a susceptibility gene for pancreatic cancer as well. While subsequently confirmed,136,137 one group was unable to find an association between polymorphisms in PALB2 and increased pancreatic cancer risk.139
G. Palladin
Palladin was identified to be mutated in a family with inherited pancreatic cancer.140 The protein product is relatively large and is thought to act as a scaffold for other proteins to bind actin. Thus, palladin is the first oncogene in PDAC to encode for a cytoskeletal protein.141 Palladin has also been found to be important for cell motility and differentiation.142 While previous work focused on the role that such a mutation could play in pancreatic cancer cells, later work identified the bulk of palladin to be expressed in the stroma, and, specifically, in cancer-associated fibroblasts (CAFs; Ref. 143). In the setting of PDAC, CAFs were found to overexpress specific isoforms of palladin, not otherwise seen in normal pancreata. These data, combined with the observation that palladin is not consistently upregulated in cancer cells, suggest a novel, but yet unconfirmed, cell-nonautonomous mechanism for carcinogenesis. Thus, further studies dissecting the role of the stroma, and specifically CAFs, in PDAC formation will likely be very insightful.
H. Temporal Accumulation of Signature Mutations During PanIN-to-PDAC Progression
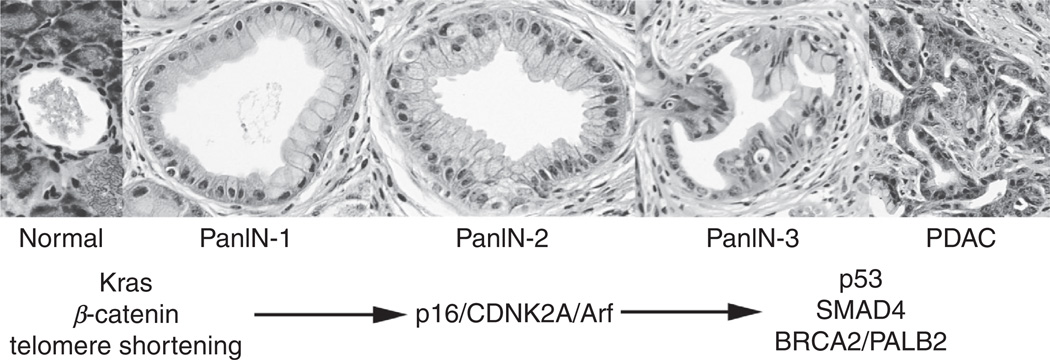
Three neoplastic, noninvasive precursor lesions to PDAC have been identified: (1) IPMN, (2) mucinous cystic neoplasm (MCN), and (3) PanIN. PanINs are the most prevalent and have been identified in up to 16–28% of individuals based on autopsy data.144,145 The prevalence and severity of PanIN lesions increases with age, and, as touched upon previously, with certain medical conditions such as chronic pancreatitis.144 Consensus conferences have classified PanINs according to grades of increasing abnormality (Fig. 4; Refs. 146,147). PanIN1 lesions exhibit flat (PanIN1A) or papillary (PanIN1B) features, though both are characterized as having a columnar appearance with accumulation of mucinous material and nuclei uniformly located along the basal aspects of the cell. PanIN2 lesions are classified by their loss of cellular polarity and some nuclear pleomorphism. PanIN3 lesions display complex and heterogenous structures within a single lesion, including papillary and cribiform-like structures. Nuclei in these lesions are extremely pleiomorphic, randomly oriented, and show obvious mitotic figures.
FIG. 4.
PanIN-carcinoma sequence. Most PDAC is thought to arise from the sequential progression of PanIN lesions, as depicted. The earliest event underlying PanIN formation is thought to be an oncogenic mutation in Kras. Recent evidence suggests that Wnt-β-catenin is required for this early step. PanIN2 formation is thought to coincide with deactivating mutations in the p16/p19 tumor suppressor locus. Late stage PanINs exhibit mutations in p53, SMAD4, BRCA2, and PALB2.
The molecular basis of PanIN formation has been partially revealed through the creation of genetically engineered models of PDAC, in which PanIN progression can be observed to occur in a step-wise manner, reflecting the accumulation of signature mutations that are also seen in human samples. To a rough approximation, activating mutations in Kras and telomere shortening characterize PanIN1 lesions, loss of Ink4a correlates with PanIN2 formation, while p53, SMAD4, BRCA2, and LKB1 loss are associated with the development of late stage PanIN lesions and frank PDAC.123,148 Thus, based on available evidence, PanIN progression to PDAC follows a somewhat predictable genetic-histological sequence, similar to the well-described polyp-to-carcinoma progression scheme described for colorectal cancer.149
Another important trend during tumor development is the dramatic buildup of stroma as PanIN lesions progress to frank carcinoma. Indeed, one of the distinguishing hallmarks of pancreatic cancer is its exuberant desmoplastic response. The stromal infiltrate is composed of a number of cell types, including fibroblasts, macrophages, and endothelial cells. As the interstitium is flooded with these cells, matrix and fibrotic material become highly abundant, leading to significant changes in the physical characteristics of the pancreas itself. As tumors form, hardened nodules arise as a result of local hyperactivation of matrix-secreting stroma adjacent to cells that have undergone malignant transformation. It is likely that this desmoplastic response is required for the accelerated nature of tumor growth and metastasis seen in this disease and that intercompartmental cross-talk through prominent developmental pathways is required for this conditioning of the neoplastic microenvironment.
V. Activation of Developmental Pathways Driving Cancer Progression
Studies utilizing genetically engineered mouse models have revealed that several developmentally relevant signal transduction pathways that are quiescent in the adult pancreas reemerge during cancer progression. The activation of these pathways seems to be shared by the vast majority of pancreatic tumors, in mice and humans.
A. Hedgehog Signaling
While the suppression of Hedgehog signaling is critical for the development of the embryonic pancreas, sustained activation of this pathway is an important contributor to the pathogenesis of PDAC. Thayer and colleagues were the first to describe the aberrant expression of Shh ligand, starting in early PanIN lesions and continuing through carcinoma in human tissue specimens,150 and this finding has been confirmed by others.123,148,151 The involvement of the Hedgehog pathway in PDAC is also supported by the work of Jones and colleagues,152 in which genetic alterations in Hedgehog pathway genes were found in a large fraction of pancreatic tumors. In a mouse model in which constitutive expression of Hedgehog signaling via Gli2 was combined with an oncogenic mutation in Kras (KrasG12D), PanIN formation and PDAC development were accelerated compared to Kras mutation alone.153 Furthermore, Hedgehog signaling was shown to act synergistically with Kras signaling by protecting pancreatic epithelial cells from apoptosis via PI3 kinase signaling.151 Finally, pharmacologic and/or genetic inhibition of Hedgehog signaling attenuates the growth and metastasis in a variety of mouse models of PDAC.154–157 Thus, Hedgehog signaling seems to be consistently activated in PDAC, where it plays an important functional role in tumor growth.
The distribution of Hedgehog signaling within the neoplastic pancreas has been a point of controversy, but it is likely that both stromal and epithelial compartments are subject to pathologic Hedgehog signaling in cancer. As stated previously, Hedgehog signaling during development occurs in a paracrine manner whereby epithelially expressed ligand acts upon the mesenchyme. Consistent with this, Shh ligand is exclusively secreted by the epithelial compartment in both PanIN and PDAC lesions.150,151,158 Furthermore, in vivo reporter assays have shown that “canonical” Hedgehog signals act exclusively in the stromal (mesenchymal) compartment in PanINs and PDACs.158 These results support the notion that paracrine Hedgehog signaling leads to profound phenotypic changes in the stroma, including diversification and activation of matrix production (Bailey, 2008). Indeed, inhibition of Hedgehog-mediated conditioning of the microenvironment has been associated with the restriction of growth and metastasis in orthotopic transplantation models of PDAC.154,156,157
Even so, other studies suggest that downstream components of the Hedgehog pathway are activated in the neoplastic epithelium as well.123,148,150,151,159 Nolan-Stevaux and colleagues showed that genetic depletion of Smo in a genetically engineered mouse model (p48-Cre; LSL-KrasG12D/+; Trp53F/+; SmoF/F) developed PDAC with the same rate and time course as control mice.160 However, further analysis of the resultant tumors showed that Gli target genes continued to be expressed in ductal structures, suggesting that the downstream mediators of Hedgehog signaling—Gli transcription factors—were activated by a noncanonical, Smo-independent mechanism. Thus, it is likely that downstream components of the Hedgehog pathway are active in both the epithelium and stroma of pancreatic cancers.
An important contribution to our understanding of the role of Hedgehog signaling in the pathogenesis of PDAC comes from the work of Tuveson and colleagues.157 This group utilized the well-established mouse model of PDAC, the KPC model (Pdx-1-Cre; LSL-KrasG12D/+; Trp53F/+). In this model, tumors were highly impermeable to the delivery of fluorescently tagged drugs; indeed, impermeability may be one reason that pancreatic tumors are so exceptionally refractory to standard chemotherapeutic agents. Remarkably, upon treatment with a small molecule inhibitor of Smo (IPI-926), the stromal compartment reorganized and condensed, leading to increased vascularization. While tumor angiogenesis is commonly viewed as having a protumorigenic effect, in this experimental model the expanded vasculature permitted enhanced delivery of the chemotherapeutic agent gemcytabine, resulting in some tumor cell death. These data provide an important proof of principle that targeting stromal Hedgehog signaling can enhance chemotherapeutic delivery and efficacy in PDAC.
B. FGF Signaling
The FGF pathway is one of the first signaling pathways to be associated with PDAC, beginning with observations from Murray Korc’s laboratory. FGFs are upregulated in approximately 60% of all PDAC samples, and the degree of FGF expression correlates with advanced tumor stage and shorter patient survival at the time of diagnosis.161,162 However, the evidence supporting a functional role for FGFs in PDAC progression comes mainly from in vitro studies. PDAC cells express high levels of FGFR3b and FGFR3c, which signal through the MAP- and Jun-kinase pathways to affect changes in cell proliferation, adhesion, and/or motility.163–165 The FGFR3b isoform has also been associated with cells that secrete SPARC (secreted protein acidic and rich in cysteine), a molecule that has been implicated in proproliferative signals and aggressive phenotypes in PDAC.166 Moreover, FGF10 signaling through FGFR2 has been shown to induce cancer cell migration and invasiveness.167 A role for FGF1 and 2 signaling in modulation of epithelial-to-mesenchymal transition (EMT) and motility has also been proposed.168 It is surprising, given the high levels of FGFs in PDACs, that these tumors are relatively hypovascular, since one of the major effects of FGF is the induction of angiogenesis.169
C. Notch Signaling
Given its role in maintaining pancreatic progenitor cells in an undifferentiated state, it is not surprising the Notch pathway is active in PanIN progression and PDAC. The first description of Notch pathway activation during pancreatic carcinogenesis came from the laboratory of Steven Leach.59 In that study, comparison of 26 human PDAC specimens with 29 normal pancreas samples revealed overexpression of a variety of downstream Notch effectors and upregulation of several Notch receptors and ligands. Many of these findings have since been confirmed in other studies,170–173 suggesting that the Notch pathway is broadly activated in PDAC. More recently, global genomic analysis of 24 human pancreatic cancer specimens revealed a high frequency of mutations affecting Notch pathway constituents.152
Several studies suggest that Notch promotes PDAC cell growth through autocrine signaling. Repression of Notch signaling in cultured pancreatic cancer cells, either through pharmacologic or genetic inhibition, leads to reduced proliferation in vitro.59,171,174–176 Notch activation causes pancreatic epithelial cells to adopt a more undifferentiated, duct-like phenotype.59 Furthermore, Notch signaling exhibits synergy with the Kras pathway, and the combination of a constitutively active Notch transgene with activated Kras dramatically increased the rate of PanIN formation in genetically engineered mouse models, though these lesions did not progress to PDAC.175,177 Finally, a recent study suggests that Notch 2 signaling as a consequence of Jagged1 ligand binding was associated with the acquisition of a mesenchymal phenotype, featuring expression of the EMT markers Zeb1, Slug, and Snail.176
Given these data, investigators have sought to inhibit Notch signaling as a way to constrain cancer progression in the pancreas.178 Indeed, using the PKC model of spontaneous PDAC, treatment with a small molecule inhibitor to the gamma-secretase enzyme, an obligatory step in Notch signaling, led to arrest of PanIN progression with no carcinoma detected in any of the treated mice.174 Similar results have been seen using other gamma-secretase inhibitors in xenograft models.171
D. TGF-β Signaling
The TGF-β signaling pathway is frequently dysregulated in PDAC. Indeed, as discussed above, loss of TGF-β signaling via Smad4 loss is seen in approximately half of all PDAC tumors. Mutations in the TGF-β receptor are present in a number of patients lacking Smad4 mutations, and collectively the vast majority of PDACs exhibit genetic alterations in a component of the TGF-β pathway by global genomic analysis.152 The general effect of these mutations is the silencing of signals that suppress proliferation and maintain an epithelial phenotype.63
Given space constraints, only a few of the many studies detailing alterations in TGF-β signaling in PDAC will be described here. One of the earliest studies to examine TGF-β signaling in PDAC was performed almost 20 years ago by the laboratory of Murray Korc.179 Sixty human pancreatic cancers were analyzed, showing that the three TGF-β ligands were expressed in about 40% of all tumors. The expression pattern of the isoforms was found to differ compared to controls with preferential expression of TGF-β2 by aggressive tumors of advanced stage. The relative expression of TGF-β receptor subtypes was also altered in PDAC compared to controls in another study.180 PDAC specimens exhibited a fivefold increase in TGF-βRII transcript, which was expressed in virtually all cancer cells. However, TGF-βRIII was not expressed in cancer cells, but rather, was confined to the stromal compartment. Indeed, later studies showed that TGF-β1 binding to this receptor induced a particularly aggressive desmoplastic response in a mouse model of PDAC.181 These and other data point to the existence of both paracrine and autocrine TGF-β signaling during PDAC progression.
A number of specific effects have been documented for TGF-β signaling in the cancer cell itself. Recent work has detailed an important role for TGF-β ligand-dependent signaling in the acquisition of a mesenchymal phenotype late in cancer development. Reflecting data seen in colon cancer, TGF-β signaling led to activation of NF-κB and the downregulation of PTEN, which caused cells to proliferate and acquire motile and invasive properties.182,183 The induction of EMT in PDAC cells by TGF-β signaling was also shown to be potentiated by Ras signaling. In elegant biochemical studies, Horiguchi and colleagues revealed that cooperation between the Ras and TGF-β pathways via Smad signaling induces the EMT marker Snail and represses the epithelial marker E-cadherin.184
However, despite the aforementioned studies, the precise role for TGF-β signaling in PDAC is still quite controversial. The frequent loss of TGF-β signaling components in PDAC suggested that this pathway was acting through a tumor suppressor mechanism. This has been confirmed in studies using genetically engineered mouse models, as loss of Smad4 in mice expressing oncogenic Kras led to rapid development of PDAC through PanIN precursors in the majority of mice.185,186 Such results underscore the complexity of the effects mediated the TGF-β pathway, and it is not surprising that many other pathway components have been implicated in the development of pancreatic cancer, including the inhibition of activins,187–190 increased expression of mitogenic BMPs,191 and the induction of the GDF family member macrophage inhibitory cytokine-1.192
E. Retinoic Acid Signaling
Studies of the role of retinoid signaling in PDAC have taken a somewhat circuitous path. Early work suggested that retinoids could have antiproliferative and prodifferentiation effects on PDAC cell lines.193–195 Transgenic mice engineered to overexpress cytoplasmic retinoid binding protein (CRBP) exhibited aggressive, poorly differentiated pancreatic cancer that was associated with inhibition of retinoid signaling.196 These and other studies served as the basis for a number of clinical trials using synthetic retinoids in combination to standard therapy, with results that have not been encouraging.197
Recent work, however, has prompted a reinterpretation of previous studies of the role of retinoids in PDAC. Rasheed and colleagues examined 269 specimens of human PDAC for expression of ALDH, the rate limiting enzyme in endogenous RA synthesis.198 In their analysis, patients with tumors harboring ALDH+ cells had a significantly shorter survival. Furthermore, ALDH+ tumors were found to have a greater capacity for clonogenic growth in vitro, and ALDH+ cells were found to express classical EMTand stem cell markers. Additional analysis showed that ALDH+ cells were enriched after exposure to chemotherapeutic agents and continued to exhibit tumorigenicity,199,200 thus prompting the authors to label these cells as candidate pancreatic cancer stem cells. While more work is needed to substantiate this claim, it is perplexing that the de novo expression of the machinery required for RA synthesis is associated with PDAC progression. Interestingly, recent work from the Leach laboratory identified ALDH1 expression as a marker for centroacinar cells,60 a cell that has been theorized not only to be a putative stem cell within the pancreas, but also the cell of origin for PDAC.61
F. EGF Signaling
EGF ligands (EGF and TNF-α) and receptors (ERBB3 and EGFR) are overexpressed in PDAC cells compared to normal pancreatic epithelium.201–203 One mechanism of enhanced EGF signaling within pancreatic tumors stems from the fact that EGF endocytosed by PDAC cells tends to be recycled and secreted as opposed to degraded.204 The high level of expression of both ligands and receptors in PDAC cells would thus permit autocrine EGF signaling. Since EGF signaling is complex and leads to diverse effects, it is hard to make broad statements regarding the effect of signaling through the EGF pathway in PDAC. Nevertheless, multiple studies have shown that pharmacologic inhibition of EGF signaling inhibits cancer cell growth in vitro and tumorigenesis in implantation models,205,206 in part due to decreased angiogenesis and fibroblast activation.205–207
Interestingly, EGF signaling exhibits significant cross-talk with other pathways that have been implicated in PDAC pathogenesis. For example, transgenic overexpression of the EGF ligand TGF-α can induce acinar-to-ductal metaplasia, resulting in the formation of early PanIN lesions.208 Underlying this phenomenon is the activation of two pathways discussed above: Notch and Kras.59,209 Activation of Erk, Cdk4, and cyclin D also occur with TGF-α stimulation. 210 Based on these data, it would be tempting to predict that pharmacologic inhibition of EGF signaling would have clinical efficacy against PDAC. However, administration of biologics (cetuximab, erlotinib) to antagonize EGF signaling have failed to produce any durable response in clinical trials.211
G. Wnt Signaling
Abnormal Wnt/β-catenin signaling has been documented in PanIN and PDAC lesions in humans.212–214 Indeed, Wnt signaling was one of four signaling pathways in which genetic alterations were found in a large percentage of PDAC specimens by global genomic analysis.152 While numerous papers have been published on the subject, a study from the Hebrok group provides unique insight into how β-catenin is involved in PanIN and PDAC formation.215 As reviewed previously, genetically engineered mice that express oncogenic Kras in the pancreatic epithelium will form PanIN lesions at a slow rate, but few, if any, tumors will form.100 Treating such animals with cerulein induces experimental pancreatitis that leads to rapid PanIN formation and eventual progression to PDAC in a large fraction of mice.216 During regeneration after injury, Wnt signaling, as measured by Axin2 and Lef1 expression, is transiently increased in control mice, and signaling is completely abolished after regeneration. However, in mice with oncogenic Kras, Wnt-β-catenin signaling continues to be active as PanINs form. To test whether β-catenin signaling was required for Kras-induced PanIN formation, compound mutants were bred to express oncogenic Kras and a transgenic allele of β-catenin that was resistant to degradation. When pancreatitis was induced in these mice, PanIN lesions did not form. Thus it was shown that PanIN formation required β-catenin signaling in concert with oncogenic Kras expression.215 Moreover, Morris and colleagues provide a possible molecular explanation for the clinical observation that chronic pancreatitis is associated with a significant increase in the risk for PDAC.111 Wnt/β-catenin signaling has been shown to interact with other developmental pathways during neoplastic progression. TGF-β signaling through Smads has been shown to lead to decreased β-catenin levels.217 Similarly, Notch signaling serves to lower β-catenin protein levels.218 Finally, β-catenin signaling, in itself, is known to activate the Gli transcription factors, the downstream effectors of the Hedgehog pathway.219 Thus, the Wnt/β-catenin pathway acts as a sort of node interconnecting multiple developmental pathways that have reemerged during PanIN and PDAC formation.
VI. Concluding Remarks
In addition to their lethal efficiency, most pancreatic cancers share major molecular pathways that drive their formation. As illustrated here, a handful signaling pathways are reproducibly activated during the initiation of PanIN formation and progression to carcinoma. However, these molecular pathways elicit broad effects that combine to produce the pathology unique to PDAC. Thus, while some of these deregulated pathways are shared by other tumors, the unique developmental history of the pancreas accounts for the distinct behavior and natural history of this malignancy. Some of these signals relate to developmental pathways utilized to maintain the undifferentiated state (e.g., Notch) while others are involved in the growth of the tissue (e.g., EGFs and FGFs). Innovative approaches to therapy will likely stem from targeting of those pathways that have little role in normal adult tissues and which are specifically utilized by cancer cells as a result of their exploitation of developmental signals.
REFERENCES
- 1.SEER N. 2009 http://seercancergov/
- 2.Winter JM, Cameron JL, Campbell KA, Arnold MA, Chang DC, Coleman J, Hodgin MB, Sauter PK, Hruban RH, Riall TS. 1423 pancreaticoduodenectomies for pancreatic cancer: a single-institution experience. J Gastrointest Surg. 2006;10:1199–1210. doi: 10.1016/j.gassur.2006.08.018. discussion 1210–1191. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Algul H, Treiber M, Lesina M, Schmid RM. Mechanisms of disease: chronic inflammation and cancer in the pancreas—a potential role for pancreatic stellate cells? Nat Clin Pract Gastroenterol Hepatol. 2007;4:454–462. doi: 10.1038/ncpgasthep0881. [DOI] [PubMed] [Google Scholar]
- 5.Masamune A, Watanabe T, Kikuta K, Shimosegawa T. Roles of pancreatic stellate cells in pancreatic inflammation and fibrosis. Clin Gastroenterol Hepatol. 2009;7:S48–S54. doi: 10.1016/j.cgh.2009.07.038. [DOI] [PubMed] [Google Scholar]
- 6.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 7.Stafford D, Prince VE. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr Biol. 2002;12:1215–1220. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- 8.Tiso N, Filippi A, Pauls S, Bortolussi M, Argenton F. BMP signalling regulates anteroposterior endoderm patterning in zebrafish. Mech Dev. 2002;118:29–37. doi: 10.1016/s0925-4773(02)00252-6. [DOI] [PubMed] [Google Scholar]
- 9.Pictet RL, Clark WR, Williams RH, Rutter WJ. An ultrastructural analysis of the developing embryonic pancreas. Dev Biol. 1972;29:436–467. doi: 10.1016/0012-1606(72)90083-8. [DOI] [PubMed] [Google Scholar]
- 10.Wessells N, Cohen J. Early pancreas organogenesis: morphogenesis, tissue interactions, and mass effects. Dev Biol. 1967;15:237–270. doi: 10.1016/0012-1606(67)90042-5. [DOI] [PubMed] [Google Scholar]
- 11.Kim S, Hebrok M, Melton D. Notochord to endoderm signaling is required for pancreas development. Development. 1997;124:4243–4252. doi: 10.1242/dev.124.21.4243. [DOI] [PubMed] [Google Scholar]
- 12.Hogan B. Morphogenesis. Cell. 1999;96:225–233. doi: 10.1016/s0092-8674(00)80562-0. [DOI] [PubMed] [Google Scholar]
- 13.Manova K, De Leon V, Angeles M, Kalantry S, Giarre M, Attisano L, Wrana J, Bachvarova RF. mRNAs for activin receptors II and IIB are expressed in mouse oocytes and in the epiblast of pregastrula and gastrula stage mouse embryos. Mech Dev. 1995;49:3–11. doi: 10.1016/0925-4773(94)00295-x. [DOI] [PubMed] [Google Scholar]
- 14.Verschueren K, Dewulf N, Goumans MJ, Lonnoy O, Feijen A, Grimsby S, Vandi Spiegle K, ten Dijke P, Moren A, Vanscheeuwijck P, et al. Expression of type I and type IB receptors for activin in midgestation mouse embryos suggests distinct functions in organogenesis. Mech Dev. 1995;52:109–123. doi: 10.1016/0925-4773(95)00395-h. [DOI] [PubMed] [Google Scholar]
- 15.Hebrok M, Kim S, Melton D. Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes Dev. 1998;12:1705–1713. doi: 10.1101/gad.12.11.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Apelqvist A, Ahlgren U, Edlund H. Sonic hedgehog directs specialised mesoderm differentiation in the intestine and pancreas. Curr Biol. 1997;7:801–804. doi: 10.1016/s0960-9822(06)00340-x. [DOI] [PubMed] [Google Scholar]
- 17.Hebrok M, Kim S, St-Jacques B, McMahon A, Melton D. Regulation of pancreas development by hedgehog signaling. Development. 2000;127:4905–4913. doi: 10.1242/dev.127.22.4905. [DOI] [PubMed] [Google Scholar]
- 18.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 19.Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 20.Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- 21.Hart A, Baeza N, Apelqvist A, Edlund H. Attenuation of FGF signalling in mouse b-cells leads to diabetes. Nature. 2000;408:864–868. doi: 10.1038/35048589. [DOI] [PubMed] [Google Scholar]
- 22.Norgaard GA, Jensen JN, Jensen J. FGF10 signaling maintains the pancreatic progenitor cell state revealing a novel role of Notch in organ development. Dev Biol. 2003;264:323–338. doi: 10.1016/j.ydbio.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 23.Lammert E, Cleaver O, Melton D. Induction of pancreatic differentiation by signals from blood vessels. Science. 2001;294:564–567. doi: 10.1126/science.1064344. [DOI] [PubMed] [Google Scholar]
- 24.Lammert E, Gu G, McLaughlin M, Brown D, Brekken R, Murtaugh LC, Gerber HP, Ferrara N, Melton DA. Role of VEGF-A in vascularization of pancreatic islets. Curr Biol. 2003;13:1070–1074. doi: 10.1016/s0960-9822(03)00378-6. [DOI] [PubMed] [Google Scholar]
- 25.Jacquemin P, Yoshitomi H, Kashima Y, Rousseau GG, Lemaigre FP, Zaret KS. An endothelial-mesenchymal relay pathway regulates early phases of pancreas development. Dev Biol. 2006;290:189–199. doi: 10.1016/j.ydbio.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 26.Krapp A, Knofler M, Frutiger S, Hughes GJ, Hagenbuchle O, Wellauer PK. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. EMBO J. 1996;15:4317–4329. [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshitomi H, Zaret KS. Endothelial cell interactions initiate dorsal pancreas development by selectively inducing the transcription factor Ptf1a. Development. 2004;131:807–817. doi: 10.1242/dev.00960. [DOI] [PubMed] [Google Scholar]
- 28.Kallman F, Grobstein C. Fine structure of differentiating mouse pancreatic exocrine cells in transfilter culture. J Cell Biol. 1964;20:399–413. doi: 10.1083/jcb.20.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munger BL. A light and electron microscopic study of cellular differentiation in the pancreatic islets of the mouse. Am J Anat. 1958;103:275–311. doi: 10.1002/aja.1001030207. [DOI] [PubMed] [Google Scholar]
- 30.Slack J. Developmental biology of the pancreas. Development. 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- 31.Theunissen JW, de Sauvage FJ. Paracrine hedgehog signaling in cancer. Cancer Res. 2009;69:6007–6010. doi: 10.1158/0008-5472.CAN-09-0756. [DOI] [PubMed] [Google Scholar]
- 32.Smith SB, Watada H, Scheel DW, Mrejen C, German MS. Autoregulation and maturity onset diabetes of the young transcription factors control the human PAX4 promoter. J Biol Chem. 2000;275:36910–36919. doi: 10.1074/jbc.M005202200. [DOI] [PubMed] [Google Scholar]
- 33.Ornitz D, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2(3005):3001–3012. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldfarb M. Signaling by fibroblast growth factors: the inside story. Sci STKE. 2001:pe37. doi: 10.1126/stke.2001.106.pe37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manfroid I, Delporte F, Baudhuin A, Motte P, Neumann CJ, Voz ML, Martial JA, Peers B. Reciprocal endoderm–mesoderm interactions mediated by fgf24 and fgf10 govern pancreas development. Development. 2007;134:4011–4021. doi: 10.1242/dev.007823. [DOI] [PubMed] [Google Scholar]
- 36.Miralles F, Lamotte L, Couton D, Joshi RL. Interplay between FGF10 and Notch signalling is required for the self-renewal of pancreatic progenitors. Int J Dev Biol. 2006;50:17–26. doi: 10.1387/ijdb.052080fm. [DOI] [PubMed] [Google Scholar]
- 37.Dichmann DS, Miller CP, Jensen J, Scott Heller R, Serup P. Expression and misexpression of members of the FGF and TGFbeta families of growth factors in the developing mouse pancreas. Dev Dyn. 2003;226:663–674. doi: 10.1002/dvdy.10270. [DOI] [PubMed] [Google Scholar]
- 38.LeBras S, Czernichow P, Scharfmann R. A search for tyrosine kinase receptors expressed in the rat embryonic pancreas. Diabetologia. 1998;41:1474–1481. doi: 10.1007/s001250051094. [DOI] [PubMed] [Google Scholar]
- 39.Pulkkinen MA, Spencer-Dene B, Dickson C, Otonkoski T. The IIIb isoform of fibroblast growth factor receptor 2 is required for proper growth and branching of pancreatic ductal epithelium but not for differentiation of exocrine or endocrine cells. Mech Dev. 2003;120:167–175. doi: 10.1016/s0925-4773(02)00440-9. [DOI] [PubMed] [Google Scholar]
- 40.Deutsch G, Jung J, Zheng M, Lora J, Zaret K. A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development. 2001;128:871–881. doi: 10.1242/dev.128.6.871. [DOI] [PubMed] [Google Scholar]
- 41.Celli G, LaRochelle WJ, Mackem S, Sharp R, Merlino G. Soluble dominant-negative receptor uncovers essential roles for fibroblast growth factors in multi-organ induction and patterning. EMBO J. 1998;17:1642–1655. doi: 10.1093/emboj/17.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elghazi L, Cras-Meneur C, Czernichow P, Scharfmann R. Role for FGFR2IIIb-mediated signals in controlling pancreatic endocrine progenitor cell proliferation. Proc Natl Acad Sci USA. 2002;99:3884–3889. doi: 10.1073/pnas.062321799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye F, Duvillie B, Scharfmann R. Fibroblast growth factors 7 and 10 are expressed in the human embryonic pancreatic mesenchyme and promote the proliferation of embryonic pancreatic epithelial cells. Diabetologia. 2005;48:277–281. doi: 10.1007/s00125-004-1638-6. [DOI] [PubMed] [Google Scholar]
- 44.Bhushan A, Itoh N, Kato S, Thiery J, Czernichow P, Bellusci S, Scharfmann R. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development. 2001;128:5109–5117. doi: 10.1242/dev.128.24.5109. [DOI] [PubMed] [Google Scholar]
- 45.Hart A, Papadopoulou S, Edlund H. Fgf10 maintains notch activation, and blocks differentiation of pancreatic epithelial cells. Dev Dyn. 2003;228:185–193. doi: 10.1002/dvdy.10368. [DOI] [PubMed] [Google Scholar]
- 46.Arnaud-Dabernat S, Kritzik M, Kayali AG, Zhang YQ, Liu G, Ungles C, Sarvetnick N. FGFR3 is a negative regulator of the expansion of pancreatic epithelial cells. Diabetes. 2007;56:96–106. doi: 10.2337/db05-1073. [DOI] [PubMed] [Google Scholar]
- 47.Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 48.Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 49.Hrabe de Angelis M, McIntyre J, 2nd, Gossler A. Maintenance of somite borders in mice requires the Delta homologue DII1. Nature. 1997;386:717–721. doi: 10.1038/386717a0. [DOI] [PubMed] [Google Scholar]
- 50.Beres TM, Masui T, Swift GH, Shi L, Henke RM, MacDonald RJ. PTF1 is an organ-specific and Notch-independent basic helix-loop-helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol Cell Biol. 2006;26:117–130. doi: 10.1128/MCB.26.1.117-130.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Georgia S, Soliz R, Li M, Zhang P, Bhushan A. p57 and Hes1 coordinate cell cycle exit with self-renewal of pancreatic progenitors. Dev Biol. 2006;298:22–31. doi: 10.1016/j.ydbio.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida T, Tokunaga A, Nakao K, Okano H. Distinct expression patterns of splicing isoforms of mNumb in the endocrine lineage of developing pancreas. Differentiation. 2003;71:486–495. doi: 10.1046/j.1432-0436.2003.7108006.x. [DOI] [PubMed] [Google Scholar]
- 53.Esni F, Ghosh B, Biankin AV, Lin JW, Albert MA, Yu X, MacDonald RJ, Civin CI, Real FX, Pack MA, et al. Notch inhibits Ptf1 function and acinar cell differentiation in developing mouse and zebrafish pancreas. Development. 2004;131:4213–4224. doi: 10.1242/dev.01280. [DOI] [PubMed] [Google Scholar]
- 54.Hald J, Hjorth JP, German MS, Madsen OD, Serup P, Jensen J. Activated Notch1 prevents differentiation of pancreatic acinar cells and attenuate endocrine development. Dev Biol. 2003;260:426–437. doi: 10.1016/s0012-1606(03)00326-9. [DOI] [PubMed] [Google Scholar]
- 55.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci USA. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu Y, Wang S, Zhang J, Zhao A, Stanger BZ, Gu G. The fringe molecules induce endocrine differentiation in embryonic endoderm by activating cMyt1/cMyt3. Dev Biol. 2006;297:340–349. doi: 10.1016/j.ydbio.2006.04.456. [DOI] [PubMed] [Google Scholar]
- 57.Johansson KA, Dursun U, Jordan N, Gu G, Beermann F, Gradwohl G, Grapin-Botton A. Temporal control of neurogenin3 activity in pancreas progenitors reveals competence windows for the generation of different endocrine cell types. Dev Cell. 2007;12:457–465. doi: 10.1016/j.devcel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 58.Golson ML, Le Lay J, Gao N, Bramswig N, Loomes KM, Oakey R, May CL, White P, Kaestner KH. Jagged1 is a competitive inhibitor of Notch signaling in the embryonic pancreas. Mech Dev. 2009;126:687–699. doi: 10.1016/j.mod.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, Sriuranpong V, Iso T, Meszoely IM, Wolfe MS, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 60.Rovira M, Scott SG, Liss AS, Jensen J, Thayer SP, Leach SD. Isolation and characterization of centroacinar/terminal ductal progenitor cells in adult mouse pancreas. Proc Natl Acad Sci USA. 2010;107:75–80. doi: 10.1073/pnas.0912589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stanger BZ, Dor Y. Dissecting the cellular origins of pancreatic cancer. Cell Cycle. 2006;5:43–46. doi: 10.4161/cc.5.1.2291. [DOI] [PubMed] [Google Scholar]
- 62.Gittes GK. Developmental biology of the pancreas: a comprehensive review. Dev Biol. 2009;326:4–35. doi: 10.1016/j.ydbio.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 63.Rane SG, Lee JH, Lin HM. Transforming growth factor-beta pathway: role in pancreas development and pancreatic disease. Cytokine Growth Factor Rev. 2006;17:107–119. doi: 10.1016/j.cytogfr.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 64.Crisera CA, Maldonado TS, Kadison AS, Li M, Alkasab SL, Longaker MT, Gittes GK. Transforming growth factor-beta 1 in the developing mouse pancreas: a potential regulator of exocrine differentiation. Differentiation. 2000;65:255–259. doi: 10.1046/j.1432-0436.2000.6550255.x. [DOI] [PubMed] [Google Scholar]
- 65.Thompson NL, Flanders KC, Smith JM, Ellingsworth LR, Roberts AB, Sporn MB. Expression of transforming growth factor-beta 1 in specific cells and tissues of adult and neonatal mice. J Cell Biol. 1989;108:661–669. doi: 10.1083/jcb.108.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crisera CA, Rose MI, Connelly PR, Li M, Colen KL, Longaker MT, Gittes GK. The ontogeny of TGF-beta1, -beta2, -beta3, and TGF-beta receptor-II expression in the pancreas: implications for regulation of growth and differentiation. J Pediatr Surg. 1999;34:689–693. doi: 10.1016/s0022-3468(99)90357-3. discussion 684–693. [DOI] [PubMed] [Google Scholar]
- 67.Bottinger EP, Jakubczak JL, Roberts IS, Mumy M, Hemmati P, Bagnall K, Merlino G, Wakefield LM. Expression of a dominant-negative mutant TGF-beta type II receptor in transgenic mice reveals essential roles for TGF-beta in regulation of growth and differentiation in the exocrine pancreas. EMBO J. 1997;16:2621–2633. doi: 10.1093/emboj/16.10.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Furukawa M, Nobusawa R, Shibata H, Eto Y, Kojima I. Initiation of insulin secretion in glucose-free medium by activin A. Mol Cell Endocrinol. 1995;113:83–87. doi: 10.1016/0303-7207(95)03617-g. [DOI] [PubMed] [Google Scholar]
- 69.Maldonado TS, Kadison AS, Crisera CA, Grau JB, Alkasab SL, Longaker MT, Gittes GK. Ontogeny of activin B and follistatin in developing embryonic mouse pancreas: implications for lineage selection. J Gastrointest Surg. 2000;4:269–275. doi: 10.1016/s1091-255x(00)80075-x. [DOI] [PubMed] [Google Scholar]
- 70.Miralles F, Czernichow P, Scharfmann R. Follistatin regulates the relative proportions of endocrine versus exocrine tissue during pancreatic development. Development. 1998;125:1017–1024. doi: 10.1242/dev.125.6.1017. [DOI] [PubMed] [Google Scholar]
- 71.Ritvos O, Tuuri T, Eramaa M, Sainio K, Hilden K, Saxen L, Gilbert SF. Activin disrupts epithelial branching morphogenesis in developing glandular organs of the mouse. Mech Dev. 1995;50:229–245. doi: 10.1016/0925-4773(94)00342-k. [DOI] [PubMed] [Google Scholar]
- 72.Sulzbacher S, Schroeder IS, Truong TT, Wobus AM. Activin A-induced differentiation of embryonic stem cells into endoderm and pancreatic progenitors-the influence of differentiation factors and culture conditions. Stem Cell Rev. 2009;5:159–173. doi: 10.1007/s12015-009-9061-5. [DOI] [PubMed] [Google Scholar]
- 73.Ahnfelt-Ronne J, Ravassard P, Pardanaud-Galvieux C, Scharfmann R, Serup P. Mesenchymal Bmp signalling is required for normal pancreas development. Diabetes. 2010 doi: 10.2337/db09-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goulley J, Dahl U, Baeza N, Mishina Y, Edlund H. BMP4-BMPR1A signaling in beta cells is required for and augments glucose-stimulated insulin secretion. Cell Metab. 2007;5:207–219. doi: 10.1016/j.cmet.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 75.Harmon EB, Apelqvist AA, Smart NG, Gu X, Osborne DH, Kim SK. GDF11 modulates NGN3+ islet progenitor cell number and promotes beta-cell differentiation in pancreas development. Development. 2004;131:6163–6174. doi: 10.1242/dev.01535. [DOI] [PubMed] [Google Scholar]
- 76.Jacquemin P, Durviaux SM, Jensen J, Godfraind C, Gradwohl G, Guillemot F, Madsen OD, Carmeliet P, Dewerchin M, Collen D, et al. Transcription factor hepatocyte nuclear factor 6 regulates pancreatic endocrine cell differentiation and controls expression of the proendocrine gene ngn3. Mol Cell Biol. 2000;20:4445–4454. doi: 10.1128/mcb.20.12.4445-4454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dichmann DS, Yassin H, Serup P. Analysis of pancreatic endocrine development in GDF11- deficient mice. Dev Dyn. 2006;235:3016–3025. doi: 10.1002/dvdy.20953. [DOI] [PubMed] [Google Scholar]
- 78.McGrane MM. Vitamin A regulation of gene expression: molecular mechanism of a prototype gene. J Nutr Biochem. 2007;18:497–508. doi: 10.1016/j.jnutbio.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 79.Martin M, Gallego-Llamas J, Ribes V, Kedinger M, Niederreither K, Chambon P, Dolle P, Gradwohl G. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev Biol. 2005;284:399–411. doi: 10.1016/j.ydbio.2005.05.035. [DOI] [PubMed] [Google Scholar]
- 80.Stafford D, White RJ, Kinkel MD, Linville A, Schilling TF, Prince VE. Retinoids signal directly to zebrafish endoderm to specify insulin-expressing beta-cells. Development. 2006;133:949–956. doi: 10.1242/dev.02263. [DOI] [PubMed] [Google Scholar]
- 81.Molotkov A, Molotkova N, Duester G. Retinoic acid generated by Raldh2 in mesoderm is required for mouse dorsal endodermal pancreas development. Dev Dyn. 2005;232:950–957. doi: 10.1002/dvdy.20256. [DOI] [PubMed] [Google Scholar]
- 82.Kobayashi H, Spilde TL, Bhatia AM, Buckingham RB, Hembree MJ, Prasadan K, Preuett BL, Imamura M, Gittes GK. Retinoid signaling controls mouse pancreatic exocrine lineage selection through epithelial–mesenchymal interactions. Gastroenterology. 2002;123:1331–1340. doi: 10.1053/gast.2002.35949. [DOI] [PubMed] [Google Scholar]
- 83.Huotari MA, Miettinen PJ, Palgi J, Koivisto T, Ustinov J, Harari D, Yarden Y, Otonkoski T. ErbB signaling regulates lineage determination of developing pancreatic islet cells in embryonic organ culture. Endocrinology. 2002;143:4437–4446. doi: 10.1210/en.2002-220382. [DOI] [PubMed] [Google Scholar]
- 84.Cras-Meneur C, Elghazi L, Czernichow P, Scharfmann R. Epidermal growth factor increases undifferentiated pancreatic embryonic cells in vitro: a balance between proliferation and differentiation. Diabetes. 2001;50:1571–1579. doi: 10.2337/diabetes.50.7.1571. [DOI] [PubMed] [Google Scholar]
- 85.Sanvito F, Herrera PL, Huarte J, Nichols A, Montesano R, Orci L, Vassalli JD. TGF-beta 1 influences the relative development of the exocrine and endocrine pancreas in vitro. Development. 1994;120:3451–3462. doi: 10.1242/dev.120.12.3451. [DOI] [PubMed] [Google Scholar]
- 86.McLin VA, Rankin SA, Zorn AM. Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development. 2007;134:2207–2217. doi: 10.1242/dev.001230. [DOI] [PubMed] [Google Scholar]
- 87.Heller RS, Dichmann DS, Jensen J, Miller C, Wong G, Madsen OD, Serup P. Expression patterns of Wnts, Frizzleds, sFRPs, and misexpression in transgenic mice suggesting a role for Wnts in pancreas and foregut pattern formation. Dev Dyn. 2002;225:260–270. doi: 10.1002/dvdy.10157. [DOI] [PubMed] [Google Scholar]
- 88.Dessimoz J, Bonnard C, Huelsken J, Grapin-Botton A. Pancreas-specific deletion of beta-catenin reveals Wnt-dependent and Wnt-independent functions during development. Curr Biol. 2005;15:1677–1683. doi: 10.1016/j.cub.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 89.Murtaugh LC, Law AC, Dor Y, Melton DA. Beta-catenin is essential for pancreatic acinar but not islet development. Development. 2005;132:4663–4674. doi: 10.1242/dev.02063. [DOI] [PubMed] [Google Scholar]
- 90.Wells JM, Esni F, Boivin GP, Aronow BJ, Stuart W, Combs C, Sklenka A, Leach SD, Lowy AM. Wnt/beta-catenin signaling is required for development of the exocrine pancreas. BMC Dev Biol. 2007;7:4. doi: 10.1186/1471-213X-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heiser PW, Lau J, Taketo MM, Herrera PL, Hebrok M. Stabilization of beta-catenin impacts pancreas growth. Development. 2006;133:2023–2032. doi: 10.1242/dev.02366. [DOI] [PubMed] [Google Scholar]
- 92.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 93.Campbell SL, Khosravi-Far R, Rossman KL, Clark GJ, Der CJ. Increasing complexity of Ras signaling. Oncogene. 1998;17:1395–1413. doi: 10.1038/sj.onc.1202174. [DOI] [PubMed] [Google Scholar]
- 94.Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 95.Sclabas GM, Fujioka S, Schmidt C, Evans DB, Chiao PJ. NF-kappaB in pancreatic cancer. Int J Gastrointest Cancer. 2003;33:15–26. doi: 10.1385/IJGC:33:1:15. [DOI] [PubMed] [Google Scholar]
- 96.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 97.Klimstra DS, Longnecker DS. K-ras mutations in pancreatic ductal proliferative lesions. Am J Pathol. 1994;145:1547–1550. [PMC free article] [PubMed] [Google Scholar]
- 98.Rozenblum E, Schutte M, Goggins M, Hahn SA, Panzer S, Zahurak M, Goodman SN, Sohn TA, Hruban RH, Yeo CJ, et al. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res. 1997;57:1731–1734. [PubMed] [Google Scholar]
- 99.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 101.Moskaluk CA, Hruban RH, Kern SE. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 1997;57:2140–2143. [PubMed] [Google Scholar]
- 102.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. discussion 3080. [DOI] [PubMed] [Google Scholar]
- 103.Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 104.Fleming JB, Shen GL, Holloway SE, Davis M, Brekken RA. Molecular consequences of silencing mutant K-ras in pancreatic cancer cells: justification for K-ras-directed therapy. Mol Cancer Res. 2005;3:413–423. doi: 10.1158/1541-7786.MCR-04-0206. [DOI] [PubMed] [Google Scholar]
- 105.Hirano T, Shino Y, Saito T, Komoda F, Okutomi Y, Takeda A, Ishihara T, Yamaguchi T, Saisho H, Shirasawa H. Dominant negative MEKK1 inhibits survival of pancreatic cancer cells. Oncogene. 2002;21:5923–5928. doi: 10.1038/sj.onc.1205643. [DOI] [PubMed] [Google Scholar]
- 106.Omer CA, Kohl NE. CA1A2X-competitive inhibitors of farnesyltransferase as anti-cancer agents. Trends Pharmacol Sci. 1997;18:437–444. doi: 10.1016/s0165-6147(97)01129-2. [DOI] [PubMed] [Google Scholar]
- 107.Van Cutsem E, van de Velde H, Karasek P, Oettle H, Vervenne WL, Szawlowski A, Schoffski P, Post S, Verslype C, Neumann H, et al. Phase III trial of gemcitabine plus tipifarnib compared with gemcitabine plus placebo in advanced pancreatic cancer. J Clin Oncol. 2004;22:1430–1438. doi: 10.1200/JCO.2004.10.112. [DOI] [PubMed] [Google Scholar]
- 108.Luttges J, Schlehe B, Menke MA, Vogel I, Henne-Bruns D, Kloppel G. The K-ras mutation pattern in pancreatic ductal adenocarcinoma usually is identical to that in associated normal, hyperplastic, and metaplastic ductal epithelium. Cancer. 1999;85:1703–1710. [PubMed] [Google Scholar]
- 109.Carriere C, Young AL, Gunn JR, Longnecker DS, Korc M. Acute pancreatitis markedly accelerates pancreatic cancer progression in mice expressing oncogenic Kras. Biochem Biophys Res Commun. 2009;382:561–565. doi: 10.1016/j.bbrc.2009.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gidekel Friedlander SY, Chu GC, Snyder EL, Girnius N, Dibelius G, Crowley D, Vasile E, DePinho RA, Jacks T. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell. 2009;16:379–389. doi: 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lowenfels AB, Maisonneuve P, Cavallini G, Ammann RW, Lankisch PG, Andersen JR, Dimagno EP, Andren-Sandberg A, Domellof L. Pancreatitis and the risk of pancreatic cancer, International pancreatitis study group. N Engl J Med. 1993;328:1433–1437. doi: 10.1056/NEJM199305203282001. [DOI] [PubMed] [Google Scholar]
- 112.Duan RD, Zheng CF, Guan KL, Williams JA. Activation of MAP kinase kinase (MEK) and Ras by cholecystokinin in rat pancreatic acini. Am J Physiol. 1995;268:G1060–G1065. doi: 10.1152/ajpgi.1995.268.6.G1060. [DOI] [PubMed] [Google Scholar]
- 113.Ji B, Tsou L, Wang H, Gaiser S, Chang DZ, Daniluk J, Bi Y, Grote T, Longnecker DS, Logsdon CD. Ras activity levels control the development of pancreatic diseases. Gastroenterology. 2009;137:1072–1082. e1071–e1076. doi: 10.1053/j.gastro.2009.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hustinx SR, Leoni LM, Yeo CJ, Brown PN, Goggins M, Kern SE, Hruban RH, Maitra A. Concordant loss of MTAP and p16/CDKN2A expression in pancreatic intraepithelial neoplasia: evidence of homozygous deletion in a noninvasive precursor lesion. Mod Pathol. 2005;18:959–963. doi: 10.1038/modpathol.3800377. [DOI] [PubMed] [Google Scholar]
- 115.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 116.Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van Heek NT, Meeker AK, Kern SE, Yeo CJ, Lillemoe KD, Cameron JL, Offerhaus GJ, Hicks JL, Wilentz RE, Goggins MG, et al. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am J Pathol. 2002;161:1541–1547. doi: 10.1016/S0002-9440(10)64432-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Goldstein AM, Fraser MC, Struewing JP, Hussussian CJ, Ranade K, Zametkin DP, Fontaine LS, Organic SM, Dracopoli NC, Clark WH, Jr, et al. Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4 mutations. N Engl J Med. 1995;333:970–974. doi: 10.1056/NEJM199510123331504. [DOI] [PubMed] [Google Scholar]
- 119.Whelan AJ, Bartsch D, Goodfellow PJ. Brief report: a familial syndrome of pancreatic cancer and melanoma with a mutation in the CDKN2 tumor-suppressor gene. N Engl J Med. 1995;333:975–977. doi: 10.1056/NEJM199510123331505. [DOI] [PubMed] [Google Scholar]
- 120.Sharpless NE, DePinho RA. Cancer: crime and punishment. Nature. 2005;436:636–637. doi: 10.1038/436636a. [DOI] [PubMed] [Google Scholar]
- 121.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 123.Maitra A, Adsay NV, Argani P, Iacobuzio-Donahue C, De Marzo A, Cameron JL, Yeo CJ, Hruban RH. Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod Pathol. 2003;16:902–912. doi: 10.1097/01.MP.0000086072.56290.FB. [DOI] [PubMed] [Google Scholar]
- 124.Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci USA. 2006;103:5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Boschman CR, Stryker S, Reddy JK, Rao MS. Expression of p53 protein in precursor lesions and adenocarcinoma of human pancreas. Am J Pathol. 1994;145:1291–1295. [PMC free article] [PubMed] [Google Scholar]
- 126.Harada T, Okita K, Shiraishi K, Kusano N, Furuya T, Oga A, Kawauchi S, Kondoh S, Sasaki K. Detection of genetic alterations in pancreatic cancers by comparative genomic hybridization coupled with tissue microdissection and degenerate oligonucleotide primed polymerase chain reaction. Oncology. 2002;62:251–258. doi: 10.1159/000059573. [DOI] [PubMed] [Google Scholar]
- 127.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 128.Wilentz RE, Iacobuzio-Donahue CA, Argani P, McCarthy DM, Parsons JL, Yeo CJ, Kern SE, Hruban RH. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60:2002–2006. [PubMed] [Google Scholar]
- 129.Baas AF, Smit L, Clevers H. LKB1 tumor suppressor protein: PARtaker in cell polarity. Trends Cell Biol. 2004;14:312–319. doi: 10.1016/j.tcb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 130.Bardeesy N, Sinha M, Hezel AF, Signoretti S, Hathaway NA, Sharpless NE, Loda M, Carrasco DR, DePinho RA. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature. 2002;419:162–167. doi: 10.1038/nature01045. [DOI] [PubMed] [Google Scholar]
- 131.Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DG, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci USA. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Giardiello FM, Brensinger JD, Tersmette AC, Goodman SN, Petersen GM, Booker SV, Cruz-Correa M, Offerhaus JA. Very high risk of cancer in familial Peutz-Jeghers syndrome. Gastroenterology. 2000;119:1447–1453. doi: 10.1053/gast.2000.20228. [DOI] [PubMed] [Google Scholar]
- 134.Sahin F, Maitra A, Argani P, Sato N, Maehara N, Montgomery E, Goggins M, Hruban RH, Su GH. Loss of Stk11/Lkb1 expression in pancreatic and biliary neoplasms. Mod Pathol. 2003;16:686–691. doi: 10.1097/01.MP.0000075645.97329.86. [DOI] [PubMed] [Google Scholar]
- 135.Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, Lin JC, Palmisano E, Brune K, Jaffee EM, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Slater EP, Langer P, Niemczyk E, Strauch K, Butler J, Habbe N, Neoptolemos J, Greenhalf W, Bartsch DK. PALB2 mutations in European familial pancreatic cancer families. Clin Genet. 2010 doi: 10.1111/j.1399-0004.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- 137.Tischkowitz MD, Sabbaghian N, Hamel N, Borgida A, Rosner C, Taherian N, Srivastava A, Holter S, Rothenmund H, Ghadirian P, et al. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology. 2009;137:1183–1186. doi: 10.1053/j.gastro.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.McWilliams RR, Bamlet WR, de Andrade M, Rider DN, Couch FJ, Cunningham JM, Matsumoto ME, Rabe KG, Hammer TJ, Petersen GM. Polymorphic variants in hereditary pancreatic cancer genes are not associated with pancreatic cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18:2549–2552. doi: 10.1158/1055-9965.EPI-09-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pogue-Geile KL, Chen R, Bronner MP, Crnogorac-Jurcevic T, Moyes KW, Dowen S, Otey CA, Crispin DA, George RD, Whitcomb DC, et al. Palladin mutation causes familial pancreatic cancer and suggests a new cancer mechanism. PLoS Med. 2006;3:e516. doi: 10.1371/journal.pmed.0030516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Parast MM, Otey CA. Characterization of palladin, a novel protein localized to stress fibers and cell adhesions. J Cell Biol. 2000;150:643–656. doi: 10.1083/jcb.150.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Otey CA, Rachlin A, Moza M, Arneman D, Carpen O. The palladin/myotilin/myopalladin family of actin-associated scaffolds. Int Rev Cytol. 2005;246:31–58. doi: 10.1016/S0074-7696(05)46002-7. [DOI] [PubMed] [Google Scholar]
- 143.Goicoechea SM, Bednarski B, Stack C, Cowan DW, Volmar K, Thorne L, Cukierman E, Rustgi AK, Brentnall T, Hwang RF, et al. Isoform-specific upregulation of palladin in human and murine pancreas tumors. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0010347. e10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Andea A, Sarkar F, Adsay VN. Clinicopathological correlates of pancreatic intraepithelial neoplasia: a comparative analysis of 82 cases with and 152 cases without pancreatic ductal adenocarcinoma. Mod Pathol. 2003;16:996–1006. doi: 10.1097/01.MP.0000087422.24733.62. [DOI] [PubMed] [Google Scholar]
- 145.Cubilla AL, Fitzgerald PJ. Morphological lesions associated with human primary invasive nonendocrine pancreas cancer. Cancer Res. 1976;36:2690–2698. [PubMed] [Google Scholar]
- 146.Hruban RH, Adsay NV, Albores-Saavedra J, Anver MR, Biankin AV, Boivin GP, Furth EE, Furukawa T, Klein A, Klimstra DS, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res. 2006;66:95–106. doi: 10.1158/0008-5472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- 147.Hruban RH, Adsay NV, Albores-Saavedra J, Compton C, Garrett ES, Goodman SN, Kern SE, Klimstra DS, Kloppel G, Longnecker DS, et al. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol. 2001;25:579–586. doi: 10.1097/00000478-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 148.Prasad NB, Biankin AV, Fukushima N, Maitra A, Dhara S, Elkahloun AG, Hruban RH, Goggins M, Leach SD. Gene expression profiles in pancreatic intraepithelial neoplasia reflect the effects of Hedgehog signaling on pancreatic ductal epithelial cells. Cancer Res. 2005;65:1619–1626. doi: 10.1158/0008-5472.CAN-04-1413. [DOI] [PubMed] [Google Scholar]
- 149.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 150.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernandez-del Castillo C, Yajnik V, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Morton JP, Mongeau ME, Klimstra DS, Morris JP, Lee YC, Kawaguchi Y, Wright CV, Hebrok M, Lewis BC. Sonic hedgehog acts at multiple stages during pancreatic tumorigenesis. Proc Natl Acad Sci USA. 2007;104:5103–5108. doi: 10.1073/pnas.0701158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pasca di Magliano M, Sekine S, Ermilov A, Ferris J, Dlugosz AA, Hebrok M. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes Dev. 2006;20:3161–3173. doi: 10.1101/gad.1470806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bailey JM, Mohr AM, Hollingsworth MA. Sonic hedgehog paracrine signaling regulates metastasis and lymphangiogenesis in pancreatic cancer. Oncogene. 2009;28:3513–3525. doi: 10.1038/onc.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, Mullendore M, Karikari C, Alvarez H, Iacobuzio-Donahue C, Jimeno A, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–2196. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Feldmann G, Habbe N, Dhara S, Bisht S, Alvarez H, Fendrich V, Beaty R, Mullendore M, Karikari C, Bardeesy N, et al. Hedgehog inhibition prolongs survival in a genetically engineered mouse model of pancreatic cancer. Gut. 2008;57:1420–1430. doi: 10.1136/gut.2007.148189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tian H, Callahan CA, DuPree KJ, Darbonne WC, Ahn CP, Scales SJ, de Sauvage FJ. Hedgehog signaling is restricted to the stromal compartment during pancreatic carcinogenesis. Proc Natl Acad Sci USA. 2009;106:4254–4259. doi: 10.1073/pnas.0813203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Strobel O, Rosow DE, Rakhlin EY, Lauwers GY, Trainor AG, Alsina J, Fernandez-Del Castillo C, Warshaw AL, Thayer SP. Pancreatic duct glands are distinct ductal compartments that react to chronic injury and mediate Shh-induced metaplasia. Gastroenterology. 2010;138:1166–1177. doi: 10.1053/j.gastro.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nolan-Stevaux O, Lau J, Truitt ML, Chu GC, Hebrok M, Fernandez-Zapico ME, Hanahan D. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 2009;23:24–36. doi: 10.1101/gad.1753809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kuwahara K, Sasaki T, Kuwada Y, Murakami M, Yamasaki S, Chayama K. Expressions of angiogenic factors in pancreatic ductal carcinoma: a correlative study with clinicopathologic parameters and patient survival. Pancreas. 2003;26:344–349. doi: 10.1097/00006676-200305000-00006. [DOI] [PubMed] [Google Scholar]
- 162.Yamanaka Y, Friess H, Buchler M, Beger HG, Uchida E, Onda M, Kobrin MS, Korc M. Overexpression of acidic and basic fibroblast growth factors in human pancreatic cancer correlates with advanced tumor stage. Cancer Res. 1993;53:5289–5296. [PubMed] [Google Scholar]
- 163.Kornmann M, Ishiwata T, Matsuda K, Lopez ME, Fukahi K, Asano G, Beger HG, Korc M. IIIc isoform of fibroblast growth factor receptor 1 is overexpressed in human pancreatic cancer and enhances tumorigenicity of hamster ductal cells. Gastroenterology. 2002;123:301–313. doi: 10.1053/gast.2002.34174. [DOI] [PubMed] [Google Scholar]
- 164.Liu Z, Neiss N, Zhou S, Henne-Bruns D, Korc M, Bachem M, Kornmann M. Identification of a fibroblast growth factor receptor 1 splice variant that inhibits pancreatic cancer cell growth. Cancer Res. 2007;67:2712–2719. doi: 10.1158/0008-5472.CAN-06-3843. [DOI] [PubMed] [Google Scholar]
- 165.Wagner M, Lopez ME, Cahn M, Korc M. Suppression of fibroblast growth factor receptor signaling inhibits pancreatic cancer growth in vitro and in vivo. Gastroenterology. 1998;114:798–807. doi: 10.1016/s0016-5085(98)70594-3. [DOI] [PubMed] [Google Scholar]
- 166.Chen G, Tian X, Liu Z, Zhou S, Schmidt B, Henne-Bruns D, Bachem M, Kornmann M. Inhibition of endogenous SPARC enhances pancreatic cancer cell growth: modulation by FGFR1-III isoform expression. Br J Cancer. 2010;102:188–195. doi: 10.1038/sj.bjc.6605440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nomura S, Yoshitomi H, Takano S, Shida T, Kobayashi S, Ohtsuka M, Kimura F, Shimizu H, Yoshidome H, Kato A, et al. FGF10/FGFR2 signal induces cell migration and invasion in pancreatic cancer. Br J Cancer. 2008;99:305–313. doi: 10.1038/sj.bjc.6604473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.El-Hariry I, Pignatelli M, Lemoine NR. FGF-1 and FGF-2 regulate the expression of E-cadherin and catenins in pancreatic adenocarcinoma. Int J Cancer. 2001;94:652–661. doi: 10.1002/ijc.1515. [DOI] [PubMed] [Google Scholar]
- 169.Pistol-Tanase C, Raducan E, Dima SO, Albulescu L, Alina I, Marius P, Cruceru LM, Codorean E, Neagu TM, Popescu I. Assessment of soluble angiogenic markers in pancreatic cancer. Biomark Med. 2008;2:447–455. doi: 10.2217/17520363.2.5.447. [DOI] [PubMed] [Google Scholar]
- 170.Habbe N, Shi G, Meguid RA, Fendrich V, Esni F, Chen H, Feldmann G, Stoffers DA, Konieczny SF, Leach SD, et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci USA. 2008;105:18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mullendore ME, Koorstra JB, Li YM, Offerhaus GJ, Fan X, Henderson CM, Matsui W, Eberhart CG, Maitra A, Feldmann G. Ligand-dependent Notch signaling is involved in tumor initiation and tumor maintenance in pancreatic cancer. Clin Cancer Res. 2009;15:2291–2301. doi: 10.1158/1078-0432.CCR-08-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Stanger BZ, Stiles B, Lauwers GY, Bardeesy N, Mendoza M, Wang Y, Greenwood A, Cheng KH, McLaughlin M, Brown D, et al. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell. 2005;8:185–195. doi: 10.1016/j.ccr.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 173.Zhu L, Shi G, Schmidt CM, Hruban RH, Konieczny SF. Acinar cells contribute to the molecular heterogeneity of pancreatic intraepithelial neoplasia. Am J Pathol. 2007;171:263–273. doi: 10.2353/ajpath.2007.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Plentz R, Park JS, Rhim AD, Abravanel D, Hezel AF, Sharma SV, Gurumurthy S, Deshpande V, Kenific C, Settleman J, et al. Inhibition of gamma-secretase activity inhibits tumor progression in a mouse model of pancreatic ductal adenocarcinoma. Gastroenterology. 2009;136(1741–1749):e1746. doi: 10.1053/j.gastro.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shi G, Zhu L, Sun Y, Bettencourt R, Damsz B, Hruban RH, Konieczny SF. Loss of the acinar-restricted transcription factor Mist1 accelerates Kras-induced pancreatic intraepithelial neoplasia. Gastroenterology. 2009;136:1368–1378. doi: 10.1053/j.gastro.2008.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang Z, Li Y, Kong D, Banerjee S, Ahmad A, Azmi AS, Ali S, Abbruzzese JL, Gallick GE, Sarkar FH. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 2009;69:2400–2407. doi: 10.1158/0008-5472.CAN-08-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.De La OJ, Emerson LL, Goodman JL, Froebe SC, Illum BE, Curtis AB, Murtaugh LC. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci USA. 2008;105:18907–18912. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ristorcelli E, Lombardo D. Targeting Notch signaling in pancreatic cancer. Expert Opin Ther Targets. 2010;14:541–552. doi: 10.1517/14728221003769895. [DOI] [PubMed] [Google Scholar]
- 179.Friess H, Yamanaka Y, Buchler M, Ebert M, Beger HG, Gold LI, Korc M. Enhanced expression of transforming growth factor beta isoforms in pancreatic cancer correlates with decreased survival. Gastroenterology. 1993;105:1846–1856. doi: 10.1016/0016-5085(93)91084-u. [DOI] [PubMed] [Google Scholar]
- 180.Friess H, Yamanaka Y, Buchler M, Berger HG, Kobrin MS, Baldwin RL, Korc M. Enhanced expression of the type II transforming growth factor beta receptor in human pancreatic cancer cells without alteration of type III receptor expression. Cancer Res. 1993;53:2704–2707. [PubMed] [Google Scholar]
- 181.Lohr M, Schmidt C, Ringel J, Kluth M, Muller P, Nizze H, Jesnowski R. Transforming growth factor-beta1 induces desmoplasia in an experimental model of human pancreatic carcinoma. Cancer Res. 2001;61:550–555. [PubMed] [Google Scholar]
- 182.Chow JY, Ban M, Wu HL, Nguyen F, Huang M, Chung H, Dong H, Carethers JM. TGF-beta downregulates PTEN via activation of NF-kappaB in pancreatic cancer cells. Am J Physiol Gastrointest Liver Physiol. 2010;298:G275–G282. doi: 10.1152/ajpgi.00344.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Maier HJ, Schmidt-Strassburger U, Huber MA, Wiedemann EM, Beug H, Wirth T. NF-kappaB promotes epithelial–mesenchymal transition, migration and invasion of pancreatic carcinoma cells. Cancer Lett. 2010 doi: 10.1016/j.canlet.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 184.Horiguchi K, Shirakihara T, Nakano A, Imamura T, Miyazono K, Saitoh M. Role of Ras signaling in the induction of snail by transforming growth factor-beta. J Biol Chem. 2009;284:245–253. doi: 10.1074/jbc.M804777200. [DOI] [PubMed] [Google Scholar]
- 185.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, Hezel AF, Horner J, Lauwers GY, Hanahan D, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ijichi H, Chytil A, Gorska AE, Aakre ME, Fujitani Y, Fujitani S, Wright CV, Moses HL. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev. 2006;20:3147–3160. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hempen PM, Zhang L, Bansal RK, Iacobuzio-Donahue CA, Murphy KM, Maitra A, Vogelstein B, Whitehead RH, Markowitz SD, Willson JK, et al. Evidence of selection for clones having genetic inactivation of the activin A type II receptor (ACVR2) gene in gastrointestinal cancers. Cancer Res. 2003;63:994–999. [PubMed] [Google Scholar]
- 188.Ryu B, Kern SE. The essential similarity of TGFbeta and activin receptor transcriptional responses in cancer cells. Cancer Biol Ther. 2003;2:10164–10170. doi: 10.4161/cbt.2.2.276. [DOI] [PubMed] [Google Scholar]
- 189.Su GH, Bansal R, Murphy KM, Montgomery E, Yeo CJ, Hruban RH, Kern SE. ACVR1B (ALK4, activin receptor type 1B) gene mutations in pancreatic carcinoma. Proc Natl Acad Sci USA. 2001;98:3254–3527. doi: 10.1073/pnas.051484398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ungefroren H, Schniewind B, Groth S, Chen WB, Muerkoster SS, Kalthoff H, Fandrich F. Antitumor activity of ALK1 in pancreatic carcinoma cells. Int J Cancer. 2007;120:1641–1651. doi: 10.1002/ijc.22393. [DOI] [PubMed] [Google Scholar]
- 191.Kayed H, Kleeff J, Keleg S, Jiang X, Penzel R, Giese T, Zentgraf H, Buchler MW, Korc M, Friess H. Correlation of glypican-1 expression with TGF-beta, BMP, and activin receptors in pancreatic ductal adenocarcinoma. Int J Oncol. 2006;29:1139–1148. [PubMed] [Google Scholar]
- 192.Golkar L, Ding XZ, Ujiki MB, Salabat MR, Kelly DL, Scholtens D, Fought AJ, Bentrem DJ, Talamonti MS, Bell RH, et al. Resveratrol inhibits pancreatic cancer cell proliferation through transcriptional induction of macrophage inhibitory cytokine-1. J Surg Res. 2007;138:163–169. doi: 10.1016/j.jss.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 193.Brembeck FH, Kaiser A, Detjen K, Hotz H, Foitzik T, Buhr HJ, Riecken EO, Rosewicz S. Retinoic acid receptor alpha mediates growth inhibition by retinoids in rat pancreatic carcinoma DSL-6A/C1 cells. Br J Cancer. 1998;78:1288–1295. doi: 10.1038/bjc.1998.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Riecken EO, Rosewicz S. Retinoids in pancreatic cancer. Ann Oncol. 1999;10(Suppl 4):197–200. [PubMed] [Google Scholar]
- 195.Rosewicz S, Stier U, Brembeck F, Kaiser A, Papadimitriou CA, Berdel WE, Wiedenmann B, Riecken EO. Retinoids: effects on growth, differentiation, and nuclear receptor expression in human pancreatic carcinoma cell lines. Gastroenterology. 1995;109:1646–1660. doi: 10.1016/0016-5085(95)90655-x. [DOI] [PubMed] [Google Scholar]
- 196.Perez-Castro AV, Tran VT, Nguyen-Huu MC. Defective lens fiber differentiation and pancreatic tumorigenesis caused by ectopic expression of the cellular retinoic acid-binding protein I. Development. 1993;119:363–375. doi: 10.1242/dev.119.2.363. [DOI] [PubMed] [Google Scholar]
- 197.Michael A, Hill M, Maraveyas A, Dalgleish A, Lofts F. 13-cis-Retinoic acid in combination with gemcitabine in the treatment of locally advanced and metastatic pancreatic cancer–report of a pilot phase II study. Clin Oncol (R Coll Radiol) 2007;19:150–153. doi: 10.1016/j.clon.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 198.Rasheed ZA, Yang J, Wang Q, Kowalski J, Freed I, Murter C, Hong SM, Koorstra JB, Rajeshkumar NV, He X, et al. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J Natl Cancer Inst. 2010;102:340–351. doi: 10.1093/jnci/djp535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Dembinski JL, Krauss S. Characterization and functional analysis of a slow cycling stem cell-like subpopulation in pancreas adenocarcinoma. Clin Exp Metastasis. 2009;26:611–623. doi: 10.1007/s10585-009-9260-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jimeno A, Feldmann G, Suarez-Gauthier A, Rasheed Z, Solomon A, Zou GM, Rubio-Viqueira B, Garcia-Garcia E, Lopez-Rios F, Matsui W, et al. A direct pancreatic cancer xenograft model as a platform for cancer stem cell therapeutic development. Mol Cancer Ther. 2009;8:310–314. doi: 10.1158/1535-7163.MCT-08-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Barton CM, Hall PA, Hughes CM, Gullick WJ, Lemoine NR. Transforming growth factor alpha and epidermal growth factor in human pancreatic cancer. J Pathol. 1991;163:111–116. doi: 10.1002/path.1711630206. [DOI] [PubMed] [Google Scholar]
- 202.Friess H, Kleeff J, Korc M, Buchler MW. Molecular aspects of pancreatic cancer and future perspectives. Dig Surg. 1999;16:281–290. doi: 10.1159/000018737. [DOI] [PubMed] [Google Scholar]
- 203.Korc M, Chandrasekar B, Yamanaka Y, Friess H, Buchier M, Beger HG. Overexpression of the epidermal growth factor receptor in human pancreatic cancer is associated with concomitant increases in the levels of epidermal growth factor and transforming growth factor alpha. J Clin Invest. 1992;90:1352–1360. doi: 10.1172/JCI116001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Korc M. Growth factors and pancreatic cancer. Int J Pancreatol. 1991;9:87–91. doi: 10.1007/BF02925583. [DOI] [PubMed] [Google Scholar]
- 205.Bruns CJ, Harbison MT, Davis DW, Portera CA, Tsan R, McConkey DJ, Evans DB, Abbruzzese JL, Hicklin DJ, Radinsky R. Epidermal growth factor receptor blockade with C225 plus gemcitabine results in regression of human pancreatic carcinoma growing orthotopically in nude mice by antiangiogenic mechanisms. Clin Cancer Res. 2000;6:1936–1948. [PubMed] [Google Scholar]
- 206.Li J, Kleeff J, Giese N, Buchler MW, Korc M, Friess H. Gefitinib (“Iressa,” ZD1839), a selective epidermal growth factor receptor tyrosine kinase inhibitor, inhibits pancreatic cancer cell growth, invasion, and colony formation. Int J Oncol. 2004;25:203–210. [PubMed] [Google Scholar]
- 207.Blaine SA, Ray KC, Branch KM, Robinson PS, Whitehead RH, Means AL. Epidermal growth factor receptor regulates pancreatic fibrosis. Am J Physiol Gastrointest Liver Physiol. 2009;297:G434–G441. doi: 10.1152/ajpgi.00152.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Song SY, Gannon M, Washington MK, Scoggins CR, Meszoely IM, Goldenring JR, Marino CR, Sandgren EP, Coffey RJ, Jr, Wright CV, et al. Expansion of Pdx1-expressing pancreatic epithelium and islet neogenesis in transgenic mice overexpressing transforming growth factor alpha. Gastroenterology. 1999;117:1416–1426. doi: 10.1016/s0016-5085(99)70292-1. [DOI] [PubMed] [Google Scholar]
- 209.Wagner M, Weber CK, Bressau F, Greten FR, Stagge V, Ebert M, Leach SD, Adler G, Schmid RM. Transgenic overexpression of amphiregulin induces a mitogenic response selectively in pancreatic duct cells. Gastroenterology. 2002;122:1898–1912. doi: 10.1053/gast.2002.33594. [DOI] [PubMed] [Google Scholar]
- 210.Wagner M, Greten FR, Weber CK, Koschnick S, Mattfeldt T, Deppert W, Kern H, Adler G, Schmid RM. A murine tumor progression model for pancreatic cancer recapitulating the genetic alterations of the human disease. Genes Dev. 2001;15:286–293. doi: 10.1101/gad.184701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Duffy A, Kortmansky J, Schwartz GK, Capanu M, Puleio S, Minsky B, Saltz L, Kelsen DP, O’Reilly EM. A phase I study of erlotinib in combination with gemcitabine and radiation in locally advanced, non-operable pancreatic adenocarcinoma. Ann Oncol. 2008;19:86–91. doi: 10.1093/annonc/mdm441. [DOI] [PubMed] [Google Scholar]
- 212.Al-Aynati MM, Radulovich N, Riddell RH, Tsao MS. Epithelial-cadherin and beta-catenin expression changes in pancreatic intraepithelial neoplasia. Clin Cancer Res. 2004;10:1235–1240. doi: 10.1158/1078-0432.ccr-03-0087. [DOI] [PubMed] [Google Scholar]
- 213.Lowy AM, Fenoglio-Preiser C, Kim OJ, Kordich J, Gomez A, Knight J, James L, Groden J. Dysregulation of beta-catenin expression correlates with tumor differentiation in pancreatic duct adenocarcinoma. Ann Surg Oncol. 2003;10:284–290. doi: 10.1245/aso.2003.05.003. [DOI] [PubMed] [Google Scholar]
- 214.Zeng G, Germinaro M, Micsenyi A, Monga NK, Bell A, Sood A, Malhotra V, Sood N, Midda V, Monga DK, et al. Aberrant Wnt/beta-catenin signaling in pancreatic adenocarcinoma. Neoplasia. 2006;8:279–289. doi: 10.1593/neo.05607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Morris JPT, Cano DA, Sekine S, Wang SC, Hebrok M. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest. 2010;120:508–520. doi: 10.1172/JCI40045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Guerra C, Schuhmacher AJ, Canamero M, Grippo PJ, Verdaguer L, Perez-Gallego L, Dubus P, Sandgren EP, Barbacid M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 217.Zhao S, Ammanamanchi S, Brattain M, Cao L, Thangasamy A, Wang J, Freeman JW. Smad4-dependent TGF-beta signaling suppresses RON receptor tyrosine kinase-dependent motility and invasion of pancreatic cancer cells. J Biol Chem. 2008;283:11293–11301. doi: 10.1074/jbc.M800154200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Siveke JT, Lubeseder-Martellato C, Lee M, Mazur PK, Nakhai H, Radtke F, Schmid RM. Notch signaling is required for exocrine regeneration after acute pancreatitis. Gastroenterology. 2008;134:544–555. doi: 10.1053/j.gastro.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 219.Dennler S, Andre J, Verrecchia F, Mauviel A. Cloning of the human GLI2 Promoter: transcriptional activation by transforming growth factor-beta via SMAD3/beta-catenin cooperation. J Biol Chem. 2009;284:31523–31531. doi: 10.1074/jbc.M109.059964. [DOI] [PMC free article] [PubMed] [Google Scholar]