Abstract
Understanding the influence of macromolecular crowding and nanoparticles on the formation of in-register β-sheets, the primary structural component of amyloid fibrils, is a first step towards describing in vivo protein aggregation and interactions between synthetic materials and proteins. Using all atom molecular simulations in implicit solvent we illustrate the effects of nanoparticle size, shape, and volume fraction on oligomer formation of an amyloidogenic peptide from the transthyretin protein. Surprisingly, we find that inert spherical crowding particles destabilize in-register β-sheets formed by dimers while stabilizing β-sheets comprised of trimers and tetramers. As the radius of the nanoparticle increases crowding effects decrease, implying smaller crowding particles have the largest influence on the earliest amyloid species. We explain these results using a theory based on the depletion effect. Finally, we show that spherocylindrical crowders destabilize the ordered β-sheet dimer to a greater extent than spherical crowders, which underscores the influence of nanoparticle shape on protein aggregation.
Keywords: crowding, in vivo, early events, amyloid
Molecular crowding can have a profound effect on virtually all biological processes such as protein folding, viral capsid assembly and protein aggregation [1–10]. It is estimated that 20 to 30% of a typical cell's volume is occupied by DNA, protein, lipids and sugars [3, 11, 12]. Such a crowded environment restricts the conformations explored by biological macromolecules, thus affecting the balance of thermodynamic forces that help regulate cellular processes. More recently, it has also been realized that synthetic nanoparticles (NPs) such as quantum dots, carbon nanotubes, and gold and other colloidal particles also affect the stability and function of proteins [13–15]. Understanding nanoparticle-protein interactions are important in our ability to use NPs for drug delivery and controlling environmental toxicity [16].
In the context of aggregation of amyloidogenic peptides, which is the focus of the present study, NPs can greatly influence the stabilities of the molecular species that accumulate along the routes to fibril formation. Amyloid fibrils are experimentally characterized as cross-β structures that are rich in β-sheet content [17–19]. Formation of amyloid is linked to a number of human diseases [18, 20], and it appears that almost any polypeptide chain can form amyloid under appropriate solution conditions [18]. Even small peptides [21], such as the fragment comprised of residues 105–115 from the Transthyretin (TTR) protein, form amyloid fibrils [22, 23]. TTR fibrils are associated with senile systemic amyloidosis and familial amyloid polyneuropathy I [20].
General theoretical arguments based on the depletion effect [3, 4, 24–27] suggest that monodisperse spherical NPs should promote protein aggregation. The depletion effect, which results in non-specific entropy-induced attraction between proteins in the presence of non-adsorbing NPs, arises from the volume excluded to the NPs by the monomer subunits of aggregating proteins. Consider for example two proteins that are spherical with a radius . When two such proteins approach each other up to a distance on the order of the radius, RC, of the crowding particles, the particles can no longer fit and are therefore expelled from the gap formed by the proteins. Thus, the crowding particles exert an osmotic pressure on the associating monomers resulting in a net inter-protein attraction [24–26]. As a result, inert crowding particles promote protein-protein association, which is in accord with several experiments [13, 28–30].
Although this qualitative argument is compelling, the molecular consequences of crowding effects on oligomer formation is unknown. In particular it is unclear whether crowding particles stabilize ordered β-sheet oligomers, which might facilitate amyloid formation, or stabilize amorphous aggregates with little β-sheet content, which would likely impede or delay amyloid formation. To address these unresolved issues we have carried out molecular simulations of systems composed of either two, three, or four TTR peptides to assess the effect of size of spherical NPs on the structural and energetic properties of TTR oligomers. We find that spherical NPs destabilize ordered dimer aggregates but stabilize trimer and tetramer ordered aggregates. Increasing the radius of the NP, at a fixed crowder volume fraction (ΦC), reduces the effect of molecular crowding due to an increase in the interstitial space between NPs. In contrast, increasing the aspect ratio of the NPs with a spherocylinder shape leads to greater destabilization of the ordered dimer than a spherical crowding particle.
In order to assess the effect of crowders on oligomer formation we first carried out separate constant temperature simulations on the monomer, dimer (denoted {T T R}2), trimer ({T T R}3), and tetramer ({T T R}4) systems in the EEF1.1 implicit solvent model [31] at ΦC = 0, a temperature of 395 K, and at a protein concentration around 30 mM (SI Table 1). A high temperature was used to increase conformational sampling and yield converged results; we found that simulations carried out at temperatures less than 330 K were not converged. It should be stressed that for a complete understanding of crowding effects peptide concentration must also be varied [4] as has been recently demonstrated in a complementary study using model systems [32]. For the systems containing two or more peptides we find that due to the high protein concentration the peptides are associated. Therefore, we cannot examine the effect of NPs on the association process of these peptides. However, as mentioned previously, the more crucial aspect from the perspective of amyloid formation is determining the effect of NPs on the different aggregated species (ordered versus disordered), an issue which these simulations can directly address.
The TTR monomer is compact and devoid of persistent secondary structure at ΦC = 0. Its average radius of gyration is 7 Å, whereas a fully extended TTR structure would have a value of around 12 Å. Analysis of the secondary structure content shows that coil and turn structures are the most prevalent while helical and β-strand structures are negligibly populated (< 2%). Even in the presence of crowding agents the strand content does not change appreciably. Thus, in isolation the TTR peptide is unstructured.
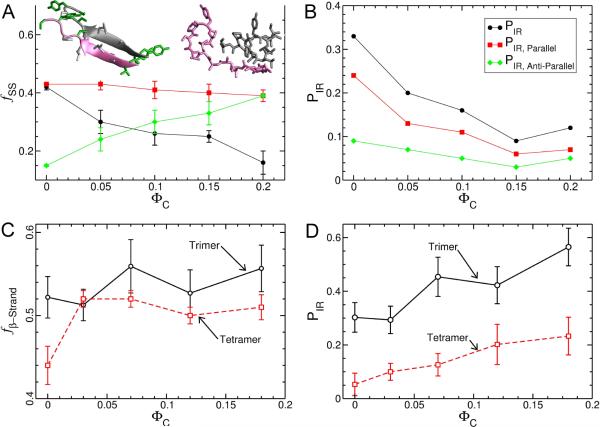
The dimer {T T R}2 has an appreciable population of both ordered and disordered aggregates. In the ordered state the peptides prefer parallel in-register β-sheets over anti-parallel structures. The fraction of β-strand content in {T T R}2 is 0.42 (Fig. 1a). The value of PIR, the probability that the two strands are in-register, when both are in a parallel arrangement is nearly 3-fold greater than the probability of forming in-register anti-parallel structures (Fig. 1b). Expanded peptide conformations have several more in-register contacts on average, make more inter-peptide backbone hydrogen bonds, and are more likely to have β-strand content than compact peptide conformations. For example, the radius of gyration 〈Rg〉 of the individual peptides in the ordered {T T R}2 (`ordered' being defined as having greater than eight in-register contacts out of a possible eleven) is 10.8 Å A while 〈Rg〉 of the peptide in the disordered {T T R}2 conformations (which have less than 5 in-register contacts) is 6.7 Å.
FIG. 1.
Effect of spherical crowding on {T T R}n as a function ΦC. (A) and (B) are for the {T T R}2. (A) Secondary structure content (β-strand (black), random coil (red), or turn(green)) versus ΦC. Example structures from the ordered aggregate and disordered aggregate are displayed on the upper left and right respectively. (B) The fraction of in-register contacts (PIR, see Eq. 4 in the SI) versus ΦC. (C) and (D) correspond to data from the {T T R}3 and {T T R}4 systems. (C) The ΦC-dependent changes in the β-strand content for the trimer (solid black line with circles) and the tetramer (dashed red line with squares).(D) Same as (C) except the ordinate is the probability of finding an in-register aggregate (including both parallel and anti-parallel configurations).
Independent simulations of the {T T R}3 and {T T R}4 at ΦC = 0 reveal they also exist as either disordered aggregates or ordered β-sheets. However, the probabilities of β-sheet structure, which includes out-of-register β-strands, for {T T R}3 and {T T R}4 are 0.52 and 0.44, respectively (Fig. 1c). The probability that {T T R}3 and {T T R}4 form ordered in-register parallel and anti-parallel strand arrangements are 0.30 and 0.06, respectively (Fig. 1d). The decrease in ordered in-register β-sheets, relative to the {T T R}2 system, is due to an increase in the number of low energy out-of-register disordered conformations that are accessible as the number of peptides in the aggregate increases. Thus, the landscape of TTR oligomers has a number of distinct basins of attraction with conformationally heterogeneous structures (see also [33]).
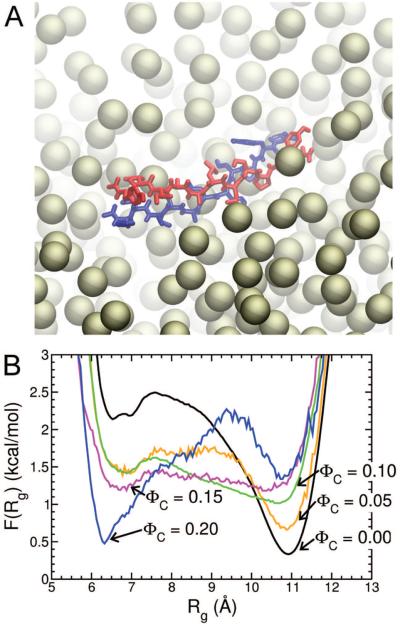
We next examined the influence of inert spherical NPs with RC = 3.5 Å (ΦC > 0, Fig. 2a). For the {T T R}2 system the presence of spherical NPs decreases the stability of the expanded (conformations with Rg > 10 Å) dimer structures monotonically as ΦC increases, while the stability of the collapsed structures (i.e., Rg < 7.5 Å) increases (Fig. 2b). As a consequence, the average solvent accessible surface area and the molecular volume of the dimer decreases as ΦC increases (data not shown). In addition, the β-strand content (Fig. 1a) and the probability of finding polymerization competent in-register dimers (PIR) decreases with increasing ΦC (Fig. 1b). Thus, relative to ΦC = 0, crowding destabilizes in-register β-sheet aggregates causing the equilibrium to shift towards collapsed associated dimer structures at all values of ΦC (see the structures in Fig. 1a).
FIG. 2.
Effect of spherical crowding on the size of the {T T R}2 dimer. (A) Snapshot of a single configuration of the NPs during the simulations. (B) The free energy profile as a function of the radius-of-gyration of the monomers in the dimer at various ΦC values.
Surprisingly, we find the opposite result for the {TTR}3 and {TTR}4, whose ordered structures are stabilized as ΦC increases. The β-strand content in {TTR}3 remains the same as ΦC increases, whereas there is a slight increase for {TTR}4 (Fig. 1c). In addition, PIR increases for the trimer and tetramer with increasing ΦC (Fig. 1d). The observed variations in crowding-induced changes in the stabilities of the ordered β-sheet aggregates may be relevant to in vivo amyloid formation as β-sheets are the primary structural component found in mature fibrils [21, 34–36]. In particular, NPs may inhibit the ordered dimer structures but stabilize higher order oligomers.
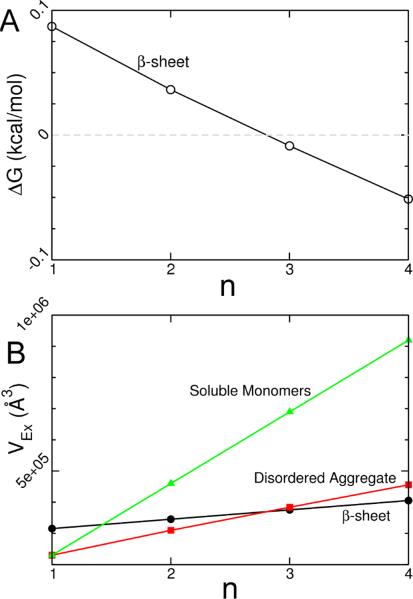
This non-monotonicity in the n-dependence of the stability of the ordered aggregate of {TTR}n (n = 2, 3, 4) at various ΦC can be explained using a theory based on depletion forces. Scaled Particle Theory [37–39] suggests that a factor influencing crowder effects is the free energy cost of creating a void within the configuration of NPs to accommodate {TTR}n when ΦC 6= 0. The free energy is a function of volume of {TTR}n's volume, which increases as n becomes larger. Clearly, the probability of finding a volume to accommodate {TTR}n decreases as n increases. Depletion-induced changes in the free energy of the ordered aggregate relative to the disordered aggregate, denoted ΔGn, is therefore proportional to ln(VOA − VDA), where VOA (VDA) is the volume excluded to the NP's center-of-mass (also known as the `depletion volume') by the ordered (disordered) aggregate. Peptide conformational entropy changes also likely contribute to the behavior of ΔGn with n. However, it is diffcult to accurately estimate these entropy changes in the ordered and disordered aggregates due to crowding. In addition to conformational entropy difference, stability is also determined by favorable enthalpic inter-peptide interactions. Enthalpic stabilization occurs for both the ordered and disordered aggregates as n increases because of an increase in the number of inter-peptide contacts. The specific value of n which stabilizes the ordered form should depend on the size and sequence of the peptide. For {TTR}n it is only when n becomes larger than 3 that the ordered aggregate structures become more stable in the presence of spherical NPs (Figs. 1b and 1d).
If depletion forces are a major factor in the non-monotonic behavior of ΔΔGn with n then we can account for the simulation results using the changes in the volume excluded to the NPs. From the Asakura-Oosawa (AO) theory [24] and a microscopic formulation [25], as well as several other approaches [37–39], it follows that the difference in free energy between species i and j is , where P = kBTΦC/VC, is the osmotic 4pressure under ideal solution conditions, kB is Boltzmann's constant, VC is the molecular volume of a nanoparticle, and is the volume excluded to the spherical nanoparticle by species i. There are three relevant species in the {TTR}n system; the soluble non-associated monomers (SM), the disordered aggregate (DA), and the ordered aggregate (OA). We calculate by assuming that the soluble monomers and disordered aggregate are hard spheres whose radii depend on n. In this case
| (1) |
| (2) |
where Naa is the number of amino acids in the peptide and the exponent of 0.6 is the Flory scaling exponent that characterizes the size of a protein in a good solvent [40]. The shape of the ordered β-sheet aggregate is taken to be a stacked rectangular parallelepiped, where each parallelepiped corresponds to one TTR peptide. For this species Vex is computed using
| (3) |
where l, w, and h correspond to the length, width and height of a β-sheet made up of n peptides, respectively. Therefore, we define l = 3.2 Naa Å, corresponding to the length of an extend β-strand made up of Naa amino-acids;W=8 Å which corresponds to the distance that the side chains stick out above and below the β-sheet; h = 4.85n Å, where 4.85 Å corresponds to the experimentally determined distance between the neighboring strands in a β-sheet.
Using Eqs. 1–3 it is straightforward to compute the Vex for each species at different ΦC, and thereby estimate ΔGi(n) at a given ΦC. We find that indeed for a range of RC values this model displays non-monotonic behavior (Fig. 3a) consistent with the trends observed in the simulations; the ordered dimer is destabilized by spherical crowders, but as more peptides are added the NPs stabilize the ordered β-sheet aggregates. The physical origin of this unusual behavior arises because disordered dimer aggregate excludes less volume than the ordered dimeric β-sheet (Fig. 3b), but as additional peptides are added to the system the volume per peptide increases to a greater extent in the disordered aggregate than the ordered β-sheet (the same observation was made in ref. [41]). As a consequence, the free energy of the system is minimized when the dimer is disordered whereas NP-induced ordered trimer and tetramer structures are more stable. Recapitulation of the qualitative trends observed in the simulation model suggests that the depletion effect explains the changes in nanoparticle-induced changes in stability as n varies.
FIG. 3.
(A) Theoretical prediction of the stability of the ordered β-sheet aggregate relative to the disordered aggregate as a function of the number of TTR peptides in the system. The solution conditions in this theoretical treatment are T = 395 K, ΦC=0.15, and RC=30 Å. (B) Volume excluded (i.e., the depletion volume) to the NPs center of mass of the soluble monomers (green triangles), disordered aggregate (red squares) and ordered β-sheet aggregate (black circles) calculated from Eqs. 1–3
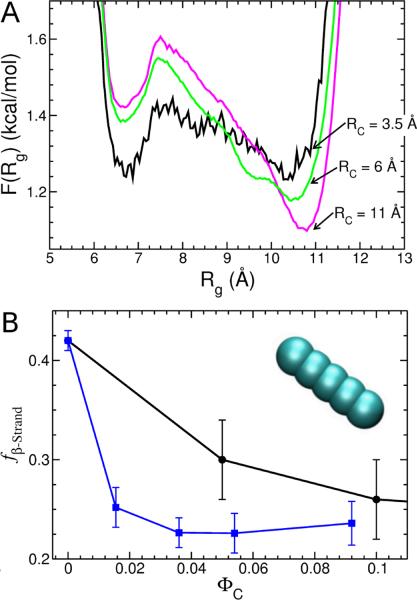
There are a number of other parameters besides ΦC that also influence oligomerization. What happens when the radius of the NPs increases? For a fixed peptide concentration ([CP]) and ΦC, larger NPs exclude less volume to the protein than smaller crowders because the interstitial space between crowders increases with crowder size. It can be shown that the strength of the depletion force is proportional to . Therefore, larger crowding particles should have a smaller effect in altering the stability of {TTR}n. Explicit simulations that we ran of {TTR}2 at ΦC = 0.15 using three spherical NP sizes (RC = 3.5, 6 and 11 Å) show that indeed as RC increases larger crowders shift the equilibrium towards expanded structures just as observed in bulk simulations (Fig. 4). However, in the simulations we observe that the average β-sheet content does not exhibit an increase as the size of the NP increases and the probability (≈ 0.06) of finding ordered in-register dimers is also unchanged. Thus, increasing RC of spherical NPs decreases the impact of crowding - a result consistent with other theoretical and simulation modeling [42].
FIG. 4.
(A) Effect of spherical NP size on {T T R}2 at ΦC = 0.15. Changes in the free energy profile as a function of Rg of the monomer (within the dimer) as the radius RC of the spherical NP is varied. (B) Average β-sheet content of the {T T R}2 system versus ΦC for spherical crowders (black line and circles), and spherocylindrical crowders with an aspect ratio of 3.3 (blue line and squares). The inset shows a spherocylinder used in the simulations.
Another variable that controls the extent or ordered oligomer formation is the shape of the crowding particles. Indeed, in vivo most naturally occurring molecular crowders are unlikely to be spherical. Analysis of protein structures in the Protein Data Bank [43] show that even in the folded state proteins adopt anisotropic shapes. To explore the effect of NP shape on dimer stability we simulated the {TTR}2 system in the presence of spherocylindrical NPs in the isotropic phase whose aspect ratio (=L/D+1, where L is the spherocylinder length and D is its diameter) is 3.3. We find that for the same ΦC value (below ΦC=0.1, which is below the isotropic to nematic transition point) spherocylindrical NPs cause a greater loss of β-strand content in the dimer than spherical NPs. Thus, spherocylindrical NPs destabilize ordered oligomers. This suggests that anisotropic NPs may have a greater impact on amyloid formation than spherical NPs, and that crowder shape is another important variable in understanding amyloid formation in vivo. To further explore realistic models of oligomerization in the cellular context it would be important to consider a soup of proteins as was done recently to explore folding and diffusion in models of E. Coli. [44].
Although the systems investigated in this study are caricatures of cellular crowding they illustrate the complexity of peptide aggregation under in vivo conditions. While generic arguments suggest that depletion forces should promote protein association, our work shows that the structures of the oligomers can be dramatically altered by the size, shape, and volume fraction of the nanoparticles. Our study shows that an interplay of a number of factors determines the equilibrium between the ordered and disordered oligomeric structures. For spherical NPs the variables that determine the depletion forces, and hence the strength and range of the entropically-induced inter peptide attraction, are (where Rg is the size of the peptide monomer), [CP] the peptide concentration, and ΦC.
This predicted complexity in the crowding-induced diagram of states of the amyloidogenic peptide is not surprising given that in the field of polymer physics it was discovered previously that the phase diagram of a mixture of hard sphere colloidal particles and self-avoiding polymers in an athermal solvent is complicated, and depends on polymer concentration, ΦC and q [26]. For the TTR peptides and RC values examined here q ranges in value from 0.6 up to 2 (Note that Rg ≈7 Å for the peptide monomer). It follows from our work that larger q values can promote extended peptide conformations with high β-strand content in aggregates consisting of more than two peptides. Conversely, in the extreme case of q << 1 (in the so called colloid limit [26]) we predict that the influence of crowding effects will be decreased, and the equilibrium between ordered and disordered species may not be significantly perturbed relative to bulk (ΦC=0).
The results presented here show that understanding of NP effects on protein aggregation requires determination of a global phase diagram in terms of a number of variables. The non-monotonicity in NP-induced changes in stability should be contrasted with the effect of crowding on monomeric protein folding stability, which monotonically increases in the presence of non-adsorbing crowding particles [4]. Further insights into in vivo aggregation will require extension of the present work by taking crowder phase and polydispersity into account perhaps along the lines suggested recently [12]. In such a complex environment polydispersity and concentration fluctuations could drive phase separation in the macromolecules [45], which could add additional complications in the process of protein aggregation.
Methods
Models for peptide and solvent
We chose the peptide fragment (sequence Tyr-Thr-Ile-Ala-Ala-Leu-Leu-Ser-Pro-Tyr-Ser) from the Transthyretin protein (TTR) corresponding to residues 105–115. The structure of these 12 residues in an amyloid fibril has been determined (PDB ID 1RVS) using solid state NMR [22, 23]. We cap the peptide's N-terminus with an acetyl blocking group and its C-terminus with an amine blocking group (NH2). For the peptide we use an all-atom representation except for non-polar hydrogen atoms, which are omitted in the calculations. The EEF1.1 implicit solvent model [46] is used to account for solvent effects in conjunction with the CHARMM19 force-field [31].
The EEF1.1 implicit solvent model
The parameterization of the EEF1.1 [31] force-field requires the use of specific cutoff distances for non-bonded interactions. The Lennard-Jones (LJ) and electrostatic interactions are truncated at 9 Å, with a switch function applied to the LJ term starting at 7 Å. The Lorentz-Berthelot mixing rules [47] are used to determine the undefined σ-values between the atomic centers in the peptide. A distance dependent dielectric constant is used for electrostatic interactions.
The LJ interactions in the EFF1.1 protein parameters are restricted to a distance less than 9 Å, which prevents us from studying crowders with RC values greater than 4.5 Å in the standard Charmm code. We modified the CHARMM code to allow the LJ cutoff to be dependent on the identity of the interacting atomic centers, thereby allowing us to use larger crowding particles. For protein-protein interactions nonbonded interactions are truncated at 9 Å, but for crowder-crowder and crowder-protein interactions we use a cutoff of 2RC+2 Å and RC+2 Å, respectively. These procedures allow us to maintain the EEF1.1 nonbond requirements for protein-protein interactions while allowing for larger crowding particles to be simulated.
Simulation details
Simulations were carried out in the NVT ensemble at 395 K using Langevin dynamics with a friction coefficient of 1 ps−1. This high temperature allowed the the dimer simulations to reach equilibrium, whereas simulations at 330 K were found to not have converged on our simulated time scales (data not shown). The SHAKE algorithm was used to fix the bond lengths of covalently bonded hydrogen, allowing the use of a 2 fs time-step.
Typically, we generated ten independent trajectories for each crowder size and ΦC. At least half of the trajectories were started with peptide conformations taken from equilibrated bulk simulations at ΦC = 0.0. When possible the other five starting conformations were taken from solution conditions that were closest to those of interest. For example, when simulating crowders with RC = 6 and 11 Å, half of the initial protein conformations were from the equilibrium simulations of crowders with RC = 3.5 Å and ΦC = 0.15.
A box length of 60.0 Å to 80.0 Å is used for all spherical NP conditions, which gives a peptide concentration in the range from 15 mM to 31 mM. At ΦC = 0.05, 0.10, 0.15, 0.20 there are 60, 120, 180 and 240 crowders in the periodic box, respectively. Because correlations in the pair distribution function between crowders do not persist for more than half the box length, finite size effects are minimized (Fig. S1). A summary of the simulation details can be found in Table S1.
Models for spherical particles
Spherical NPs are modeled as Lennard-Jones particles. The interaction between sites i and j on two distinct NPs is
| (4) |
The choice of ∈ = 10−15 kcal/mol makes Eq. 4 essentially repulsive, and the condition VLJ(rij = 2RC) = kBT, allows us to solve for σ. We consider three values for RC = 3.5, 6, and 11 Å, which leads to σ= 108.7, 186.3, and 341.6 Å, respectively.
Model for spherocylinders and crowder-protein interactions
We define all protein-crowder σ-values such that when a crowding particle interaction site is (RC + 1) Å from a protein atomic center VLJ(RC + 1) = kBT. The resulting σ values are 139.7, 217.4 and 372.6 Å for RC = 3.5, 6 and 11 Å, respectively.
The covalent bond between the tethered spheres that comprise the spherocylinder was modeled using
| (5) |
where KB = 30 kcal/(mol·Å2) and ro = 4 Å is the equilibrium bond length. The rigidity of the spherocylinder is maintained using a bond angle potential
| (6) |
with θo = 180° and KA = 0.6092 kcal/degree2. An important characteristic of a spherocylinder is the aspect ratio λ. A value of λ = 1 corresponds to a spherical crowder with diameter D = 2RC. In our study all spherocylinders have RC = 3.5 Å, and LC, which is proportional to the number of spheres (NB) in the spherocylinder. LC = (NB − 1)4.0 Å. We used λ = 3.3, corresponding to a spherocylinder made up of 5 beads.
Supplementary Material
Acknowledgments
This work was supported by a NIH grant (R01 GM076688-080) to JES and DT, a NSF postdoctoral Fellowship to EO, and the intramural program at the National Heart Lung and Blood Institute of the NIH to BB.
Footnotes
Supporting Information Analysis details. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- [1].Hofrichter J, Ross PD, Eaton WA. Supersaturation in Sickle-cell Hemoglobin Solutions. Proc. Natl. Acad. Sci. USA. 1976;73:3035–3039. doi: 10.1073/pnas.73.9.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ross PD, Minton AP. Analysis of Non-ideal Behavior in Concentrated Hemoglobin Solutions. J. Molec. Biol. 1977;112:437–452. doi: 10.1016/s0022-2836(77)80191-5. [DOI] [PubMed] [Google Scholar]
- [3].Zhou HX, Rivas GN, Minton AP. Macromolecular Crowding and Confinement: Biochemical, Biophysical, and Potential Physiological Consequences. Ann. Rev. Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cheung MS, Klimov D, Thirumalai D. Molecular Crowding Enhances Native State Stability and Refolding Rates of Globular Proteins. Proc. Natl. Acad. Sci. USA. 2005;102:4753–4758. doi: 10.1073/pnas.0409630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].del Álamao M, Rivas G, Mateu MG. Effect of Macromolecular Crowding Agents on Human Immunodeficiency Virus Type 1 Capsid Protein Assembly In Vitro. J. Virol. 2005;79:14271–14281. doi: 10.1128/JVI.79.22.14271-14281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Straub JE, Thirumalai D. Principles Governing Oligomer Formation in Amyloidogenic Peptides. Curr. Opin. Struc. Biol. 2010;20:187–195. doi: 10.1016/j.sbi.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Stagg L, Zhang SQ, Cheung MS, Wittung-Stafshede P. Molecular Crowding Enhances Native Structure and Stability of Alpha/Beta Protein Flavodoxin. Proc. Natl. Acad. Sci. USA. 2007;104:18976–18981. doi: 10.1073/pnas.0705127104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schreiber G, Haran G, Zhou HX. Fundamental Aspects of Protein-Protein Association Kinetics. Chem. Rev. 2009;109:839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dhar A, Samiotakis A, Ebbinghaus S, Nienhaus L, Homouz D, Gruebele M, Cheung MS. Structure, Function and Folding of Phosphoglycerate Kinase are Strongly Perturbed by Macromolecular Crowding. Proc. Natl. Acad. Sci. USA. 2010;107:17586–17591. doi: 10.1073/pnas.1006760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wu C, Shea JE. Coarse-grained Models for Protein Aggregation. Curr. Opin. Struc. Biol. 2010;21:1–12. doi: 10.1016/j.sbi.2011.02.002. [DOI] [PubMed] [Google Scholar]
- [11].Zimmerman SB, Minton AP. Macromolecular Crowding: Biochemical, Biophysical and Physiological Consequences. Annu. Rev. Biophys. Biomol. Struct. 1993;22:27–75. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- [12].Elcock AH. Models of Macromolecular Crowding Effects and the Need for Quantitative Comparisons with Experiment. Curr. Opin. Struc. Biol. 2010;20:196–206. doi: 10.1016/j.sbi.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Linse S, Cabaleiro-Lago C, Xue WF, Lynch I, Lindman S, Thulin E, Radford SE, Dawson KA. Nucleation of Protein Fibrillation by Nanoparticles. Proc. Natl. Acad. Sci. 2007;104:8691–8696. doi: 10.1073/pnas.0701250104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Auer S, Trovato A, Vendruscolo M. A Condensation-Ordering Mechanism in Nanoparticle-Catalyzed Peptide Aggregation. Plos Comp. Biol. 2009;5:e1000458. doi: 10.1371/journal.pcbi.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Colvin VL, Kulinowski KM. Nanoparticles as Catalysts for Protein Fibrillation. Proc. Natl. Acad. Sci. 2007;104:8679–8680. doi: 10.1073/pnas.0703194104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Colvin VL. The Potential Environmental Impact of Engineered Nanomaterials. Nat. Biotech. 2003;21:1166–1170. doi: 10.1038/nbt875. [DOI] [PubMed] [Google Scholar]
- [17].Chan JCC, Oyler NA, Yau WM, Tycko R. Parallel beta-sheets and Polar Zippers in Amyloid Fibrils Formed by Residues 10–39 of the Yeast Prion Protein Ure2p. Biochem. 2005;44:10669–10680. doi: 10.1021/bi050724t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chiti F, Dobson CM. Protein Misfolding, Functional Amyloid, and Human Disease. Ann. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- [19].Nelson R, Eisenberg D. Recent Atomic Models of Amyloid Fibril Structure. Curr. Opin. Struc. Biol. 006;16:260–265. doi: 10.1016/j.sbi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- [20].Sacchettini JC, Kelly JW. Therapeutic Strategies for Human Amyloid Diseases. Nature Rev. Drug Disc. 2002;1:267–275. doi: 10.1038/nrd769. [DOI] [PubMed] [Google Scholar]
- [21].Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Structure of the Cross-beta Spine of Amyloid-like Fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jaroniec CP, MacPhee CE, Astrof NS, Dobson CM, Griffin RG. Molecular Conformation of a Peptide Fragment of Transthyretin in an Amyloid Fibril. Proc. Natl. Acad. Sci. USA. 2002;99:16748–16753. doi: 10.1073/pnas.252625999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. High-resolution Molecular Structure of a Peptide in an Amyloid Fibril Determined by Magic Angle Spinning NMR Spectroscopy. Proc. Natl. Acad. Sci. USA. 2004;101:711–716. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Asakura S, Oosawa F. On Interaction Between 2 Bodies Immersed in a Solution of Macromolecules. J. Chem. Phys. 1954;22:1255–1256. [Google Scholar]
- [25].Shaw MR, Thirumalai D. Free Polymer in a Colloidal Solution. Phys. Rev. A. 1991;44:4797–4800. doi: 10.1103/physreva.44.r4797. [DOI] [PubMed] [Google Scholar]
- [26].Fleer GJ, Tuinier R. Analytical Phase Diagrams for Colloids and Non-adsorbing Polymer. Adv. Colloid. Int. Sci. 2008;143:1–47. doi: 10.1016/j.cis.2008.07.001. [DOI] [PubMed] [Google Scholar]
- [27].Cunha S, Woldringh CL, Odijk T. Polymer-mediated Compaction and Internal Dynamics of Isolated Escherichia coli Nucleoids. J. Struc. Biol. 2001;136:53–66. doi: 10.1006/jsbi.2001.4420. [DOI] [PubMed] [Google Scholar]
- [28].Behe M, Englander SW. Sickle Hemoglobin Gelation: Reaction Order and Critical Nucleus Size. Biophys. J. 1978;23:129–145. doi: 10.1016/S0006-3495(78)85438-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lindner RA, Ralston GB. Macromolecular Crowding: Effects on Actin Polymerization. Biophys. Chem. 1997;66:57–66. doi: 10.1016/s0301-4622(97)00011-2. [DOI] [PubMed] [Google Scholar]
- [30].Tellam RL, Sculley MJ, Nichol LW, Wills PR. The Influence of Polyethylene Glycol 6000 on the Properties of Skeletal-muscle Actin. Biochem. J. 1983;213:6551–6559. doi: 10.1042/bj2130651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lazaridis T, Karplus M. Effective Energy Function for Proteins in Solution. Prot. Struc. Func. Gene. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- [32].Magno A, Caflisch A, Pellarin R. Crowding Effects on Amyloid Aggregation Kinetics. J. Phys. Chem. Lett. 2010;1:3027–3032. [Google Scholar]
- [33].Nguyen PH, Li MS, Stock G, Straub JE, Thiruamalai D. Monomer Adds to Preformed Structured Oligomers of A beta-peptides by a Two-stage Dock-lock Mechanism. Proc. Natl. Acad. Sci. USA. 2007;104:111–116. doi: 10.1073/pnas.0607440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Petty SA, Decatur SM. Intersheet Rearrangement of Polypeptides During Nucleation of beta-sheet Aggregates. Proc. Natl. Acad. Sci. USA. 2005;102:14272–14277. doi: 10.1073/pnas.0502804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tycko R. Characterization of Amyloid Structures at the Molecular Level by Solid State Nuclear Magnetic Resonance Spectroscopy. Meth. Enzym. 2006;413:103–122. doi: 10.1016/S0076-6879(06)13006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tarus B, Straub JE, Thirumalai D. Dynamics of Asp23-Lys28 Salt-bridge Formation in A beta(10–35) Monomer. J. Amer. Chem. Soc. 2006;128:16159–16168. doi: 10.1021/ja064872y. [DOI] [PubMed] [Google Scholar]
- [37].Cotter MA, Martire DE. Statistical Mechanics of Rodlike Particles .2. A Scaled Particle Investigation of Aligned-Isotropic Transition in a Fluid of Rigid Spherocylinders. J. Chem. Phys. 1970;52:1909–1919. [Google Scholar]
- [38].Pierotti RA. Scaled Particle Theory of Aqueous and Non-Aqueous Solutions. Chem. Rev. 1976;76:717–726. [Google Scholar]
- [39].Hall D, Minton AP. Macromolecular Crowding: Qualitative and Semiquantitative Successes, Quantitative Challenges. Biochim. Biophys. Acta Prot. 2003;1649:127–139. doi: 10.1016/s1570-9639(03)00167-5. [DOI] [PubMed] [Google Scholar]
- [40].Flory PJ. Principles of Polymer Chemistry. 2nd ed. Cornell University Press; Ithaca, NY: 1957. [Google Scholar]
- [41].Rohrig UF, Laio A, Tantalo N, Parrinello M, Petronzio R. Stability and Structure of Oligomers of the Alzheimer Peptide A beta(16–22): From the Dimer to the 32-mer. Biophys. J. 2006;91:3217–3229. doi: 10.1529/biophysj.106.088542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Batra J, Xu K, Qin S, Zhou HX. Effect of Macromolecular Crowding on Protein Binding Stability: Modest Stabilization and Significant Biological Consequences. Biophys. J. 2009;97:906–911. doi: 10.1016/j.bpj.2009.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dima RI, Thirumalai D. Asymmetry in the Shapes of Folded and Denatured States of Proteins. J. Phys. Chem. B. 2004;108:6564–6570. [Google Scholar]
- [44].McGuffee SR, Elcock AH. Diffusion, Crowding & Protein Stability in a Dynamic Molecular Model of the Bacterial Cytoplasm. Plos Comp. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000694. year. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Walter H, Brooks DE. Phase Separation in Cytoplasm, Due to Macromolecular Crowding, is the Basis for Microcompartmentation. FEBS Lett. 1995;361:135–139. doi: 10.1016/0014-5793(95)00159-7. [DOI] [PubMed] [Google Scholar]
- [46].Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM - A Program for Macromolecular Energy, Minimization, and Dynamics Calculations. J. Comp. Chem. 1983;4:187–217. [Google Scholar]
- [47].Allen MP, Tildesley DJ. Computer Simulations of Liquids. 9th ed. Oxford Univ. Press; Oxford, NY: 1987. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.