Abstract
Despite twin studies showing that 50–70% of variation in DSM-IV cannabis dependence is attributable to heritable influences, little is known of specific genotypes that influence vulnerability to cannabis dependence. We conducted a genomewide association study of DSM-IV cannabis dependence. Association analyses of 708 DSM-IV cannabis dependent cases with 2,346 cannabis exposed nondependent controls was conducted using logistic regression in PLINK. None of the 948,142 SNPs met genomewide significance (p < E−8). The lowest p-values were obtained for polymorphisms on chromosome 17 (rs1019238 and rs1431318, p-values at E−7) in the ANKFN1 gene. While replication is required, this study represents an important first step towards clarifying the biological underpinnings of cannabis dependence.
Cannabis dependence is the third leading contributor to admissions to chemical dependency treatment settings (Treatment Episode Data Set, 2003) and rates of past year cannabis dependence in the U.S. population have increased by 18% since 1991–1992 (Compton et al., 2004). Several studies report strong heritable influences on cannabis dependence [h2 of 50–70%] (Agrawal and Lynskey, 2006). In an effort to identify contributors to this composite heritability, multiple linkage and association studies have been conducted. Several promising linkage regions (e.g. chromosome 3) exist. Candidate gene studies have largely focused on variants in the gene encoding the cannabinoid receptor CNR1 – results remain equivocal (Agrawal and Lynskey, 2009). To our knowledge, there are currently no published genomewide association studies (GWAS) of cannabis dependence. The goal of this investigation is to conduct association analysis between 948,142 single nucleotide polymorphisms (SNPs) and lifetime DSM-IV cannabis dependence using data on 708 cases with DSM-IV cannabis dependence and 2346 controls who report lifetime cannabis use but do not meet criteria for DSM-IV dependence.
Data for this study come from the Study of Addiction: Genes and Environment (SAGE) (Bierut et al., 2010), which was one of the 8 Phase 1 studies of the Gene Environment Association (GENEVA) consortium (Cornelis et al., 2010). The study was designed primarily to study DSM-IV alcohol dependence as well as other correlated addiction phenotypes. All phenotypic data were collected using the Semi-Structured Assessment for the Genetics of Alcoholism Interview (SSAGA) (Bucholz et al., 1994b). Reliability of SSAGA diagnoses of substance use disorders is good (kappas of 0.7 and higher)(Bucholz et al., 1994a). Case status for the analyses reported here was defined as a lifetime history of DSM-IV cannabis dependence, modified to include cannabis withdrawal (i.e. 3 or more of 7 criteria clustering within a 12-month period). Controls had used cannabis at least once in their lifetime but did not meet criteria for DSM-IV dependence criteria (however, those meeting criteria for dependence on other psychoactive substances, including alcohol, were not excluded). Those who had never used cannabis even once in their lifetime (N=946) were omitted (however, re-doing analyses including them as unaffected showed similar results).
DNA samples were genotyped on the Illumina Human 1M beadchip by the Center for Inherited Diseases Research (CIDR) at Johns Hopkins University. 948,658 SNPs passed data cleaning procedures – further within-sample filtering for autosomal and X-chromosome markers yielded 948,142 markers. HapMap genotyping controls, duplicates, related subjects, and outliers were removed from the sample set. Two thousand and nineteen subjects reported being European American and 1,035 reported being African American. Further details are available in a related publication (Bierut et al., 2010).
Logistic regressions analyses were conducted in PLINK (Purcell et al., 2007), controlling for sex, age (defined, using quartiles, as 3 dummy measures representing 34 years or younger, 35–39 years, 40–44 years, with 45 years and older as the reference group), study source (see Bierut et al., 2010 for details) and two principal components (computed in EIGENSTRAT/EIGENSOFT, (Price et al., 2006)) indexing continuous variation in ethnicity. The overall genomic inflation factor was 1.014 after controlling for principal components representing ethnicity, suggesting minimal stratification effects. Genotypes (917,694 autosomal SNPs and 30,448 SNPs on the X-chromosome) were coded as 0, 1 or 2 copies of the reference allele so that risk associated with genotype was modified with each additional copy of the reference allele.
DSM-IV cannabis dependent cases were significantly more likely to be males (68% vs. 43%) and were less likely to report completion of high school (18% vs 10% who did not complete). Consistent with ascertainment, cases were significantly more likely than controls to meet criteria for alcohol (94% vs 46%), nicotine (78% vs 46%) and cocaine (73% vs 21%) dependence, greater use of other illicit drugs (88% vs 47%) as well as report a younger age at initiation of cannabis use (14.5 vs 17.4 years) and more recent (44% vs 19% using in the past year) cannabis involvement. Thus, cases show marked phenotypic similarity to the highly heritable TypeII/B cluster of cannabis dependent subjects (Ehlers et al., 2009).
The lowest p-values were obtained for polymorphisms on chromosome 17 (rs1019238 and rs1431318, p-values < E−7; rs8065311, p-value < E−6; rs9894332 and rs10521290, p-values < E−5). The top SNP lies in the ankyrin-repeat and fibronectin type III domain containing 1 (ANKFN1) gene, which was previously identified in a genomic study of general vulnerability to substance use disorders (Johnson et al., 2009). The genomic control p-value for rs1019238 was 7.3 × 10−7 suggesting adequate control for ethnic variation. The remaining SNPs (except rs10521290) are non-genic but are in strong LD (r2 of 0.75–0.88) with SNPs in ANKFN1. Fibronectin repeats are important constituents of a variety of proteins (Bloom and Calabro, 2009) and ankyrin repeats mediate protein interactions (Li et al., 2006). While there are links between ANKFN1 and PTEN (phosphatase and tensin homolog) in tumorigenesis (Syed et al., 2010), the specific role that ANKFN1 might play in the etiology of cannabis dependence is unknown.
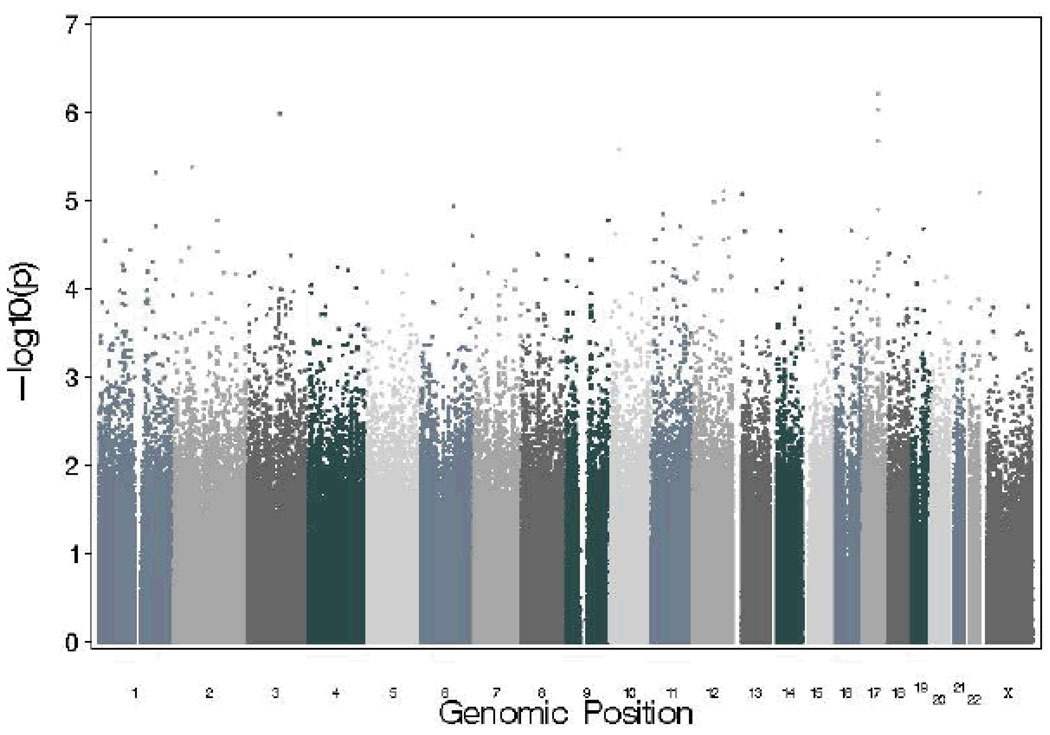
Table 1 presents odds-ratios for the full sample, as well as results stratified by ethnicity, for SNPs that yielded the most promising results. A number of genes on chromosome 12 (including the p- and q-arm) were associated, including carbohydrate (chondroitin 4) sulfotransferase 11 (CHST11). None of the SNPs that emerged as our top findings have known functional significance. Association results across the full autosomal genome and the X chromosome are shown in Figure 1.
Table 1.
Association results of top 30 SNPs from a genomewide association study of 708 DSM-IV cannabis dependence cases and 2346 controls from SAGE. (All allele frequencies and odds-ratios reflect analysis of increasing risk associated with the reference allele).
| Chromosome | SNP | Gene | Function | Overall (N=3054): |
European-American (N=2019): | African-American (N=1035): |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Odds Ratio |
Lower 95% |
Upper 95% |
P-value | Allele frequency |
O.R. | P-value | Allele frequency |
O.R. | P-value | ||||
| 1 | rs9427573 | UCHL5 | Intron | 2.557 | 1.71 | 3.822 | 4.75E-06 | 0.001 | 1.94 | 0.6318 | 0.06 | 2.47 | 0.00001 |
| 1 | rs9427836 | UCHL5 | Intron | 2.411 | 1.611 | 3.61 | 1.91E-05 | 0.001 | 1.94 | 0.631 | 0.06 | 2.35 | 0.00002 |
| 2 | rs17552189 | Nongenic | 1.397 | 1.212 | 1.61 | 4.12E-06 | 0.38 | 1.33 | 0.00124 | 0.17 | 1.58 | 0.0005 | |
| 2 | rs2381356 | KYNU | Intron | 0.637 | 0.518 | 0.782 | 1.70E-05 | 0.06 | 0.67 | 0.04543 | 0.29 | 0.63 | 0.00014 |
| 3 | rs12491921 | Nongenic | 1.385 | 1.215 | 1.578 | 1.03E-06 | 0.48 | 1.27 | 0.00471 | 0.36 | 1.63 | 0.00001 | |
| 6 | rs1462146 | AIM1 | Intron | 0.514 | 0.381 | 0.692 | 1.16E-05 | 0.003 | - | - | 0.2 | 0.54 | 0.00004 |
| 6 | rs16900973 | RPS6KA2 | Intron | 1.595 | 1.284 | 1.982 | 2.48E-05 | 0.06 | 1.39 | 0.04809 | 0.12 | 1.71 | 0.00031 |
| 9 | rs10858373 | Nongenic | 1.4 | 1.201 | 1.632 | 1.68E-05 | 0.27 | 1.4 | 0.00027 | 0.11 | 1.43 | 0.02477 | |
| 10 | rs11007350 | Nongenic | 0.731 | 0.641 | 0.833 | 2.68E-06 | 0.55 | 0.68 | 0.00001 | 0.36 | 0.89 | 0.0892 | |
| 10 | rs1007264 | STAM | Intron | 1.512 | 1.248 | 1.831 | 2.34E-05 | 0.09 | 1.67 | 0.00021 | 0.15 | 1.4 | 0.01665 |
| 11 | rs2169487 | Nongenic | 1.636 | 1.31 | 2.042 | 1.41E-05 | 0.01 | 1.46 | 0.2442 | 0.27 | 1.57 | 0.00013 | |
| 11 | rs2068909 | Nongenic | 2.238 | 1.546 | 3.239 | 1.96E-05 | 0.0004 | - | - | 0.07 | 1.99 | 0.0002 | |
| 11 | rs1352414 | Nongenic | 2.736 | 1.721 | 4.349 | 2.07E-05 | 0.0004 | - | - | 0.05 | 2.53 | 0.00006 | |
| 11 | rs1609930 | MICAL2 | Intron | 0.713 | 0.609 | 0.835 | 2.77E-05 | 0.17 | 0.71 | 0.0026 | 0.37 | 0.69 | 0.00102 |
| 12 | rs12811699 | CHST11 | Intron | 1.737 | 1.363 | 2.213 | 7.88E-06 | 0.02 | 2.03 | 0.02759 | 0.18 | 1.6 | 0.00034 |
| 12 | rs12828809 | CHST11 | Intron | 1.724 | 1.355 | 2.195 | 9.61E-06 | 0.01 | 2.04 | 0.02659 | 0.1832 | 1.59 | 0.00039 |
| 12 | rs1280605 | LGR5 | Intron | 0.699 | 0.596 | 0.819 | 1.04E-05 | 0.15 | 0.68 | 0.00163 | 0.61 | 0.71 | 0.00115 |
| 12 | rs7313862 | DKFZP779 | Intron | 1.327 | 1.163 | 1.514 | 2.61E-05 | 0.32 | 1.31 | 0.003 | 0.47 | 1.33 | 0.00593 |
| 12 | rs555404 | ACADS | Intron | 0.758 | 0.667 | 0.863 | 2.66E-05 | 0.54 | 0.78 | 0.00255 | 0.34 | 0.71 | 0.00207 |
| 13 | rs9507041 | Nongenic | 1.549 | 1.278 | 1.878 | 8.47E-06 | 0.11 | 1.53 | 0.00115 | 0.12 | 1.63 | 0.00137 | |
| 13 | rs7317962 | KATNAL1 | Intron | 0.735 | 0.638 | 0.847 | 2.23E-05 | 0.39 | 0.72 | 0.00033 | 0.23 | 0.78 | 0.04643 |
| 14 | rs17102248 | Nongenic | 4.291 | 2.19 | 8.405 | 2.19E-05 | 0.0002 | - | - | 0.02 | 4 | 0.00004 | |
| 16 | rs11865530 | FTO | Intron | 2.404 | 1.603 | 3.603 | 2.18E-05 | 0.0004 | - | - | 0.06 | 2.28 | 0.00005 |
| 17 | rs1019238 | ANKFN1 | Intron | 1.453 | 1.254 | 1.682 | 6.12E-07 | 0.4 | 1.43 | 0.00003 | 0.1 | 1.53 | 0.00828 |
| 17 | rs1431318 | Nongenic | 0.708 | 0.616 | 0.812 | 9.14E-07 | 0.44 | 0.68 | 0.00001 | 0.25 | 0.8 | 0.06522 | |
| 17 | rs8065311 | Nongenic | 1.432 | 1.234 | 1.661 | 2.10E-06 | 0.4 | 1.4 | 0.00007 | 0.09 | 1.55 | 0.01125 | |
| 17 | rs9894332 | Nongenic | 0.748 | 0.657 | 0.852 | 1.25E-05 | 0.47 | 0.74 | 0.00032 | 0.36 | 0.81 | 0.04837 | |
| 17 | rs10521290 | Nongenic | 1.331 | 1.165 | 1.522 | 2.69E-05 | 0.72 | 0.73 | 0.0002 | 0.28 | 0.83 | 0.104 | |
| 19 | rs8111749 | Nongenic | 1.572 | 1.277 | 1.936 | 2.05E-05 | 0.05 | 2.06 | 0.00008 | 0.19 | 1.35 | 0.02105 | |
| 22 | rs28372448 | PIM3 | Intron | 1.681 | 1.338 | 2.112 | 8.00E-06 | 0.06 | 1.82 | 0.00015 | 0.08 | 1.54 | 0.01367 |
Figure 1.
Manhattan plot showing genomewide association results for autosomal SNPs and SNPs on the X chromosome for DSM-IV cannabis dependence in SAGE.
While our primary analyses accounted for population stratification, we also show, in Table 1, findings within each ethnic group. A number of the SNPs showed ethnic variations in allele frequency. In a majority of instances, odds-ratios were comparable across the European- and African-American samples. However, 5 SNPs were found to have exceedingly low MAFs in the European-American sample suggesting that their overall significance in the full sample may be attributable to their effects in the smaller African-American sample.
A limitation of this study is that SAGE was ascertained for alcohol dependence. This led to a high level of comorbidity in the cannabis dependent cases and exposed controls. However, the use of controls with other forms of substance dependence, but not cannabis dependence, protected against signals that may have been less specific. In addition, a series of analyses was conducted that supported the specificity of these findings. First, SNPs in ANKFN1 were not significant when case status was alcohol (also nicotine and cocaine) dependence without comorbid cannabis dependence. Second, we created a polydrug dependence diagnosis, and results were not significant when cannabis dependent individuals were excluded from those with other polydrug dependence. While a sample ascertained for cannabis dependence would be ideal, there are challenges associated with the recruitment of such a sample (Agrawal and Lynskey, 2009), and to our knowledge, no such samples currently exist. Further, while cannabis dependence is highly heritable, none of our association signals reach genomewide significance, nor do the composite of the top 30 signals explain a considerable proportion of this heritable variance in cannabis dependence. The Benjamini-Hochberg False Discovery Rate (Benjamini and Hochberg, 1995) for the top signal was .325. Power computations reveal that at MAFs ranging from 15–40%, association signals with odds-ratios exceeding 1.45, such as our top SNP, would be detected. Thus, both replication and meta-analysis are required to confirm or refute these findings.
Characterizing the genetic underpinnings of cannabis dependence remains elusive – to this end, results from GWAS might provide clues into alternative biological pathways that shape the etiology of cannabis dependence.
ACKNOWLEDGEMENTS
Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract "High throughput genotyping for studying the genetic contributions to human disease" (HHSN268200782096C). Other support includes DA23668 (A.A.), DA18860 & DA18267 (M.T.L.).
Footnotes
| Author | Phenotype expertise |
Genotype Expertise |
Data collection/cleaning/QC |
Analyzed Data |
Wrote Paper |
|---|---|---|---|---|---|
| Agrawal | x | x | x | x | |
| Lynskey | x | x | x | ||
| Hinrichs | x | x | x | x | |
| Grucza | x | x | x | x | |
| Saccone | x | x | x | ||
| Krueger | x | x | x | ||
| Neuman | x | x | x | ||
| Howells | x | x | |||
| Fisher | x | x | x | ||
| Fox | x | x | x | ||
| Cloninger | x | x | |||
| Dick | x | x | x | ||
| Doheny | x | x | x | ||
| Edenberg | x | x | x | x | |
| Goate | x | x | x | ||
| Hesselbrock | x | x | x | ||
| Johnson | x | x | x | ||
| Kramer | x | x | x | ||
| Kuperman | x | x | x | ||
| Nurnberger | x | x | x | x | |
| Pugh | x | x | x | ||
| Schuckit | x | x | x | x | |
| Tischfield | x | x | x | ||
| GENEVA | x | x | x | x | x |
| Rice | x | x | x | ||
| Bucholz | x | x | x | ||
| Bierut | x | x | x | x | x |
Financial Disclosures
Drs. LJ Bierut, J. Rice, A. Goate and S Saccone are listed as inventors on the patent "Markers for Addiction" (US 20070258898): covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. Dr. Bierut has acted as a consultant for Pfizer, Inc. in 2008. All other authors report no competing interests.
Reference List
- Agrawal A, Lynskey M. The Genetic Epidemiology of Cannabis Use, Abuse and Dependence: A Review. Addiction. 2006;101:801–812. doi: 10.1111/j.1360-0443.2006.01399.x. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT. Candidate genes for cannabis use disorders: findings, challenges and directions. Addiction. 2009;104:518–532. doi: 10.1111/j.1360-0443.2009.02504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- Bierut L, Agrawal A, Bucholz K, Doheny KF, Laurie CC, Pugh EW, Fisher S, Fox L, Howells B, Bertelsen S, Hinrichs A, Almasy L, Breslau N, Culverhouse R, Dick D, Edenberg H, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson E, Kramer J, Krueger R, Kuperman S, Lynskey MT, Mann K, Neuman R, Nothen M, Nurnberger J, Jr, Porjesz B, Ridinger M, Saccone NL, Schuckit M, Tischfield J, Wang JC, Rietschel M, Goate A, Rice JP. A Genome-wide Association Study of Alcohol Dependence. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0911109107. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom L, Calabro V. FN3: a new protein scaffold reaches the clinic. Drug Discov Today. 2009;14:949–955. doi: 10.1016/j.drudis.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994a;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret RJ, Cloninger RC, Dinwiddie SH, Hesselbrock V, Nurnberger JI, Reich T, Schmidt I, Schuckit MA. A New, Semi-Structured Psychiatric Interview For Use In Genetic Linkage Studies. J Stud Alcohol. 1994b;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Compton WM, Grant BF, Colliver JD, Glantz MD, Stinson FS. Prevalence of marijuana use disorders in the United States: 1991–1992 and 2001–2002. JAMA. 2004;291:2114–2121. doi: 10.1001/jama.291.17.2114. [DOI] [PubMed] [Google Scholar]
- Cornelis MC, Agrawal A, Cole JW, Hansel NN, Barnes KC, Beaty TH, Bennett SN, Bierut LJ, Boerwinkle E, Doheny KF, Feenstra B, Feingold E, Fornage M, Haiman CA, Harris EL, Hayes MG, Heit JA, Hu FB, Kang JH, Laurie CC, Ling H, Manolio TA, Marazita ML, Mathias RA, Mirel DB, Paschall J, Pasquale LR, Pugh EW, Rice JP, Udren J, van Dam RM, Wang X, Wiggs JL, Williams K, Yu K. The gene, environment association studies consortium (GENEVA): maximizing the knowledge obtained from GWAS by collaboration across studies of multiple conditions. Genet Epidemiol. 2010 doi: 10.1002/gepi.20492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Gizer IR, Wilhelmsen KC. Heritability and a genome-wide linkage analysis of a Type II/B cluster construct for cannabis dependence in an American Indian community. Addict Biol. 2009;14:338–348. doi: 10.1111/j.1369-1600.2009.00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Drgon T, McMahon FJ, Uhl GR. Convergent genome wide association results for bipolar disorder and substance dependence. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:182–190. doi: 10.1002/ajmg.b.30900. [DOI] [PubMed] [Google Scholar]
- Li J, Mahajan A, Tsai MD. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed AS, D'Antonio M, Ciccarelli FD. Network of Cancer Genes: a web resource to analyze duplicability, orthology and network properties of cancer genes. Nucleic Acids Res. 2010;38:D670–D675. doi: 10.1093/nar/gkp957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treatment Episode Data Set. Substance Abuse Treatment Admissions by Primary Substance of Abuse, According to Sex, Age Group, Race, and Ethnicity, funded by the Substance Abuse and Mental Health Services Administration, DHHS. 2003 The latest data are available at 800-729-6686 or online at www.samhsa.gov.