Abstract
Raman spectroscopy has the potential to differentiate among the various stages leading to high-grade cervical cancer such as normal, squamous metaplasia, and low-grade cancer. For Raman spectroscopy to successfully differentiate among the stages, an applicable statistical method must be developed. Algorithms like linear discriminant analysis (LDA) are incapable of differentiating among three or more types of tissues. We developed a novel statistical method combining the method of maximum representation and discrimination feature (MRDF) to extract diagnostic information with sparse multinomial logistic regression (SMLR) to classify spectra based on nonlinear features for multiclass analysis of Raman spectra. We found that high-grade spectra classified correctly 95% of the time; low-grade data classified correctly 74% of the time, improving sensitivity from 92 to 98% and specificity from 81 to 96% suggesting that MRDF with SMLR is a more appropriate technique for categorizing Raman spectra. SMLR also outputs a posterior probability to evaluate the algorithm’s accuracy. This combined method holds promise to diagnose subtle changes leading to cervical cancer.
Keywords: Raman spectroscopy, optical diagnosis, cervix, dysplasia
Introduction
Raman spectroscopy has been used for many years to probe into the biochemistry of various biological molecules.[1] It is a molecule-specific technique that can be used as a biochemical tool to provide differential diagnosis of precancers and cancers. Several biological molecules such as nucleic acids, proteins and lipids have distinctive Raman features that yield molecule-specific structural and environmental information. Results indicate that molecular and cellular changes that occur in precancerous tissues as well as in benign abnormalities, such as inflammation, yield characteristic Raman features that allow their differentiation. For example, one of the more prominent changes that occur with cancerous and precancerous conditions is increased cellular nucleic acid content; extensive DNA studies indicate that it may be possible to detect this change using Raman spectroscopy.[2] On the basis of these biochemical differences, several groups have studied the potential of vibrational spectroscopy for cancer diagnosis in various organ sites.[1] These studies have shown that features of the vibrational spectra can be related to molecular and structural changes associated with neoplastic transformation. Accordingly, Raman spectroscopy has been applied to in vitro detection of cancers of epithelial and mesenchymal origin such as breast, colon, esophagus and gynecologic tissues.[3] While many challenges have prevented the widespread application of Raman spectroscopy for disease detection, recent developments in detector and source technologies have resulted in acquisition of Raman spectra from tissue in 1–3 s. Several fiber optic probes have also been developed that are capable of measuring Raman spectra in vivo, making it possible to apply this technique in a clinical setting.[4] There have been an increased number of reports published on applying Raman spectroscopy for detecting cancers in vivo, such as in the cervix, skin, breast and the gastrointestinal (GI) tract with high sensitivities and specificities.[5 – 9]
In order to achieve such high sensitivities and specificities, appropriate statistical algorithms must be used to tease out important information from the Raman data. A variety of statistical methods have been developed to classify the tissue as normal or abnormal. For example, many research groups have normalized peak intensities to the four common Raman bands and then performed a Student’s t-test to identify the peak ratios corresponding to the most significant difference between tissue types.[9,10] Logistic regression algorithms have also been utilized to distinguish between cancerous and noncancerous tissue on the basis of Raman spectra. This algorithm was developed by nonlinearly transforming the traditional linear regression so that the outcome is only 0 (normal) or 1 (cancerous).[11] After normalizing peak ratios, multiple analyses of variance (ANOVA) have sometimes been used to identify the most diagnostically significant peaks.[12]
Other attempts to analyze data have included the use of principal component analysis (PCA) to establish differences among and decrease data from Raman spectra.[7,12,13] Principal components are a set of virtual spectra; the use of weighted linear combinations (scores) results in the real, measured spectra with a specified percentage variance. The scores provide information on how the spectra are correlated. Sometimes, this scoring is followed by other statistical analyses such as probabilistic artificial neural networks, which can then be used to train the input Raman spectra to correlate with known outputs or pathological categories; this network can then be used to predict the pathology of a new input of Raman spectra. Alternatively, after the spectra are broken down using PCA, linear discriminant analysis (LDA) can be used to maximize differences between pathology groups and minimize differences within groups.[14] Other approaches utilize Fisher discriminant analyses (FDA) to classify the spectra following PCA to search for nonlinear correlations.[13] The majority of these algorithms undergo cross-validation analysis using the leave-one-out method to assess their validity.[13,14]
Cluster analysis is one method where similarities between genes are described mathematically, by measuring the Euclidean distance, angle or dot products of two n-dimensional vectors from a series of n measurements of genetic information.[15] This algorithm can be similarly applied to determine subtle changes in Raman data. Another process is decision tree learning with genetic algorithms to determine optimal subsets of discriminatory features for pattern recognition.[16,17] A linear decision binary tree can be used for binary and multiclass, such as Pap smear cell classification.[16] A few drawbacks of these algorithms are that they require a significant amount of time to develop and, once developed, they are only applicable to one type of data set.
The major limitation of these previous applications is that the discrimination algorithms are binary, which are not capable of determining which class the tissue could belong to. Tissue is also not homogeneous–there could be multiple tissue types present in a single tissue sample. Therefore, some of these algorithms are run a second and third time to further classify the outcome.[11] More recently, Widjaja et al.,[18] have combined support vector machines (SVM) with PCA to classify colonic tissues as normal, hyperplastic polyps, or adenocarcinomas However, conventional SVM techniques are used to solve problems with binary solutions. Their modified SVM is able to perform multiclass classification, but still relies on initial binary classification with an incorporated one-against-one strategy to train the model based on probabilities.
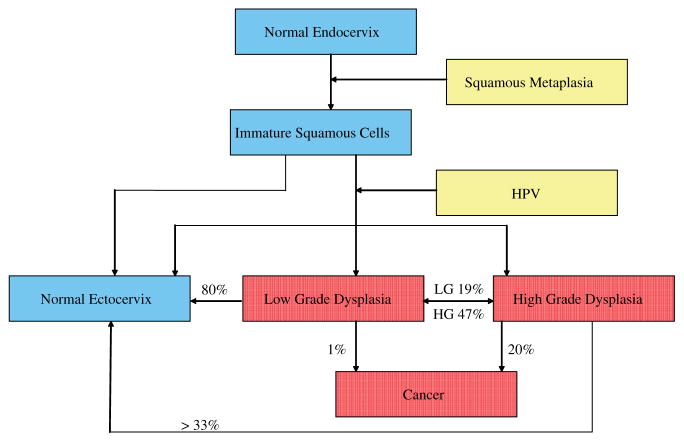
Here we present a multiclass approach algorithm based on novel nonlinear statistical methods: maximum representation and discrimination feature (MRDF) combined with sparse multinomial logistic regression (SMLR). We will demonstrate this multiclass method in the case of cervical dysplasia. Cervical dysplasia is a problem in both the United States and throughout the world. Cervical cancer is the second most common malignancy among women worldwide, with more than 490 000 cases diagnosed and 274 000 deaths each year.[19] In the United States alone, it is estimated that 3870 deaths will occur from this disease in 2008 and 11 070 new cases of invasive cervical cancer will be diagnosed.[20] The mortality rate in the US has been greatly reduced owing to effective screening using the Pap smear and effective treatment of precancers (dysplasia).[21] Due to the complicated manner in which the disease progresses and regresses (shown in Fig. 1), diagnosing the correct grade and progress of cervical dysplasia is very important in treating the disease. Cervical dysplasia is usually classified as one of two groups: (1) low-grade dysplasia which includes human papillomavirus (HPV) and cervical squamous intraepithelial neoplasia 1 (CIN1) and (2) high-grade dysplasia which includes CIN2, CIN3, and carcinoma in situ (CIS).
Figure 1.
A schematic of the progression of normal endocervix cells after squamous metaplasia begins. The cells either transform into normal ectocervix or if infected with HPV may become dysplastic. This figure is available in colour online at www.interscience.wiley.com/journal/jrs.
The progression of the disease is shown in Fig. 1; typically, low-grade dysplasia is followed but not treated since approximately 80% of low-grade dysplasia regresses without treatment and less than 1% will develop into cancer.[22] Conversely, 20% of high-grade dysplasia will develop into cancer and only one-third will regress to a normal state without treatment.[20,22] Therefore, an algorithm than can differentiate cervical tissue into at least three categories is essential: (1) benign cervix, normal, metaplasia and inflammation, (2) low grade, and (3) high grade. Metaplasia is often misclassified as dysplastic and therefore an additional category that classifies metaplasia could be beneficial.
In this paper, we demonstrate that by combining Raman spectroscopy data with a more sophisticated statistical method for classification will lead to an enhanced real-time diagnostic tool for cervical dysplasia. First, we will show previous data analyzed with old algorithms. Then, we will establish our new statistical method and use it on the data to show an improvement in specificity and sensitivity. Finally, we will show that we can match or improve classification by Raman versus colposcopy.
Methods
Data collection and instrumentation
A total of 90 patients participated in this study. Measurements were taken from either a procedure that removed diseased cervical tissue or a hysterectomy. The same procedure was followed for data collection regardless of the procedure being performed.
Thirty-three patients undergoing a colposcopy-guided biopsy or loop electrosurgical excision procedure (LEEP) were recruited to participate in the study as approved by the Vanderbilt and Copernicus Group Institutional Review Boards (IRBs). Informed consent was obtained from each patient prior to the procedure. The cervix was exposed and visually examined by the doctor. Acetic acid was applied to the cervix to turn abnormal areas white, followed by an application of iodine to clean the tissue and reveal the location of squamous epithelium. Any abnormal tissue was removed and histopathology was performed. Raman spectra were acquired after the application of acetic acid but before the application of iodine and the removal of tissue. Spectra were measured from each visually abnormal area (one to six measurements) and one visually normal area. The patient’s age, date of last period, abnormal Pap smear result and menopausal status were noted upon chart review.
Additionally, 33 patients undergoing hysterectomy were recruited to participate in the study as approved by Vanderbilt IRB. Informed consent was obtained from each patient prior to the procedure. The cervix was then exposed and visually examined by the doctor. Acetic acid was applied to the cervix to keep the procedure similar to that performed in dysplasia patients. If the cervix was visually normal, spectra were measured from multiple normal areas of tissue. Measured areas were marked, the hysterectomy then proceeded as required and the removed tissue was subjected to histopathology.
Raman spectra were acquired using a portable Raman spectroscopy system consisting of a 785 nm diode laser (Process Instruments, Inc., Salt Lake City, UT), 7 (300 μm) around 1 (400 μm) beam-steered fiber optic probe (Visionex Inc.), imaging spectrograph (Kaiser Optical Systems, Inc., Ann Arbor, MI) and back-illuminated, deep-depletion, charge coupled device (CCD) camera (Princeton Instruments, Princeton, NJ), all controlled with a laptop computer. For this study, the fiber optic probe delivered 80 mW of incident light onto the tissue and collected the scattered light for 5 s. In all cases, the overhead fluorescent lights and colposcope light were turned off during the measurements. Any luminescent lights were left on but turned away from the measurement site.
Data processing
The wavenumber axis was calibrated using a neon–argon lamp, acetaminophen and naphthalene standards each day. The signal from the Raman spectrum was binned along the vertical axis to create a single spectrum per measurement site. Prior to any signal processing, the spectrum was truncated to include only the region from about 990 to 1850 cm−1 to eliminate the Raman peaks due to the silica present in the fiber optic probe. The spectrum was then binned along the wavenumber axis in 3.5 cm−1 intervals and noise-smoothed with a second-order Savitzky–Golay filter. Additionally, the fluorescence background was removed using an automated, modified polynomial fitting method that utilizes a fifth degree polynomial to fit the fluorescence baseline.[23] Once noise smoothing and fluorescence subtraction were done, the spectra were normalized to their mean spectral intensity across all Raman bands and were used for subsequent data analysis.
Statistical analysis
To compare and contrast the binary versus multinomial class techniques, two different algorithms have been developed to classify cervical data. The first is a binary algorithm that is based on peak ratios and logistic regression. The second is a multiclass probabilistic algorithm that is based on machine support vectors and nonlinear logistic regression. Both algorithms are described in detail below.
Statistical analysis – binary
The first step in using Raman spectra is to develop a basic algorithm to discriminate between abnormal and normal tissues. First, the mean and standard deviation at each wavenumber of the spectra within each pathology group was calculated to characterize the overall spectral trends for each group. A Student’s t-test was performed at each wavenumber between individual pairs of pathology groups to identify regions of spectral distinction between two different pathologies. Any major peak that showed statistical differences at the level of p < 0.01 between normal ectocervix spectra and high-grade dysplasia spectra was chosen as an input for the algorithm. Thus, the inputs to the algorithm are the normalized intensity values at 1006, 1055, 1244, 1305, 1324, 1450, 1550, and 1657 cm−1. The classification model was constructed to automatically classify spectra into one of two categories (high-grade dysplasia or benign cervix) using a two-tiered logistic regression model.[11] The first algorithm was developed to distinguish normal pathology from all other pathologies (metaplasia, high-grade dysplasia), and the second algorithm discriminated high-grade dysplasia from other pathologies (metaplasia).
The first algorithm was trained using a training set to classify a spectrum as either normal ectocervix (score = 0) or high-grade dysplasia (score = 1); the algorithm was then tested using a separate validation set. The training and validation sets were randomly generated by dividing the normal ectocervix and high-grade dysplasia data sets into a training set (two-thirds of the patients) and a validation set (one-third of the patients). The training and validation sets were divided by patients, not by individual spectra, such that all spectra from one patient were either in the training set or the validation set, but not both.
The algorithm then outputs a score, which represents the likelihood that the input data represents high-grade dysplasia. Data from squamous metaplasia were also included as part of the validation set for the model even though no data from this category were included in the training set to examine the possibility that a single algorithm could discriminate all spectra of benign pathology from dysplasia spectra (See Section on Discussion). Since the specificity of this single-algorithm model was less than satisfactory primarily because of misclassifications of squamous metaplasia spectra, a second logistic regression algorithm was developed to separate high-grade dysplasia from squamous metaplasia to increase the specificity of the overall model.
Any spectra from the test set with a score > 0.5 from the first algorithm formed the test set into the second algorithm (thus there were eight high-grade dysplasia, eight squamous metaplasia, and four normal ectocervix spectra). The training set for the second algorithm was formed using only the high-grade dysplasia spectra (29 spectra, score = 1) and squamous metaplasia (29 spectra, score = 0) spectra, as there were not enough spectra to create separate training and validation sets. The same Raman bands from the first algorithm were also used as inputs in the second algorithm, but the output was a value (score) that represents the probability that the spectra were measured from an area of high-grade dysplasia when compared with squamous metaplasia. While the data did classify well using these algorithms, it was clear that we were losing some information in the Raman spectra by only looking at binary classification.
Statistical analysis – multiclass
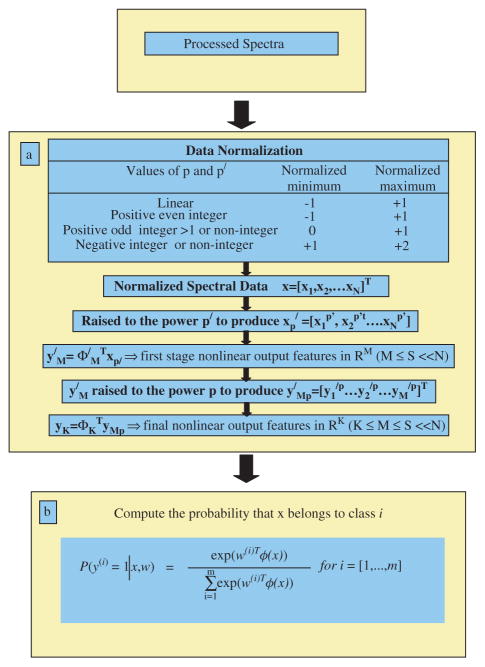
MRDF combined with SMLR was used to develop a multiclass diagnostic algorithm.[24] This algorithm is a two-step process: (1) extraction of diagnostic features from spectra using nonlinear MRDF and (2) classification based on these nonlinear features into corresponding tissue categories using SMLR. Figure 2 shows a flow chart of this algorithm.
Figure 2.
Flow chart of the multiclass discrimination algorithm. This figure is available in colour online at www.interscience.wiley.com/journal/jrs.
MRDF is a method of feature extraction; it maximally extracts the diagnostic information otherwise hidden in a set of measured spectral data by reducing its dimensionality through a set of mathematical transforms. Given a set of input data comprised of spectra from different classes with a given dimensionality, nonlinear MRDF determines a set of nonlinear transformations of the input data that optimally discriminates between the different classes in a reduced dimensionality space. It invokes nonlinear transforms (restricted order polynomial mappings of the input data) in two successive stages. In the first stage, the input spectral data x = [x1, x2, … xN]T (intensities corresponding to Raman shifts of the spectra) from each tissue type are raised to the power p′ to produce the associated nonlinear input vectors . These vectors are then subject to a transform such that y′M = ΦM′T xp′ and are the first-stage output features in the nonlinear space of reduced dimension M ≪ N. In the second stage, the reduced M-dimensional output features y′M for each tissue type are further transformed nonlinearly to the power p to produce higher-order features y′MP = [y1′p, y2′p, …, yM′p], and a second transform ΦK is computed so as to yield the final output features in the nonlinear space of dimension K (K ≤ M).[25]
SMLR is a method of supervised classification. It is a probabilistic multiclass model based on sparse Bayesian machine-learning framework of statistical pattern recognition. The central idea of SMLR is to separate a set of labeled input data into its constituent classes by predicting the posterior probabilities of their class membership. It computes the posterior probabilities (from the equations shown in Fig. 2(b)) using a multinomial logistic regression model and constructs a decision boundary that separates the data into its constituent classes on the basis of the computed posterior probabilities following Bayes’ rule (i.e. a class is assigned to a data for which its posterior probability is the highest).[25] Traditional statistical methods have focused primarily on using binary classification. However, this method is limited when looking at complicated diseases, like stages of cancer. A novel, multiclass method is more suitable for such applications.
Results
Using both algorithms, a total of 29 high-grade dysplasia (from 19 patients), six low-grade dysplasia (from five patients), 29 squamous metaplasia (from 20 patients) and 100 normal ectocervix (from 47 patients) were classified to compare the sensitivity and specificity of the two algorithms.
First, the resulting spectra were correlated with the corresponding histopathologic diagnosis to characterize the differences between various diagnostic categories. Figure 3 shows the mean spectra for the full data set for each of the different categories. Peaks were found at 1006, 1058, 1086, 1244, 1270, 1324, 1450, 1550, and 1655 cm−1 in most spectra. Although the peak shapes and locations are consistent across all pathology classifications, there are small but significant differences in peak intensities between the different pathology categories. Several spectral regions show statistically significant differences in comparing precancer to the normal ectocervix. For example, in the low-grade spectra the 1324 cm−1 peak increases as compared to that in normal ectocervix, similar to the high-grade precancer/normal ectocervix spectral comparison. But, the intensity of the 1272 and 1450 cm−1 peaks in low-grade precancer spectra seems to remain similar to that seen in normal ectocervix, unlike in the high-grade precancer spectra. These differences are very subtle, so there is a need for statistical approaches.
Figure 3.
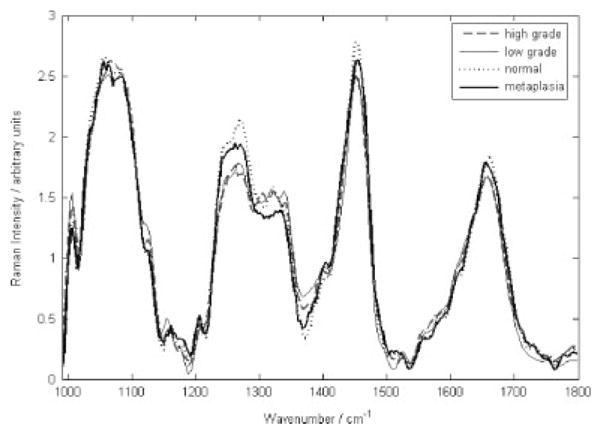
Average Raman spectra for normal ectocervix, low-grade dysplasia, high-grade dysplasia and metaplasia.
A binary algorithm based on LDA was applied; using the output of this algorithm, we were able to distinguish between high-grade precancer and benign areas of the cervix (normal ectocervix and squamous metaplasia) with 89% of and 88% specificity. Due to insufficient numbers, we did not include low-grade data in this analysis. The limitation of this particular discrimination algorithm is that it is binary that does not allow for the multiple classes that the tissue could belong to. In addition, the inputs for the algorithm were selected as normalized peak intensities, thereby throwing away other potentially useful information present in the spectra. To address these limitations, a second discrimination algorithm was developed, which is based on novel nonlinear statistical methods: MRDF combined with SMLR.
In order to determine the effectiveness of a new discrimination algorithm as compared to the one used previously, we used the same 66 patients as before and classified this data using an algorithm based on MRDF and SMLR. The diagnostic algorithm using this method is capable of simultaneously discriminating complete in vivo Raman spectra acquired from the human cervix, into the different pathological categories. Unbiased performance estimates were obtained using leave-one-patient-out cross-validation. The results indicate that Raman spectroscopy can distinguish high-grade precancer from normal ectocervix and squamous metaplasia with a higher sensitivity and specificity than the binary algorithm (sensitivity 92% and specificity 96%) as shown in Table 1. High-grade spectra were classified correctly 95% of the time, and only one was misclassified as normal. Low-grade data was never classified as high grade and was misclassified as normal 29% of the time. Very few low-grade spectra were included in the analysis but this method now has a similar sensitivity to and much higher specificity than colposcopy-guided biopsy in expert hands (sensitivity of 87% and specificity of 72%).
Table 1.
Classification using algorithm based on MRDF and SMLR
| Classification | Pathology |
|||
|---|---|---|---|---|
| Raman algorithm | High grade |
Low grade |
Metaplasia | Normal |
| High grade | 20 | 0 | 0 | 0 |
| Low grade | 0 | 5 | 0 | 0 |
| Metaplasia | 0 | 0 | 20 | 3 |
| Normal | 1 | 2 | 1 | 66 |
In order to ensure that our algorithm applies also to low-grade cervical precancers, we added 27 patients, which increased the number of samples within each category with an emphasis on low-grade lesions. Raman spectra from a total of 93 patients were analyzed using the algorithm based on MRDF and SMLR with leave-one-patient-out cross-validation. The performance of the model is reported as percentages that were classified correctly into each category in Table 2. The result of SMLR is a set of predictive values (or posterior probabilities) and these were obtained by using leave-one-patient-out cross-validation. Figure 4 plots the predictive posterior probabilities of being classified as high-grade dysplasia, low-grade dysplasia, squamous metaplasia and normal ectocervix for the normalized Raman spectra of the corresponding cervical tissue sites. Even though emphasis was placed on collecting low-grade spectra, only 22 low-grade spectra from 19 patients are represented in this study, which may be a reason for the higher misclassification rate. Even though we had some misclassifications with the low-grade data, overall, more than 88% of the data from this set classified correctly. More clinical data from low-grade cervix is needed to determine the full capability of this algorithm to differentiate between normal and low grade.
Table 2.
Classification using algorithm based on MRDF and SMLR with additional low-grade samples
| Classification | Pathology |
|||
|---|---|---|---|---|
| Raman algorithm | High grade |
Low grade |
Metaplasia | Normal |
| High grade | 24 | 1 | 1 | 3 |
| Low grade | 0 | 18 | 0 | 3 |
| Metaplasia | 0 | 0 | 19 | 10 |
| Normal | 5 | 3 | 10 | 208 |
Figure 4.
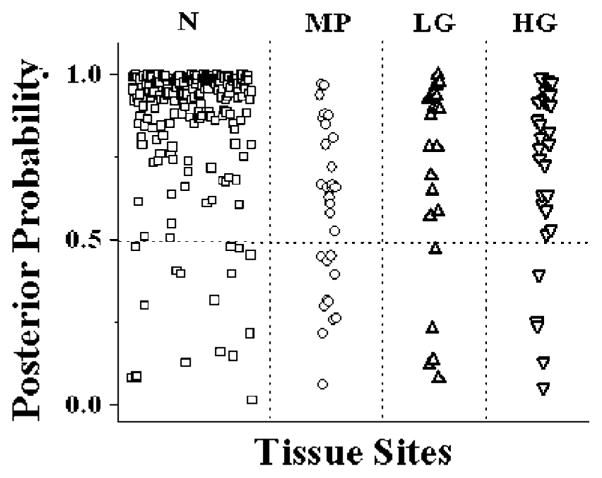
Posterior probabilities of being classified as normal ectocervix, low-grade dysplasia, high-grade dysplasia and metaplasia.
Discussion
Raman spectroscopy has the power to optically identify subtle changes in tissue that can lead to diseases such as cancer. Many statistical methods have been developed to tease out important clinical parts of Raman spectra, allowing them to be correlated to specific pathological conditions. Although binary methods have traditionally been used to classify spectroscopy data, a more sophisticated method that is able to classify multiple classes at the same time is necessary as Raman spectroscopy moves closer to the clinic. Raman spectroscopy combined with a multiclass discrimination algorithm has great potential for biological applications, especially within tissue. This paper demonstrates that when using the multiclass algorithm, we can improve the sensitivity from 92% to 98% and the specificity from 81% to 96%. Both methods (binary and multiclass) are an improvement over the current method of diagnosis – colposcopy-guided biopsy in expert hands has a sensitivity of 87% and a specificity of 72% – showing the capabilities of Raman spectroscopy.
One major concern when taking in vivo measurements is that a certain sample volume may have two different pathological classifications. The sample could be 75% metaplasia and 25% high-grade dysplasia. Therefore, a binary algorithm that separates between high grade and metaplasia would classify this sample as metaplasia since the sample is dominated by metaplasia. But with the multiclass algorithm, we may be able to show that this sample is mostly metaplasia but has spectral contributions from the high-grade dysplastic tissue. This feature would prevent misdiagnosing the tissue as normal instead of metaplasia with some high-grade regions. This is an important benefit of using the multiclass algorithm, even though the implementation of it is more complicated than a binary algorithm.
The goal of this present study was to develop a multivariate statistical algorithm capable of simultaneously classifying Raman spectral data acquired in vivo from human cervical tissues into high-grade dysplasia, low-grade dysplasia, squamous metaplasia, and normal ectocervix. The first task for the development of such an algorithm is the extraction of diagnostically relevant features from the observed spectra by reducing the dimensionality of the measured spectral variables. For good classification performance, the extracted features should contain sufficient class-discriminatory information. Most of the published reports on spectroscopic diagnostic algorithms have reported using standard linear techniques like PCA and FDA to extract diagnostic features from the measured spectra of tissue.[26 – 29] Although these linear techniques have the advantage of providing closed-form solutions, which make them relatively easy to implement, they are limited because they extract information only from the second-order correlation in the data and ignore higher-order correlations that could be useful for improved discrimination. Use of nonlinear techniques is required for this purpose.[30] There are several nonlinear methods that exist for feature extraction in pattern recognition literature; most of them are iterative and often need a priori selection of a number of parameters associated with the learning or the optimization technique used.[30] They are also limited by problems with convergence.[30] One major advantage of the nonlinear MRDF technique is that unlike the iterative nature of other nonlinear feature extraction algorithms, it provides a closed-form expression of the nonlinear transform for maximum discrimination.[31,32] Another advantage of using this method to classify spectral data is that it has the ability to separate classes that are not linearly separable. As spectral data tends to be nonsymmetric, using MRDF can lead to spectral separations with higher accuracy.
This increased sensitivity makes Raman spectroscopy superior to other types of spectroscopy and therefore ideal for detection of small changes in early dysplasia. Although the number of low-grade spectra in this study is small, we are capable of distinguishing the low-grade spectra 74% of the time. Once the diagnostic features are extracted from the measured spectral data, the final task of the algorithm is to classify these extracted features into respective tissue categories. The major advantage of using the SMLR approach for classification is that since it is based on a Bayesian framework, it is able to predict the posterior probability of class membership of the investigated tissue site. This idea is demonstrated in Fig. 4, where the predicted posterior probabilities of the different cervical tissue sites classified as high-grade dysplasia, low-grade dysplasia, squamous metaplasia and normal ectocervix are plotted. One may also note that most of the dysplastic sites have been classified with a posterior probability of greater than 80% into the corresponding tissue categories. The probabilistic approach can offer an important advantage by making it possible to further interrogate these sites, especially when the goal is to correctly identify all abnormal sites for accurate screening of cervical dysplasia. An additional advantage of the new algorithm is that it provides the posterior probability of samples belonging to the different diagnostic categories. We expect this to be extremely useful in a clinical setting because health providers could recheck samples having lower posterior probabilities of belonging to one category by using a traditional biopsy method.
Other groups have suggested that optical technologies are capable of distinguishing high-grade dysplasia or cancer from normal cervix, but have had little success at differentiating low-grade dysplasia from normal or high grade. In this paper, we have demonstrated that by using MRDF with SMLR, Raman spectroscopy is capable of picking out some of the subtle changes that occur during the early stages of dysplasia. Unfortunately, we were unable to collect a large number of low-grade dysplasia data, and future studies need to be focused on low-grade data collection. Also, these algorithms need to be usable in real-time clinical settings, which will also be developed in the future.
Conclusions
The use of a probability-based robust diagnostic algorithm capable of simultaneously discriminating in vivo Raman spectra acquired from human cervical tissues into various pathological categories improves performance compared to the more traditional methods by allowing for multiclass discrimination. The results indicate that Raman spectroscopy in conjunction with the diagnostic algorithm can distinguish dysplasia from normal ectocervix (including metaplasia) with a classification accuracy of 95%. One additional advantage of the algorithm developed in this study is that it provides the posterior probability of samples belonging to the different diagnostic categories. This is expected to be extremely useful in a clinical setting, because clinicians could recheck any sample having a lower posterior probability of belonging to one category with the conventional biopsy method. These discrimination techniques are not only applicable to the cervix but also could be used in the spectral data analysis of tissue types that require a multiclass diagnosis, like GI and skin cancers.
Acknowledgments
The authors would like to acknowledge the financial support of the NCI/NIH (R01-CA95405). They would also like to thank the doctors and staff at Vanderbilt University and at Tri-State Women’s Health for all their help.
References
- 1.Mahadevan-Jansen A, Richards-Kortum R. J Biomed Opt. 1996;1:31. doi: 10.1117/12.227815. [DOI] [PubMed] [Google Scholar]
- 2.Feld MS, Manoharan R, Salenius J, Orenstein-Carndona J, Roemer TJ, Brennan JF, III, Dasari RR, Wang Y. Proc SPIE. 1995;2388:99. [Google Scholar]
- 3.Hanlon EB, Manoharan R, Koo TW, Shafer KE, Motz JT, Fitzmaurice M, Kramer JR, Itzkan I, Dasari RR, Feld MS. Phys Med Biol. 2000;45:R1. doi: 10.1088/0031-9155/45/2/201. [DOI] [PubMed] [Google Scholar]
- 4.Utzinger U, Richards-Kortum RR. J Biomed Opt. 2003;8:121. doi: 10.1117/1.1528207. [DOI] [PubMed] [Google Scholar]
- 5.Chowdary MVP, Kumar KK, Kurien J, Mathew S, Krishna CM. Biopolymers. 2006;83:556. doi: 10.1002/bip.20586. [DOI] [PubMed] [Google Scholar]
- 6.Hata TR, Scholz TA, Ermakov IV, McClane RW, Khachik F, Gellermann W, Pershing LK. J Invest Dermatol. 2000;115:441. doi: 10.1046/j.1523-1747.2000.00060.x. [DOI] [PubMed] [Google Scholar]
- 7.Shim MG, Song L, Marcon NE, Wilson BC. Photochem Photobiol. 2000;72:146. doi: 10.1562/0031-8655(2000)072<0146:IVNIRS>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 8.Oliveira AP, Bitar RA, Silveira L, Zangaro RA, Martin AA. Photomed Laser Surg. 2006;24:348. doi: 10.1089/pho.2006.24.348. [DOI] [PubMed] [Google Scholar]
- 9.Mahadevan-Jansen A, Mitchell WF, Ramanujam N, Utzinger U, Richards-Kortum R. Photochem Photobiol. 1998;68:427. [PubMed] [Google Scholar]
- 10.Utzinger U, Heintzelman DL, Mahadevan-Jansen A, Malpica A, Follen M, Richards-Kortum R. Appl Spectrosc. 2001;55:955. [Google Scholar]
- 11.Viehoever Robichaux A, Kanter E, Shappell H, Billheimer D, Jones H, Mahadevan-Jansen A. Appl Spectrosc. 2007;61:986. doi: 10.1366/000370207781746053. [DOI] [PubMed] [Google Scholar]
- 12.Stone N, Stavroulaki P, Kendall C, Birchall M, Barr H. Laryngoscope. 2000;110:1756. doi: 10.1097/00005537-200010000-00037. [DOI] [PubMed] [Google Scholar]
- 13.Mahadevan-Jansen A, Mitchell MF, Ramanujam N, Malpica A, Thomsen S, Utzinger U, Richards-Kortum R. Photochem Photobiol. 1998;68:123. doi: 10.1562/0031-8655(1998)068<0123:nirsfv>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 14.Kendall C, Stone N, Shepherd N, Geboes K, Warren B, Bennett R, Barr H. J Pathol. 2003;200:602. doi: 10.1002/path.1376. [DOI] [PubMed] [Google Scholar]
- 15.Eisen MB, Spellman PT, Brown PO, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chai BB, Huang T, Zhuang XH, Zhao YX, Sklansky J. Pattern Recognit. 1996;29:1905. [Google Scholar]
- 17.Zhou G, Chen Y, Wang Z, Song H. Appl Opt. 1999;38:4281. doi: 10.1364/ao.38.004281. [DOI] [PubMed] [Google Scholar]
- 18.Widjaja E, Zheng W, Huang Z. Int J Oncol. 2008;32:653. [PubMed] [Google Scholar]
- 19.Parham GP, Sahasrabuddhe VV, Mwanahamuntu MH, Shepherd BE, Hicks ML, Stringer EM, Vermund SH. Gynecol Oncol. 2006;103:1017. doi: 10.1016/j.ygyno.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ACS. Cervical Cancer Resource Center: American Cancer Society. 2008 [Google Scholar]
- 21.Myers ER, McCrory DC, Subramanian S, McCall N, Nanda K, Datta S, Matchar DB. Obstet Gynecol. 2000;96:645. doi: 10.1016/s0029-7844(00)00979-0. [DOI] [PubMed] [Google Scholar]
- 22.AMA. Cervical Cancer: American Medical Association. 1999 [Google Scholar]
- 23.Lieber CA, Mahadevan-Jansen A. Appl Spectrosc. 2003;57:1363. doi: 10.1366/000370203322554518. [DOI] [PubMed] [Google Scholar]
- 24.Majumder SK, Gebhart S, Johnson MD, Thompson R, Lin WC, Mahadevan-Jansen A. Appl Spectrosc. 2007;61:548. doi: 10.1366/000370207780807704. [DOI] [PubMed] [Google Scholar]
- 25.Majumder SK, Kanter E, Viehoever AR, Jones H, Mahadevan-Jansen A. Proc SPIE. 2007;6430:64300 Q. [Google Scholar]
- 26.Ramanujam N, Mitchell MF, Mahadevan-Jansen A, Thomsen SL, Staerkel G, Malpica A, Wright T, Atkinson N, Richards-Kortum R. Photochem Photobiol. 1996;64:720. doi: 10.1111/j.1751-1097.1996.tb03130.x. [DOI] [PubMed] [Google Scholar]
- 27.Majumder SK, Mohanty SK, Ghosh N, Gupta PK, Jain DK, Khan F. Curr Sci. 2000;79:1089. [Google Scholar]
- 28.Wang CY, Chen CT, Chiang CP, Young ST, Chow SN, Chiang HK. Photochem Photobiol. 1999;69:471. [PubMed] [Google Scholar]
- 29.Atkinson EN, Mitchell MF, Ramanujam N, Richards-Kortum R. J Cell Biochem Suppl. 1995;23:125. doi: 10.1002/jcb.240590916. [DOI] [PubMed] [Google Scholar]
- 30.Jain AK, Duin RPW, Mao JC. IEEE Trans Pattern Anal Mach Intell. 2000;22:4. [Google Scholar]
- 31.Talukder A. PhD Thesis, Nonlinear Feature Extraction for Pattern Recognition Applications. Carnegie Mellon University; PA, USA: 1999. [Google Scholar]
- 32.Talukder A, Casasent D. Opt Eng. 1998;37:904. [Google Scholar]