Abstract
A milestone of molecular medicine is the identification of dystrophin gene mutation as the cause of Duchenne muscular dystrophy (DMD). Over the last 2 decades, major advances in dystrophin biology and gene delivery technology have created an opportunity to treat DMD with gene therapy. Remarkable success has been achieved in treating dystrophic mice. Several gene therapy strategies, including plasmid transfer, exon skipping, and adeno-associated virus-mediated microdystrophin therapy, have entered clinical trials. However, therapeutic benefit has not been realized in DMD patients. Bridging the gap between mice and humans is no doubt the most pressing issue facing DMD gene therapy now. In contrast to mice, dystrophin-deficient dogs are genetically and phenotypically similar to human patients. Preliminary gene therapy studies in the canine model may offer critical insights that cannot be obtained from murine studies. It is clear that the canine DMD model may represent an important link between mice and humans. Unfortunately, our current knowledge of dystrophic dogs is limited, and the full picture of disease progression remains to be clearly defined. We also lack rigorous outcome measures (such as in situ force measurement) to monitor therapeutic efficacy in dystrophic dogs. Undoubtedly, maintaining a dystrophic dog colony is technically demanding, and the cost of dog studies cannot be underestimated. A carefully coordinated effort from the entire DMD community is needed to make the best use of the precious dog resource. Successful DMD gene therapy may depend on valid translational studies in dystrophin-deficient dogs.
Keywords: Duchenne muscular dystrophy, gene therapy, dystrophin, adeno-associated virus, exon-skipping, canine model
Introduction
Duchenne muscular dystrophy (DMD) is the most common lethal muscle disease caused by dystrophin gene mutation.1–3 It affects 1–3 boys per 10,000 male birth worldwide.4,5 Patients start to lose their mobility around 2–6 years of age and are often wheelchair bound by their early teenage years. Life expectancy is shortened to one-third to half of normal as a consequence of respiratory insufficiency and/or heart failure. DMD remains an incurable disease today.
In 1987, the coding sequence of the dystrophin gene was discovered and deciphered.3 Dystrophin is a subcellular cytoskeletal protein. It scaffolds a series of transmembrane and cytosolic proteins (including dystroglycans, sarcoglycans, sarcospan, neuronal nitric oxide synthase (nNOS), syntrophin, and dystrobrevin) into a dystrophin-associated glycoprotein complex (DGC). The DGC plays the important mechanical and signaling roles in muscle cells. The cloning of the dystrophin gene led to the recognition that the loss of dystrophin expression underlies clinical manifestations of DMD.1 The discovery of the dystrophin gene has revolutionized DMD diagnosis and created hope for a cure with gene therapy. The reasoning is straightforward. If a method can be invented to deliver a functional dystrophin gene into the diseased muscle cells, the problem will be solved. Transgenic studies confirmed this hypothesis, and, it was also found that dystrophin overexpression was not toxic.6,7 Meanwhile, exciting advances were made in dystrophin biology and muscle gene transfer,8–15 heralding a new era of muscle gene therapy. To many, a possible cure of DMD by gene therapy appeared just around the corner.
Genes that can be used to treat DMD
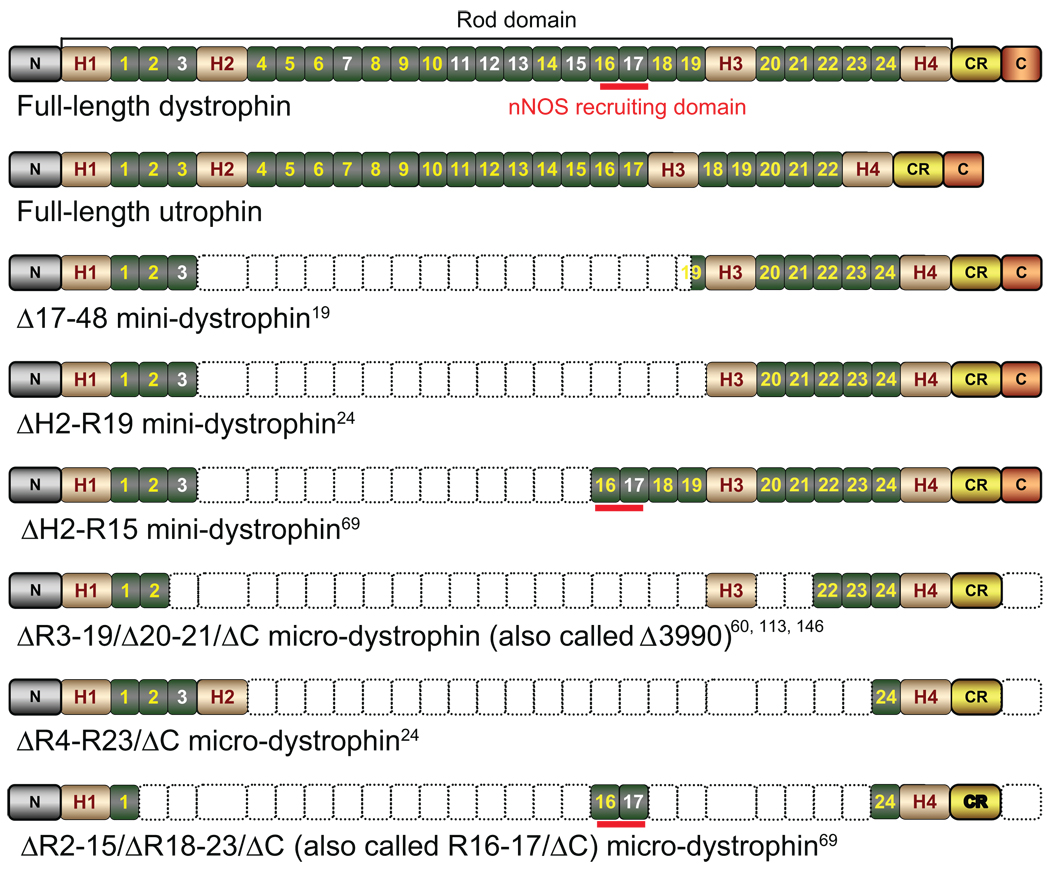
The discovery of the dystrophin gene has also raised a seemingly insurmountable challenges to the nascent field of DMD gene therapy. The gene itself is huge. The 2.5-mega base (mb) gene contains 79 exons, and it transcribes into a 14-kb cDNA.2 A vehicle that is capable of carrying the full-length dystrophin gene is beyond the reach of the current technology. A few viral vectors that may have the packaging capacity for the full-length dystrophin coding sequence (such as gutted adenovirus and herpes virus) are very inefficient in transducing whole-body muscle cells. The hurdle of gene size was partially overcome by genotype–phenotype analysis in patient populations and by spectacular case reports of individual patients.16–19 These studies suggest that deletion of a fairly large region of the dystrophin gene may not cause significant clinical consequences as long as the remaining parts are in frame and the most critical regions are retained.20–22 This reading frame theory constitutes the foundation of current DMD gene therapy. Briefly, two distinctive roads are taken. One approach attempts to generate a synthetic, minimized dystrophin gene that expresses these key components (Figure 1).23 Examples of these synthetic genes include the highly functional minidystrophin genes (6–8 kb) and the highly abbreviated microdystrophin genes (<4 kb).24 The other approach is to restore the reading frame using antisense oligonucleotide (AON)-mediated exon skipping.25
Figure 1.
Schematic illustration of full-length dystrophin, utrophin, and representative minidystrophin and microdystrophin.
Abbreviations: N, the N-terminal domain of the dystrophin protein; H1–4, hinges 1–4 in the rod domain of the dystrophin protein; numeric numbers, spectrin-like repeats in the dystrophin rod domain. Positively charged repeats are in white color. Repeats 11–17 represent the second actin-binding domain. CR, the cysteine-rich domain in the dystrophin protein; C, the C-terminal domain of the dystrophin protein; nNOS, neuronal nitric oxide synthase. Empty boxes denote the regions that are deleted in each respective minidystrophin and microdystrophin construct. Among three microgenes listed here, the ΔR3–19/Δ20–21/ΔC microdystrophin has been tested in affected dogs and human patients. However, these trials have failed to demonstrate a therapeutic efficacy. The hinge 2 region in ΔR4–R23/ΔC microdystrophin has been shown to compromise function. The potentially deleterious hinges 2 and 3 are removed in the ΔR2–15/ΔR18–23/ΔC microgene. Further, this microgene carries the nNOS localization domain.
One obvious problem associated with dystrophin replacement is the potential immune rejection of the newly introduced protein. The use of an existing gene may minimize this risk. Although intensive research is ongoing to reestablish dystrophin expression, creative means have also been developed using the alternative self-genes.26
Soon after the discovery of the dystrophin gene, the utrophin gene was identified.27–29 Utrophin shares structural and functional similarity to dystrophin (Figure 1). Although utrophin cannot carry out all the functions of dystrophin, it still provides substantial benefit to dystrophic muscle.30,31 In addition to utrophin, increased expression of several functionally relevant proteins (such as laminin, sarcoglycan, sarcospan, integrin, and nNOS) has also been shown to reduce muscular dystrophy in the mouse model.32–36
Another promising field for alternative gene therapy is the identification of genetic modifiers. Several highly promising candidate genes have emerged. Upregulation of follistatin, insulin-like growth factor 1, ADAM12, cytidine monophosphate-sialic acid hydroxylase (CMAH), sarcoplasmic reticulum calcium ATPase, or downregulation of myostatin, osteopontin, cyclophilin D, latent transforming growth factor-β binding protein 4, vascular endothelial growth factor receptor 1, and histone deacetylase have been shown to reduce dystrophinopathy in animal models.37–48 Targeting these alternative genes will likely complement dystrophin gene replacement/repair therapies.
We cured a DMD mouse
Many dystrophin-deficient mice have been generated, such as naturally occurring mdx; chemically induced mdx2cv, mdx3cv, mdx4cv, and mdx5cv; and exon 52 knockout mdx (mdx52) mice (Figure 2).49–51 Unlike human patients, dystrophin-null mice exhibit very mild symptoms until they get very old (Figure 2).52,53 To create a phenotypic model that more closely mimics human disease, a great variety of double knockout (dKO) mice were made. These include utrophin/ dystrophin dKO, myoD/dystrophin dKO, integrin/dystrophin dKO, δ-sarcoglycan/dystrophin dKO, CMAH mdx, and mdx/ mTR mice (Figure 2).32,48,54–59 Although there are genetic and/ or phenotypic differences between mice and humans, these mouse models, nevertheless, provide a great opportunity to test experimental DMD gene therapy in a live animal.
Figure 2.
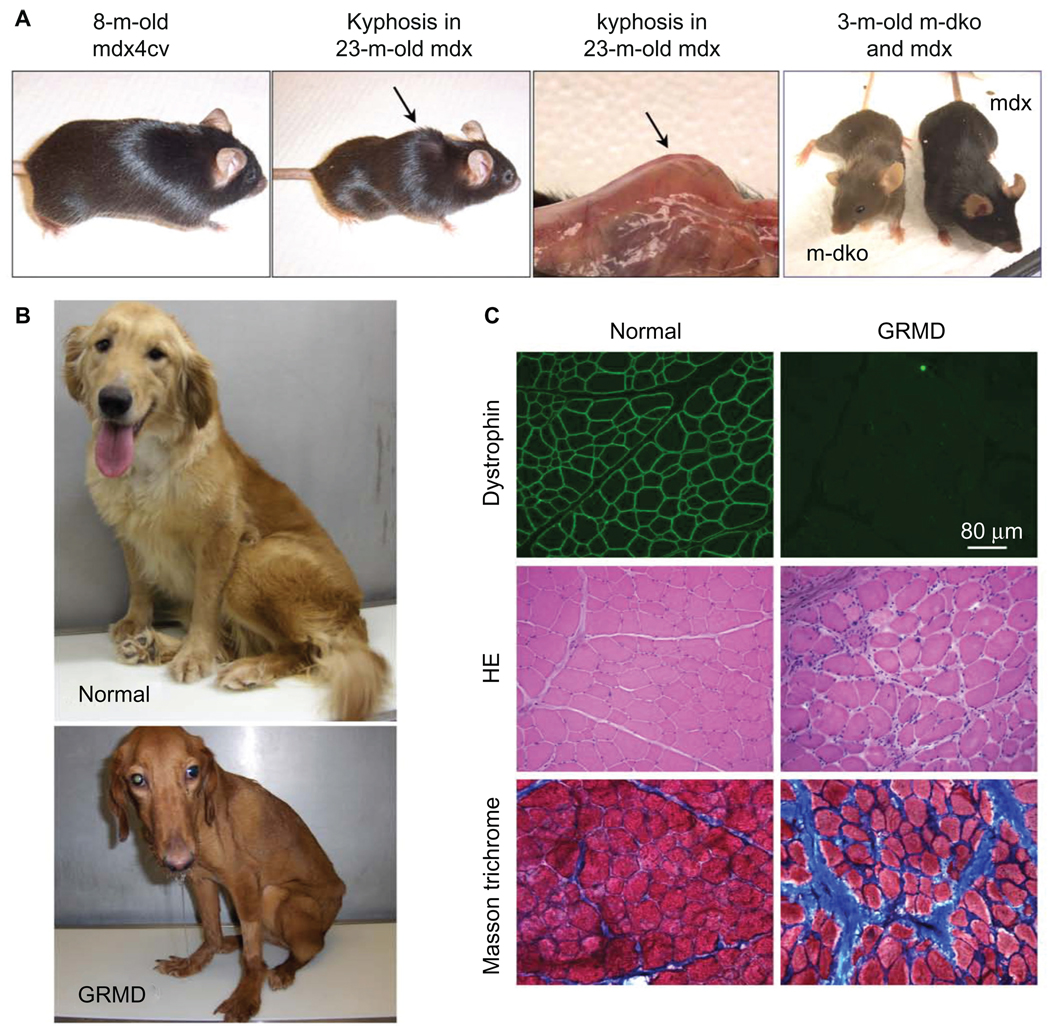
Mouse and dog models of DMD. A) Representative pictures of an 8-month-old dystrophin-deficient mdx4cv mouse (left panel), a 23-month-old mdx mouse (middle two panels), and a 3-month-old myoD/dystrophin double knockout (m-dKO) mouse and a 3-month-old mdx mouse (right panel). Adult dystrophin-null mice do not display clinical symptoms (left panel). The middle two panels show body emaciation and mouse in aged dystrophin-null mouse. (Arrow points to kyphosis seen in aged mdx mice. The left panel is a full-body view. The right-side panel represents a close view of kyphosis after the skin is removed.) The difference in the genetic background results in different fur color in m-dKO (brown) and mdx (black) mice (right panel). Findings from dKO mice may be biased by their mixed genetic background. B) Representative pictures of a 2-year-old normal golden retriever (top) and an affected GRMD (bottom) dog. The affected dog shows body and limb muscle atrophy, drooling, and ulnocarpal joints (forelimbs) dropped to the ground level. C) Representative muscle immunofluorescence staining and histopathology staining in normal (left panels) and GRMD (right panels) dogs. Dystrophin is detected by an antibody (Dys-1) specific for spectrin-like repeats 6–8 (top row). Muscle pathology is revealed by hematoxylin–eosin (HE) staining (middle row) and Masson’s trichrome staining (bottom row). HE staining shows inflammatory cell infiltration, central nucleation, and variable myofiber size. Masson’s trichrome staining shows fibrosis. Blue color in Masson’s trichrome staining images represents fibrotic tissue.
Abbreviations: DMD, Duchenne muscular dystrophy; GRMD, golden retriever muscular dystrophy.
Early gene therapy studies were performed on a single-limb muscle in mdx mice. The primary end points of these studies were dystrophin expression, DGC restoration, myofber degeneration/regeneration, sarcolemmal integrity, and muscle force. Long-term robust dystrophin expression was first demonstrated using vectors based on adeno-associated virus (AAV).24,60 AAV is a single-stranded DNA virus. In an AAV vector, all the wild-type viral genes are removed, and a therapeutic/marker gene expression cassette serves as the vector genome. The naturally occurring AAV serotypes and the molecularly engineered A AV capsids have offered essentially unlimited options for gene delivery.61,62 The major drawback of AAV is its 5 kb packaging capacity.63–65 Only the massively truncated microgenes can fit into a single AAV vector. Although a microgene only carries ~30% of the dystrophin coding sequence, local AAV microgene injection has ameliorated muscle pathology in mdx mice.24,60 Subsequent studies suggest that AAV microdystrophin vectors also improved muscle force and prevented eccentric contraction-induced injury in dystrophin-deficient mice.66,67
There remain several limitations of the AAV microgene vector. First, it cannot anchor nNOS to the sarcolemma. In normal muscle, dystrophin helps recruit nNOS to the membrane.31,68–70 In DMD, the loss of membrane-associated nNOS results in muscle ischemia.69,71,72 Further, nitrosative stress induced by delocalized nNOS inhibits muscle force generation.70 Second, the microgene cannot fully restore muscle strength to the normal level.24 The larger minigene is needed for better force recovery.24 Third, the configuration of the earlier versions of the microgene may not be ideal.24,60 In this regard, it has been suggested that the inclusion of hinge 2 and/or hinge 3 may compromise the function.73,74 The first issue was addressed recently by identification of dystrophin spectrin-like repeats 16/17 (R16–17) as the nNOS-binding domain.69 Incorporation of this domain in the microgene results in the R16–17/ΔC microgene that normalizes nNOS localization (Figure 1).69, 70 To address the second issue, innovative strategies are needed to expand the AAV packaging capacity. This is achieved using various dual-vector systems, including the trans-splicing, overlapping, and hybrid vectors.75–77 The trans-splicing vectors are based on the head-to-tail concatamerization of the AAV inverted terminal repeats. The overlapping vectors are based on homologous recombination of the transgene. Newly developed hybrid vectors integrate the advantages of these two approaches and may result in the most efficient reconstitution of a split gene.78,79 Promising minidystrophin expression has been achieved with all three dual-vector systems.78,80,81 To address the third issue, investigators have developed newer versions of microdystrophins that do not carry hinges 2 and 3. An example is the ΔR2–15/ΔR18–23/ΔC microgene (Figure 1).
Although local intramuscular gene delivery is important for proof of principle, it cannot meet the need of treating all affected muscles in the body. A cure for DMD requires systemic gene transfer. Whole-body delivery was first demonstrated with AAV serotype 6 in mdx mice by intravascular injection.82 Subsequently, it was reported that several other AAV serotypes (such as AAV-8 and AAV-9) also mediate robust body-wide gene transfer.83–87 Successful systemic dual AAV vector transduction was also achieved.80,88–90 With these great tools in hand, it did not take long to prove that we can effectively treat a dystrophic mouse with AAV microdystrophin vectors.91,92 The histopathology was ameliorated, muscle function improved, and life span prolonged.
Initial studies with exon skipping were performed with 2-O-methylated phosphorothioated (2-OMePS) AON. These studies showed efficient local restoration of dystrophin expression.93,94 More effective exon skipping has been achieved with recently developed phosphorodiamidate morpholino oligomers (PMOs), peptide-tagged PMO, and nonpeptide polymer–tagged PMO.95–99 Collectively, these studies suggest that repeated intravascular or intraperitoneal AON injection is sufficient to ameliorate muscular dystrophy in dystrophin-deficient mice. Additionally, a combination of AAV gene transfer and AON-mediated exon skipping may yield more persistent dystrophin expression.100 In summary, curing a dystrophin-deficient mouse is no longer beyond the reach of the current technology.
The status of DMD clinical trials
A total of three DMD gene therapy approaches have entered clinical trials. These include full-length dystrophin replacement with a plasmid vector, AON-mediated exon skipping, and AAV-mediated microgene therapy.
The first DMD gene therapy clinical trial was performed with a plasmid vector via direct muscle injection.101,102 Three 17- to 21-year-old DMD patients and six 30- to 50-year-old Becker muscular dystrophy (a mild form of DMD) patients received 200–1200 µg dystrophin plasmid in the extensor radialis muscle.102 This phase I study showed low and variable expression at 21 days after injection.103,104 The efficiency is clearly below the therapeutic threshold.102 Unless there is a revolutionary breakthrough in the transduction efficiency, it seems unlikely the DMD patients will benefit from this seemingly simple and straightforward gene transfer technology in the near future.
There is no doubt that exon-skipping trials have yielded unprecedented success in terms of restoring dystrophin expression.105,106 Two trials have been reported. Both trials aimed at restoring the open-reading frame by skipping exon 51. Four 8- to 16-year-old DMD patients received 800 µg (in a total volume of 800 µL) of 2-OMePS AON along a 1.5-cm long line in the tibialis anterior muscle.106 Biopsy at day 28 showed 64%–97% of dystrophin-positive fibers at the intensity of 17%–35% of control. In another trial, two DMD patients (13- and 16-year-old) received 90 µg and five DMD patients (10 to 15 years old) received 900 µg (all in the volume of 900 µL/patient) of PMO AON in the extensor digitorum brevis muscle.105 Minimal expression was detected at the low dose. However, significant dystrophin expression was detected in the high-dose group between 21 and 28 days. Myofibers in the range of 44%–79% showed increased dystrophin expression at the intensity of 22%–32% of healthy muscle. No adverse response was detected in either trial. Currently, both trials have moved to repeated systemic administration.25
Although the results of these initial exon-skipping trials are highly promising, we should be cautious. Dystrophin contains four domains, including the N-terminal, rod, cysteine-rich (CR), and the C-terminal domain (Figure 1). The rod domain can be further divided into 24 spectrin-like repeats and four hinges. The N-terminal domain and a specific region of the rod domain (repeats 11–17) provide two independent binding sites for the cytoskeleton. The CR domain mediates connection to the extracellular matrix. These domains are essential for dystrophin function. A patient with mutations in the CR domain is unlikely to benefit from a treatment that removes the CR domain. By definition, exon skipping only produces an internally truncated, but not necessarily an optimized protein.74 It is quite clear that the function of the dystrophin protein is more complex than previously thought.107 For example, exons 42–45 encode R16–17, the nNOS-binding domain.69 Skipping these exons may compromise nNOS anchoring. As a matter of fact, many patients who carry in-frame deletion in this region are still afflicted by the disease.16,108–111 Perhaps, the biggest challenge for exon skipping is the need to design the patient-specific AON. Due to the difference in the mutation location, one has to use individualized AON to skip specific exon(s) in order to restore the open reading frame. The AON tailored to one type of mutation in one patient may not be applicable to another mutation in a different patient. Since every AON has its unique composition, it will be a challenge for the regulatory authority to approve all AONs based on the success of a specific AON.112
The highly anticipated AAV microgene trial has some interesting revelations.113 Six DMD boys received 0.2 to 1 × 10e11 viral genome particles/kg body weight of an AAV serotype 2.5 ΔR3–19/ΔR20–21/ΔC microdystrophin vector in their biceps (Figure 1). The ubiquitous cytomega-lovirus promoter was used to control microgene expression. Previous studies with similar vectors have yielded a great success in the mouse model.60 Unexpectedly, biopsy at 42 and 90 days showed essentially no microdystrophin expression despite detection of the AAV genome. Further investigation revealed a potential T-cell immune response to the dystrophin epitopes that were either presented or not presented in the AAV vector.113 This result was totally unpredicted by the mouse studies.
DMD dogs, a bridge between mice and humans
A model that better resembles human patients may bridge the gap between mice and humans. In this regard, a dystrophin-deficient dog represents an ideal intermediate model. Unlike mice, dogs have a body size similar to that of affected boys. More importantly, it has been shown that the absence of dystrophin indeed causes severe muscular dystrophy in dogs (reviewed in various studies114–118). The affected pups are stunted and weaker. This soon progresses into dysphagia, fatigue, abnormal gait, joint contracture, severe muscle wasting, and premature death (Figure 2). Recent studies suggest that the phenotypic differences in mice, dogs, and humans may be rooted in the difference of cell surface sialic acids.48 Dogs and humans share similar glycol modification, but mice have a different type of glycol modification.48,119 In summary, dystrophin-deficient dogs are genetically (in terms of mutated dystrophin gene) and phenotypically (in terms of clinical manifestation) similar to human patients. They represent superior models for DMD research.
Duchenne-like muscular dystrophy has been reported in many different dog breeds (Table 1).120–138 However, few have been characterized. Currently, experimental dog colonies have only been established in Beagle, golden retriever muscular dystrophy (GRMD), and Corgi models. The GRMD model is the best studied (Figure 2).126,127 Beagle and GRMD models share the same mutation.120 In these dogs, an A-to-G transition near the end of intron 6 disrupts the normal splicing acceptor signal. Exon 7 is lost in the resulting transcript. Jumping from exon 6 to exon 8 introduces frameshift mutation and a premature stop codon.137,138 Mutations in several other canine DMD models have also been identified. In the case of German short-haired pointer, a 2.7-mb deletion in the X chromosome removes the entire dystrophin gene.125 In the case of Cavalier King Charles spaniels, a G-to-T transversion at the beginning of intron 50 results in exon 50 deletion and subsequent frameshift and premature termination.124 Smith et al have recently reported a Corgi DMD model.139 In this model, insertion of a repetitive element in intron 13 aborts dystrophin translation.139
Table 1.
Dystrophin-deficient dogs
| Breed | Mutation | Comment | References |
|---|---|---|---|
| Beagle | Intron 6 point mutation (same as in GRMD) |
Colony established. Exon skipping tested |
120,121 |
| Belgian Groenendaeler shepherds | Unknown | 122 | |
| Brittany spaniels | Unknown | 123 | |
| Cavalier King Charles spaniels | Intron 50 point mutation | Exon skipping tested in cultured myoblasts | 124 |
| Corgi | Repetitive element insertion in intron 13 |
Colony established, AAV microgene ongoing | 139 |
| German short-haired pointer | Unknown | 125 | |
| Golden retriever (GRMD) | Intron 6 point mutation | Colony established in several places. Plasmid, exon skipping, and AAV microgene tested |
121,126,127 |
| Grand Basset Griffon Vendéen | Unknown | 128 | |
| Irish terriers | Unknown | 129 | |
| Japanese spitz | Unknown | 130 | |
| Labrador retrievera | Unknown | 131 | |
| Labrador retrievera | Repetitive element insertion in intron |
166 | |
| Miniature schnauzer | Unknown | 132 | |
| Old english sheepdog | Unknown | 133 | |
| Rat terrier | Unknown | 134 | |
| Rottweiler | Point mutation in exon 52 | 167 | |
| Samoyed | Unknown | 135 | |
| Weimaraner | Unknown | 136 |
Note:
It is not clear whether the two Labrador retrievers share the same mutation.
Abbreviations: GRMD, golden retriever muscular dystrophy; AAV, adeno-associated virus.
Plasmid injection, exon-skipping, and AAV microgene therapies have all been tested in the canine model. So far, all the reported studies were performed in either GRMD or Beagle dystrophic dogs. The result of plasmid injection has been briefly mentioned in several articles, but a comprehensive report is lacking. It was suggested that plasmid injection efficiency was very poor. In general, <1% myofibers were transduced.102,104,140,141 Exon skipping was initially tested in cultured GRMD muscle cells in vitro.142 However, recent studies suggest that such an in vitro assay cannot faithfully predict the in vivo outcome.143,144 Instead of a single AON, it appears that a cocktail of several different AONs is needed to achieve efficient skipping in the Beagle dystrophy dogs.144 AAV microgene therapy has been tested in adult GRMD dogs by local injection and in newborn GRMD dogs by systemic injection.145–148 Transient immune suppression is required to achieve persistent expression following direct muscle injection in adult dystrophic dogs.147 Intravascular delivery of AAV serotype-9 ΔR3–19/ΔR20–21/ΔC microdystrophin (this microgene contains hinge 3) vector resulted in widespread expression in newborn dogs145,146 (Figure 1). Paradoxically, this therapy did not lead to expected disease amelioration. In contrast, the treated dogs showed growth delay, pelvic limb muscle atrophy, and contracture.146 The exact reason behind this peculiar finding is not clear. However, a clinical study suggests that in-frame deletion of hinge 3 is associated with a milder disease.74 From this point, a microgene without hinge 3 may represent a better option. Collectively, the preliminary results of the canine studies have probably revealed a more realistic picture of the challenges facing DMD gene therapy. Yes, we can now cure a dystrophic mouse. However, we are still far from curing a DMD boy.
To-do list in the canine model
How can we fill the gap between mice and humans? Considering the limitations of the murine models and also considering the genetic and clinical similarities between dystrophic dogs and DMD patients, a logical next step would be to test DMD gene therapy in the dog model. Our limited experience in dystrophic dogs has already offered critical insight. The low efficiency of plasmid therapy was confirmed in human trials.104 The canine studies also raised the need for applying transient immune suppression in AAV-mediated delivery.145,147,148
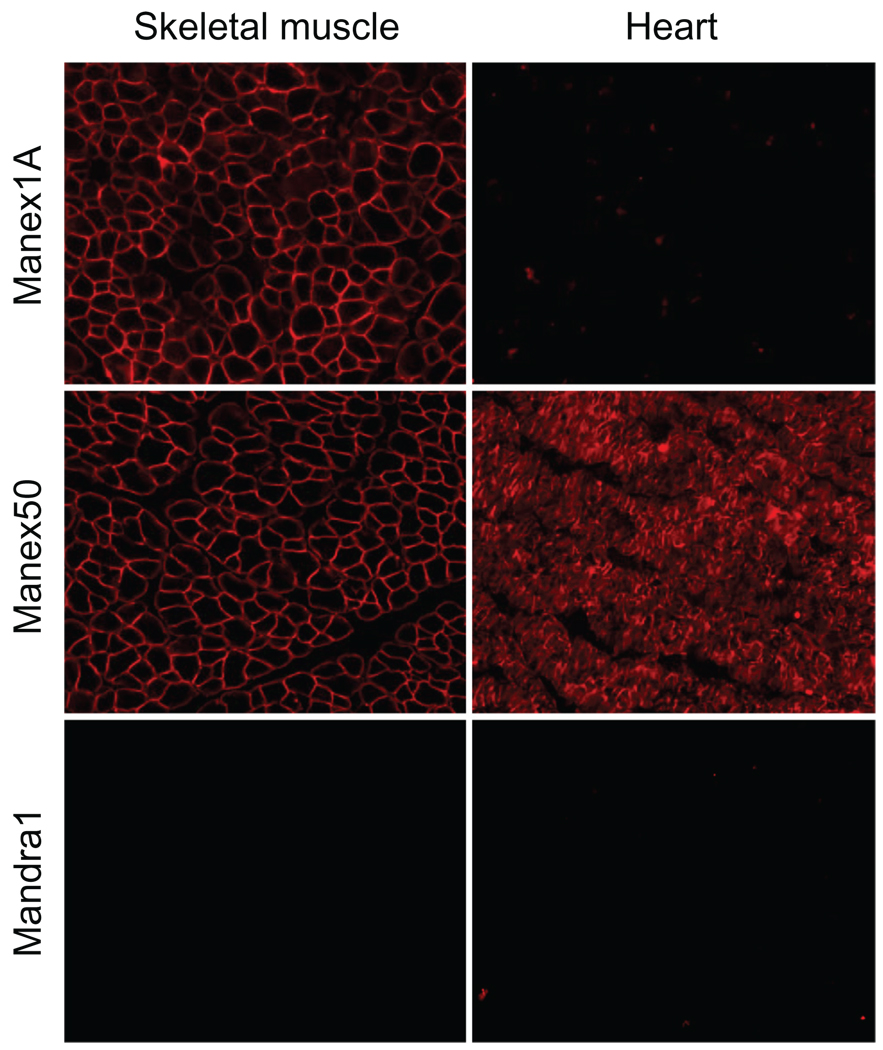
Unfortunately, there are more limitations to the canine studies than the murine studies. The cost of housing, breeding, and raising dystrophic dogs greatly exceeds that of dystrophic mice. A specialized team consisting of veterinary doctors of different disciplines, experienced technicians, and basic scientists are needed to maintain a dystrophic dog colony for translational research.149 Another critical issue is the experimental scale. Unlike mice, dogs only go in heat twice a year, and dystrophic dogs are usually not suitable for natural breeding. This signiificantly limits the number of affected dogs one can obtain for experiments. Considering the pronounced variations of disease progression among individual dogs (Fine D, Shin J-H, Duan D. Unpublished data),150,151 great caution should be taken in interpreting the data from a few dogs.152 Lack of canine-specific reagents constitutes another barrier. More than 100 epitope-specific dystrophin antibodies have been developed (see http://glennmorris.org.uk/monopubs.htm).153 However few have been characterized for canine muscle applications ( Figure 3). Perhaps, the most constraining hurdle is the lack of rigorous physiological parameters to evaluate the therapeutic outcome in dogs. Standard protocols are available to diagnose dystrophin gene mutation and to evaluate dystrophin expression at the mRNA and protein levels. A wide array of in vitro and in vivo physiological assays has also been established to monitor muscle strength changes in mice.154,155 These include force measurement in a single, intact muscle, such as in vitro assay in the extensor digitorum longus muscle and in situ assay in the tibialis anterior muscle.66,70,156 Other assays include forelimb or hind limb grip strength and treadmill exercise. However, similar assays are either not developed or the baseline values are not available for dystrophic and normal control dogs. Further, clinical relevance and reliability remain to be validated.
Figure 3.
Detection of canine dystrophin with monoclonal antibodies. Monoclonal dystrophin antibodies were used to detect dystrophin expression in a normal dog skeletal muscle and the heart. Representative immunostaining photomicrographs are shown for antibodies Manex1 A, Manex50, and Mandra1. Manex1A recognizes an epitope encoded by exon 1 (N-terminal domain). Interestingly, it only reacts with dystrophin in dog skeletal muscle. Manex50 maps to an epitope encoded by exon 50 (rod domain). This antibody can detect dog dystrophin in both skeletal and cardiac muscles. Mandra1 is supposed to react with an epitope encoded by exon 70 (C-terminal domain). However, it fails to reveal canine dystrophin in either skeletal muscle or the heart.
There are many unanswered questions. With limited resources, how can we take maximal advantage of the canine model? To prioritize our effort, it is important to point out that the core of current DMD gene therapies (such as exon-skipping therapy and AAV microgene therapy) is based on minimized dystrophin genes. Yet the reading frame theory does not always hold true for every single patient. Severe cases have been reported in patients harboring an in-frame deletion (reviewed in Yokota et al157). The gene size is particularly relevant to the microgene approach. All the mildly affected patients reported so far carry at least 50% of the dystrophin coding sequence.16,17,19–22,158,159 However, none of the microgenes contain more than 40% of the coding sequence. A patient who carried a fairly large rod-domain in-frame deletion actually developed severe DMD.160 The size of the truncated dystrophin in this severely affected patient is ~160 kDa, a size similar to that of microdystrophins.160 Although it is possible that a rationally designed microgene may be functionally superior, this clinical report indeed raises the importance of rigorously testing the synthetic microgenes prior to human trials.
Although individual investigator-initiated studies should be encouraged, focused studies in a few carefully weighed experimental therapies could be more productive for the entire field. In this regard, a platform is needed to convincingly test the therapeutic efficacy in a single, intact dog muscle at the molecular and physiological levels. Such an approach may lead to some immediate benefits in terms of improving the quality of life. It will also lay the foundation for whole-body therapy. On the other hand, we should continue our effort to expand the available colonies for statistically meaningful large sample-size studies. Further characterization of the existing dystrophic dog models and the development of new canine DMD models are also important measures for preclinical investigations.
What else?
Our current effort is mainly focused on skeletal muscle disease. However, we cannot and should not ignore other aspects of DMD. Although DMD is often referred to as a muscle disease, it actually affects multiple organ systems. Cardiac complications and central nerve system involvement are also highly relevant to the health of DMD patients.161,162 A therapy for skeletal muscle may not effectively treat other complications.163 Recent studies suggest that very old female mdx mice may model dilated cardiomyopathy in DMD patients.84,164 However, none of the current gene therapy strategies have been evaluated in this model. Reports on the cardiac changes of dystrophin-deficient dogs are rare. Age-matched electrocardiogram and echocardiography examination between normal and affected dogs may provide a valuable baseline for cardiac outcome measurement.
Most current studies are aimed at restoring dystrophin expression (yes, this is still on the very top of our list). The rapidly expanding library of the disease-modifying genes may also offer new opportunities. Considering the immense variance in clinical manifestations among certain patients who carry the same gene mutation,165 investigation in disease-modifying genes may yield novel alternative gene therapies to treat DMD.
Perspective
Tremendous progress has been achieved over the last two decades in developing novel genetic therapies for treating DMD. Our success in treating dystrophic mice suggests that gene therapy may be a successful modality. Clinical trials have been initiated but they have yet to produce convincing benefit in DMD patients. Dystrophic dogs represent an important translational bridge between mice and humans. Unfortunately, we know much less about dystrophic dogs than we do about dystrophin-deficient mice and DMD patients. There is an urgent need to expand our investigations in the canine model. This investment will allow us to perfect gene therapy protocols and minimize unnecessary detours in human trials. To make this emphasis shift requires commitment and support from the entire DMD community, including the researchers, funding agencies, and patients and their families and friends.
Acknowledgments
DMD research in the author’s lab is supported by National Institutes of Health (AR-49419, AR-57209, HL-91883 and NS-62934), Muscular Dystrophy Association, Parent Project Muscular Dystrophy and Jesse’s Journey, The Foundation for Gene and Cell Therapy. I thank Dr Robert J McDonald for critical reading of the manuscript.
Footnotes
Disclosure
The author reports no conflicts of interest in this work.
References
- 1.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 2.Kunkel LM. 2004 William Allan Award address. Cloning of the DMD gene. Am J Hum Genet. 2005;76(2):205–214. doi: 10.1086/428143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50(3):509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 4.Romitti P, Puzhankara S, Mathews K, et al. Centers for Disease Control and Prevention (CDC). Prevalence of Duchenne/Becker muscular dystrophy among males aged 5–24 years – four states, 2007. Morb Mortal Wkly Rep. 2009;58(40):1119–1122. [PubMed] [Google Scholar]
- 5.Dooley J, Gordon KE, Dodds L, MacSween J. Duchenne muscular dystrophy: a 30-year population-based incidence study. Clin Pediatr (Phila) 2010;49(2):177–179. doi: 10.1177/0009922809347777. [DOI] [PubMed] [Google Scholar]
- 6.Wells DJ, Wells KE, Walsh FS, et al. Human dystrophin expression corrects the myopathic phenotype in transgenic mdx mice. Hum Mol Genet. 1992;1(1):35–40. doi: 10.1093/hmg/1.1.35. [DOI] [PubMed] [Google Scholar]
- 7.Cox GA, Cole NM, Matsumura K, et al. Overexpression of dystrophin in transgenic mdx mice eliminates dystrophic symptoms without toxicity [see comments] Nature. 1993;364(6439):725–729. doi: 10.1038/364725a0. [DOI] [PubMed] [Google Scholar]
- 8.Dickson G, Love DR, Davies KE, Wells KE, Piper TA, Walsh FS. Human dystrophin gene transfer: production and expression of a functional recombinant DNA-based gene. Hum Genet. 1991;88(1):53–58. doi: 10.1007/BF00204929. [DOI] [PubMed] [Google Scholar]
- 9.Ragot T, Vincent N, Chafey P, et al. Efficient adenovirus-mediated transfer of a human minidystrophin gene to skeletal muscle of mdx mice. Nature. 1993;361(6413):647–650. doi: 10.1038/361647a0. [DOI] [PubMed] [Google Scholar]
- 10.Vincent N, Ragot T, Gilgenkrantz H, et al. Long-term correction of mouse dystrophic degeneration by adenovirus-mediated transfer of a minidystrophin gene. Nat Genet. 1993;5(2):130–134. doi: 10.1038/ng1093-130. [DOI] [PubMed] [Google Scholar]
- 11.Stratford-Perricaudet LD, Makeh I, Perricaudet M, Briand P. Widespread long-term gene transfer to mouse skeletal muscles and heart. J Clin Invest. 1992;90(2):626–630. doi: 10.1172/JCI115902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quantin B, Perricaudet LD, Tajbakhsh S, Mandel JL. Adenovirus as an expression vector in muscle cells in vivo. Proc Natl Acad Sci U S A. 1992;89(7):2581–2584. doi: 10.1073/pnas.89.7.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee CC, Pearlman JA, Chamberlain JS, Caskey CT. Expression of recombinant dystrophin and its localization to the cell membrane. Nature. 1991;349(6307):334–336. doi: 10.1038/349334a0. [DOI] [PubMed] [Google Scholar]
- 14.Wolff JA, Budker V. The mechanism of naked DNA uptake and expression. Adv Genet. 2005;54:3–20. doi: 10.1016/S0065-2660(05)54001-X. [DOI] [PubMed] [Google Scholar]
- 15.Acsadi G, Dickson G, Love DR, et al. Human dystrophin expression in mdx mice after intramuscular injection of DNA constructs [see comments] Nature. 1991;352(6338):815–818. doi: 10.1038/352815a0. [DOI] [PubMed] [Google Scholar]
- 16.Beggs AH, Hoffman EP, Snyder JR, et al. Exploring the molecular basis for variability among patients with Becker muscular dystrophy: dystrophin gene and protein studies. Am J Hum Genet. 1991;49(1):54–67. [PMC free article] [PubMed] [Google Scholar]
- 17.Koenig M, Beggs AH, Moyer M, et al. The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet. 1989;45(4):498–506. [PMC free article] [PubMed] [Google Scholar]
- 18.Gillard EF, Chamberlain JS, Murphy EG, et al. Molecular and phenotypic analysis of patients with deletions within the deletion-rich region of the Duchenne muscular dystrophy (DMD) gene. Am J Hum Genet. 1989;45(4):507–520. [PMC free article] [PubMed] [Google Scholar]
- 19.England SB, Nicholson LV, Johnson MA, et al. Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature. 1990;343(6254):180–182. doi: 10.1038/343180a0. [DOI] [PubMed] [Google Scholar]
- 20.Aartsma-Rus A, van Deutekom JC, Fokkema IF, van Ommen GJ, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confrm the reading-frame rule. Muscle Nerve. 2006;34(2):135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- 21.Tuffery-Giraud S, Béroud C, Leturcq F, et al. Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase. Hum Mutat. 2009;30(6):934–945. doi: 10.1002/humu.20976. [DOI] [PubMed] [Google Scholar]
- 22.Flanigan KM, Dunn DM, von Niederhausern A, et al. United Dystrophinopathy Project Consortium. Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum Mutat. 2009;30(12):1657–1666. doi: 10.1002/humu.21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chamberlain JS. Gene therapy of muscular dystrophy. Hum Mol Genet. 2002;11(20):2355–2362. doi: 10.1093/hmg/11.20.2355. [DOI] [PubMed] [Google Scholar]
- 24.Harper SQ, Hauser MA, DelloRusso C, et al. Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat Med. 2002;8(3):253–261. doi: 10.1038/nm0302-253. [DOI] [PubMed] [Google Scholar]
- 25.Lu QL, Yokota T, Takeda S, Garcia L, Muntoni F, Partridge T. The status of exon skipping as a therapeutic approach to Duchenne muscular dystrophy. Mol Ther. 2011;19(1):9–15. doi: 10.1038/mt.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heydemann A, Doherty KR, McNally EM. Genetic modifiers of muscular dystrophy: implications for therapy. Biochim Biophys Acta. 2007;1772(2):216–228. doi: 10.1016/j.bbadis.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Love DR, Hill DF, Dickson G, et al. An autosomal transcript in skeletal muscle with homology to dystrophin. Nature. 1989;339(6219):55–58. doi: 10.1038/339055a0. [DOI] [PubMed] [Google Scholar]
- 28.Tinsley JM, Blake DJ, Roche A, et al. Primary structure of dystrophin- related protein. Nature. 1992;360(6404):591–593. doi: 10.1038/360591a0. [DOI] [PubMed] [Google Scholar]
- 29.Khurana TS, Hoffman EP, Kunkel LM. Identification of a chromosome 6-encoded dystrophin-related protein. J Biol Chem. 1990;265(28):16717–16720. [PubMed] [Google Scholar]
- 30.Tinsley JM, Davies KE. Utrophin: a potential replacement for dystrophin? Neuromuscul Disord. 1993;3(5–6):537–539. doi: 10.1016/0960-8966(93)90111-v. [DOI] [PubMed] [Google Scholar]
- 31.Li D, Bareja A, Judge L, et al. Sarcolemmal nNOS anchoring reveals a qualitative difference between dystrophin and utrophin. J Cell Sci. 2010;123(Pt 12):2008–2013. doi: 10.1242/jcs.064808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li D, Long C, Yue Y, Duan D. Sub-physiological sarcoglycan expression contributes to compensatory muscle protection in mdx mice. Hum Mol Genet. 2009;18(7):1209–1220. doi: 10.1093/hmg/ddp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wehling M, Spencer MJ, Tidball JG. A nitric oxide synthase transgene ameliorates muscular dystrophy in mdx mice. J Cell Biol. 2001;155(1):123–131. doi: 10.1083/jcb.200105110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burkin DJ, Wallace GQ, Nicol KJ, Kaufman DJ, Kaufman SJ. Enhanced expression of the alpha 7 beta 1 integrin reduces muscular dystrophy and restores viability in dystrophic mice. J Cell Biol. 2001;152(6):1207–1218. doi: 10.1083/jcb.152.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peter AK, Marshall JL, Crosbie RH. Sarcospan reduces dystrophic pathology: stabilization of the utrophin-glycoprotein complex. J Cell Biol. 2008;183(3):419–427. doi: 10.1083/jcb.200808027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rooney JE, Gurpur PB, Burkin DJ. Laminin-111 protein therapy prevents muscle disease in the mdx mouse model for Duchenne muscular dystrophy. Proc Natl Acad Sci U S A. 2009;106(19):7991–7996. doi: 10.1073/pnas.0811599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verma M, Asakura Y, Hirai H, et al. Flt-1 haploinsufficiency ameliorates muscular dystrophy phenotype by developmentally increased vasculature in mdx mice. Hum Mol Genet. 2010;19(21):4145–4159. doi: 10.1093/hmg/ddq334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morine KJ, Sleeper MM, Barton ER, Sweeney HL. Overexpression of SERCA1a in the mdx diaphragm reduces susceptibility to contraction- induced damage. Hum Gene Ther. 2010;21(12):1735–1739. doi: 10.1089/hum.2010.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minetti GC, Colussi C, Adami R, et al. Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nat Med. 2006;12(10):1147–1150. doi: 10.1038/nm1479. [DOI] [PubMed] [Google Scholar]
- 40.Vetrone SA, Montecino-Rodriguez E, Kudryashova E, et al. Osteopontin promotes fibrosis in dystrophic mouse muscle by modulating immune cell subsets and intramuscular TGF-beta. J Clin Invest. 2009;119(6):1583–1594. doi: 10.1172/JCI37662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner KR, McPherron AC, Winik N, Lee SJ. Loss of myostatin attenuates severity of muscular dystrophy in mdx mice. Ann Neurol. 2002;52(6):832–836. doi: 10.1002/ana.10385. [DOI] [PubMed] [Google Scholar]
- 42.Bogdanovich S, Krag TO, Barton ER, et al. Functional improvement of dystrophic muscle by myostatin blockade. Nature. 2002;420(6914):418–421. doi: 10.1038/nature01154. [DOI] [PubMed] [Google Scholar]
- 43.Millay DP, Sargent MA, Osinska H, et al. Genetic and pharmacologic inhibition of mitochondrial-dependent necrosis attenuates muscular dystrophy. Nat Med. 2008;14(4):442–447. doi: 10.1038/nm1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heydemann A, Ceco E, Lim JE, et al. Latent TGF-beta-binding protein 4 modifies muscular dystrophy in mice. J Clin Invest. 2009;119(12):3703–3712. doi: 10.1172/JCI39845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukada S, Morikawa D, Yamamoto Y, et al. Genetic background affects properties of satellite cells and mdx phenotypes. Am J Pathol. 2010;176(5):2414–2424. doi: 10.2353/ajpath.2010.090887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barton ER, Morris L, Musaro A, Rosenthal N, Sweeney HL. Muscle-specific expression of insulin-like growth factor I counters muscle decline in mdx mice. J Cell Biol. 2002;157(1):137–148. doi: 10.1083/jcb.200108071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kronqvist P, Kawaguchi N, Albrechtsen R, et al. ADAM12 alleviates the skeletal muscle pathology in mdx dystrophic mice. Am J Pathol. 2002;161(5):1535–1540. doi: 10.1016/S0002-9440(10)64431-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chandrasekharan K, Yoon JH, Xu Y, et al. A human-specific deletion in mouse Cmah increases disease severity in the mdx model of Duchenne muscular dystrophy. Sci Transl Med. 2010;2(42) doi: 10.1126/scitranslmed.3000692. 42ra54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bulfeld G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci U S A. 1984;81(4):1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Araki E, Nakamura K, Nakao K, et al. Targeted disruption of exon 52 in the mouse dystrophin gene induced muscle degeneration similar to that observed in Duchenne muscular dystrophy. Biochem Biophys Res Commun. 1997;238(2):492–497. doi: 10.1006/bbrc.1997.7328. [DOI] [PubMed] [Google Scholar]
- 51.Chapman VM, Miller DR, Armstrong D, Caskey CT. Recovery of induced mutations for X chromosome-linked muscular dystrophy in mice. Proc Natl Acad Sci U S A. 1989;86(4):1292–1296. doi: 10.1073/pnas.86.4.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bostick B, Yue Y, Long C, Duan D. Prevention of dystrophin-deficient cardiomyopathy in twenty-one-month-old carrier mice by mosaic dystrophin expression or complementary dystrophin/utrophin expression. Circ Res. 2008;102(1):121–130. doi: 10.1161/CIRCRESAHA.107.162982. [DOI] [PubMed] [Google Scholar]
- 53.Chamberlain JS, Metzger J, Reyes M, Townsend D, Faulkner JA. Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. FASEB J. 2007;21(9):2195–2204. doi: 10.1096/fj.06-7353com. [DOI] [PubMed] [Google Scholar]
- 54.Deconinck AE, Rafael JA, Skinner JA, et al. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell. 1997;90(4):717–727. doi: 10.1016/s0092-8674(00)80532-2. [DOI] [PubMed] [Google Scholar]
- 55.Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10(10):1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 56.Rooney JE, Welser JV, Dechert MA, Flintoff-Dye NL, Kaufman SJ, Burkin DJ. Severe muscular dystrophy in mice that lack dystrophin and alpha7 integrin. J Cell Sci. 2006;119(Pt 11):2185–2195. doi: 10.1242/jcs.02952. [DOI] [PubMed] [Google Scholar]
- 57.Sacco A, Mourkioti F, Tran R, et al. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143(7):1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo C, Willem M, Werner A, et al. Absence of alpha 7 integrin in dystrophin-deficient mice causes a myopathy similar to Duchenne muscular dystrophy. Hum Mol Genet. 2006;15(6):989–998. doi: 10.1093/hmg/ddl018. [DOI] [PubMed] [Google Scholar]
- 59.Grady RM, Teng H, Nichol MC, Cunningham JC, Wilkinson RS, Sanes JR. Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: a model for Duchenne muscular dystrophy. Cell. 1997;90(4):729–738. doi: 10.1016/s0092-8674(00)80533-4. [DOI] [PubMed] [Google Scholar]
- 60.Wang B, Li J, Xiao X. Adeno-associated virus vector carrying human minidystrophin genes effectively ameliorates muscular dystrophy in mdx mouse model. Proc Natl Acad Sci U S A. 2000;97(25):13714–13719. doi: 10.1073/pnas.240335297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao G, Vandenberghe LH, Wilson JM. New recombinant serotypes of AAV vectors. Curr Gene Ther. 2005;5(3):285–297. doi: 10.2174/1566523054065057. [DOI] [PubMed] [Google Scholar]
- 62.Vandenberghe LH, Wilson JM, Gao G. Tailoring the AAV vector capsid for gene therapy. Gene Ther. 2009;16(3):311–319. doi: 10.1038/gt.2008.170. [DOI] [PubMed] [Google Scholar]
- 63.Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol Ther. 2010;18(1):80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lai Y, Yue Y, Duan D. Evidence for the failure of adeno-associated virus serotype 5 to package a viral genome ≥8.2 kb. Mol Ther. 2010;18(1):75–79. doi: 10.1038/mt.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong B, Nakai H, Xiao W. Characterization of genome integrity for oversized recombinant AAV vector. Mol Ther. 2010;18(1):87–92. doi: 10.1038/mt.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu M, Yue Y, Harper SQ, Grange RW, Chamberlain JS, Duan D. Adeno-associated virus-mediated microdystrophin expression protects young mdx muscle from contraction-induced injury. Mol Ther. 2005;11(2):245–256. doi: 10.1016/j.ymthe.2004.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watchko J, O’Day T, Wang B, et al. Adeno-associated virus vector- mediated minidystrophin gene therapy improves dystrophic muscle contractile function in mdx mice. Hum Gene Ther. 2002;13(12):1451–1460. doi: 10.1089/10430340260185085. [DOI] [PubMed] [Google Scholar]
- 68.Brenman JE, Chao DS, Xia H, Aldape K, Bredt DS. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarco- lemma in Duchenne muscular dystrophy. Cell. 1995;82(5):743–752. doi: 10.1016/0092-8674(95)90471-9. [DOI] [PubMed] [Google Scholar]
- 69.Lai Y, Thomas GD, Yue Y, et al. Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. J Clin Invest. 2009;119(3):624–635. doi: 10.1172/JCI36612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li D, Yue Y, Lai Y, Hakim CH, Duan D. Nitrosative stress elicited by nNOSµ delocalization inhibits muscle force in dystrophin-null mice. J Pathol. 2011;223(1):88–98. doi: 10.1002/path.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sander M, Chavoshan B, Harris SA, et al. Functional muscle ischemia in neuronal nitric oxide synthase-deficient skeletal muscle of children with Duchenne muscular dystrophy. Proc Natl Acad Sci U S A. 2000;97(25):13818–13823. doi: 10.1073/pnas.250379497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomas GD, Sander M, Lau KS, Huang PL, Stull JT, Victor RG. Impaired metabolic modulation of alpha-adrenergic vasoconstriction in dystrophin-deficient skeletal muscle. Proc Natl Acad Sci U S A. 1998;95(25):15090–15095. doi: 10.1073/pnas.95.25.15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Banks GB, Judge LM, Allen JM, Chamberlain JS. The polyproline site in hinge 2 influences the functional capacity of truncated dystrophins. PLoS Genet. 2010;6(5):e1000958. doi: 10.1371/journal.pgen.1000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carsana A, Frisso G, Tremolaterra MR, et al. Analysis of dystrophin gene deletions indicates that the hinge III region of the protein correlates with disease severity. Ann Hum Genet. 2005;69(Pt 3):253–259. doi: 10.1046/j.1529-8817.2005.00160.x. [DOI] [PubMed] [Google Scholar]
- 75.Duan D, Yan Z, Engelhardt JF. Expanding the capacity of AAV vectors. In: Bloom ME, Cotmore SF, Linden RM, Parrish CR, Kerr JR, editors. Parvoviruses. New York, NY: Hodder Arnold; 2006. pp. 525–532. [Google Scholar]
- 76.Duan D. From the smallest virus to the biggest gene: marching towards gene therapy for duchenne muscular dystrophy. Discov Med. 2006;6(33):103–108. [PMC free article] [PubMed] [Google Scholar]
- 77.Ghosh A, Duan D. Expanding adeno-associated viral vector capacity: a tale of two vectors. Biotechnol Genet Eng Rev. 2007;24:165–177. doi: 10.1080/02648725.2007.10648098. [DOI] [PubMed] [Google Scholar]
- 78.Ghosh A, Yue Y, Lai Y, Duan D. A hybrid vector system expands adeno-associated viral vector packaging capacity in a transgene-independent manner. Mol Ther. 2008;16(1):124–130. doi: 10.1038/sj.mt.6300322. [DOI] [PubMed] [Google Scholar]
- 79.Ghosh A, Yue Y, Duan D. Efficient transgene reconstitution with hybrid dual AAV vectors carrying the minimized bridging sequences. Hum Gene Ther. 2011;22(1):1–7. doi: 10.1089/hum.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Odom GL, Gregorevic P, Allen JM, Chamberlain JS. Gene therapy of mdx mice with large truncated dystrophins generated by recombination using rAAV6. Mol Ther. 2011;19(1):36–45. doi: 10.1038/mt.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lai Y, Yue Y, Liu M, et al. Efficient in vivo gene expression by trans-splicing adeno-associated viral vectors. Nat Biotechnol. 2005;23(11):1435–1439. doi: 10.1038/nbt1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gregorevic P, Blankinship MJ, Allen JM, et al. Systemic delivery of genes to striated muscles using adeno-associated viral vectors. Nat Med. 2004;10(8):828–834. doi: 10.1038/nm1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Z, Zhu T, Qiao C, et al. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat Biotechnol. 2005;23(3):321–328. doi: 10.1038/nbt1073. [DOI] [PubMed] [Google Scholar]
- 84.Bostick B, Ghosh A, Yue Y, Long C, Duan D. Systemic AAV-9 transduction in mice is infuenced by animal age but not by the route of administration. Gene Ther. 2007;14(22):1605–1609. doi: 10.1038/sj.gt.3303029. [DOI] [PubMed] [Google Scholar]
- 85.Inagaki K, Fuess S, Storm TA, et al. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol Ther. 2006;14(1):45–53. doi: 10.1016/j.ymthe.2006.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pacak CA, Mah CS, Thattaliyath BD, et al. Recombinant adeno- associated virus serotype 9 leads to preferential cardiac transduction in vivo. Circ Res. 2006;99(4):e3–e9. doi: 10.1161/01.RES.0000237661.18885.f6. [DOI] [PubMed] [Google Scholar]
- 87.Nakai H, Fuess S, Storm TA, Muramatsu S, Nara Y, Kay MA. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J Virol. 2005;79(1):214–224. doi: 10.1128/JVI.79.1.214-224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ghosh A, Yue Y, Shin JH, Duan D. Systemic trans-splicing adeno- associated viral delivery effciently transduces the heart of adult mdx mouse, a model for Duchenne muscular dystrophy. Hum Gene Ther. 2009;20(11):1319–1328. doi: 10.1089/hum.2009.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lostal W, Bartoli M, Bourg N, et al. Effcient recovery of dysferlin deficiency by dual adeno-associated vector-mediated gene transfer. Hum Mol Genet. 2010;19(10):1897–1907. doi: 10.1093/hmg/ddq065. [DOI] [PubMed] [Google Scholar]
- 90.Ghosh A, Yue Y, Long C, Bostick B, Duan D. Efficient whole-body transduction with trans-splicing adeno-associated viral vectors. Mol Ther. 2007;15(4):750–755. doi: 10.1038/sj.mt.6300081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gregorevic P, Blankinship MJ, Allen JM, Chamberlain JS. Systemic microdystrophin gene delivery improves skeletal muscle structure and function in old dystrophic mdx mice. Mol Ther. 2008;16(4):657–664. doi: 10.1038/mt.2008.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gregorevic P, Allen JM, Minami E, et al. rAAV6-microdystrophin preserves muscle function and extends lifespan in severely dystrophic mice. Nat Med. 2006;12(7):787–789. doi: 10.1038/nm1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mann CJ, Honeyman K, Cheng AJ, et al. Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proc Natl Acad Sci U S A. 2001;98(1):42–47. doi: 10.1073/pnas.011408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lu QL, Mann CJ, Lou F, et al. Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat Med. 2003;9(8):1009–1014. doi: 10.1038/nm897. [DOI] [PubMed] [Google Scholar]
- 95.Wu B, Li Y, Morcos PA, Doran TJ, Lu P, Lu QL. Octa-guanidine morpholino restores dystrophin expression in cardiac and skeletal muscles and ameliorates pathology in dystrophic mdx mice. Mol Ther. 2009;17(5):864–871. doi: 10.1038/mt.2009.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goyenvalle A, Babbs A, Powell D, et al. Prevention of dystrophic pathology in severely affected dystrophin/utrophin-defcient mice by morpholino-oligomer-mediated exon-skipping. Mol Ther. 2010;18(1):198–205. doi: 10.1038/mt.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yin H, Moulton HM, Betts C, et al. Functional rescue of dystrophin- deficient mdx mice by a chimeric peptide-PMO. Mol Ther. 2010;18(10):1822–1829. doi: 10.1038/mt.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alter J, Lou F, Rabinowitz A, et al. Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat Med. 2006;12(2):175–177. doi: 10.1038/nm1345. [DOI] [PubMed] [Google Scholar]
- 99.Wu B, Moulton HM, Iversen PL, et al. Effective rescue of dystrophin improves cardiac function in dystrophin-defcient mice by a modified morpholino oligomer. Proc Natl Acad Sci U S A. 2008;105(39):14814–14819. doi: 10.1073/pnas.0805676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goyenvalle A, Vulin A, Fougerousse F, et al. Rescue of dystro-phic muscle through U7 snRNA-mediated exon skipping. Science. 2004;306(5702):1796–1799. doi: 10.1126/science.1104297. [DOI] [PubMed] [Google Scholar]
- 101.Romero NB, Benveniste O, Payan C, et al. Current protocol of a research phase I clinical trial of full-length dystrophin plasmid DNA in Duchenne/Becker muscular dystrophies. Part II: clinical protocol. Neuromuscul Disord. 2002;12 Suppl 1:S45–S48. doi: 10.1016/s0960-8966(02)00081-0. [DOI] [PubMed] [Google Scholar]
- 102.Romero NB, Braun S, Benveniste O, et al. Phase I study of dystrophin plasmid-based gene therapy in Duchenne/Becker muscular dystrophy. Hum Gene Ther. 2004;15(11):1065–1076. doi: 10.1089/hum.2004.15.1065. [DOI] [PubMed] [Google Scholar]
- 103.Braun S. Muscular gene transfer using nonviral vectors. Curr Gene Ther. 2008;8(5):391–405. doi: 10.2174/156652308786070998. [DOI] [PubMed] [Google Scholar]
- 104.Duan D. Myodys, a full-length dystrophin plasmid vector for Duchenne and Becker muscular dystrophy gene therapy. Curr Opin Mol Ther. 2008;10(1):86–94. [PubMed] [Google Scholar]
- 105.Kinali M, Arechavala-Gomeza V, Feng L, et al. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8(10):918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Van Deutekom JC, Janson AA, Ginjaar IB, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357(26):2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 107.Koenig M, Kunkel LM. Detailed analysis of the repeat domain of dystrophin reveals four potential hinge segments that may confer flexibility. J Biol Chem. 1990;265(8):4560–4566. [PubMed] [Google Scholar]
- 108.Ferreiro V, Giliberto F, Muñiz GM, et al. Asymptomatic Becker muscular dystrophy in a family with a multiexon deletion. Muscle Nerve. 2009;39(2):239–243. doi: 10.1002/mus.21193. [DOI] [PubMed] [Google Scholar]
- 109.Samaha FJ, Quinlan JG. Dystrophinopathies: clarification and complication. J Child Neurol. 1996;11(1):13–20. doi: 10.1177/088307389601100103. [DOI] [PubMed] [Google Scholar]
- 110.Samaha FJ, Quinlan JG. Myalgia and cramps: dystrophinopathy with wide-ranging laboratory fndings. J Child Neurol. 1996;11(1):21–24. doi: 10.1177/088307389601100105. [DOI] [PubMed] [Google Scholar]
- 111.Sánchez-Arjona MB, Rodríguez-Uranga JJ, Giles-Lima M, et al. Spanish family with myalgia and cramps syndrome. J Neurol Neuro-surg Psychiatry. 2005;76(2):286–289. doi: 10.1136/jnnp.2004.037325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hoffman EP. Skipping toward personalized molecular medicine. N Engl J Med. 2007;357(26):2719–2722. doi: 10.1056/NEJMe0707795. [DOI] [PubMed] [Google Scholar]
- 113.Mendell JR, Campbell K, Rodino-Klapac L, et al. Dystrophin immunity in Duchenne’s muscular dystrophy. N Engl J Med. 2010;363(15):1429–1437. doi: 10.1056/NEJMoa1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Howell JM, Fletcher S, Kakulas BA, O’Hara M, Lochmuller H, Karpati G. Use of the dog model for Duchenne muscular dystrophy in gene therapy trials. Neuromuscul Disord. 1997;7(5):325–328. doi: 10.1016/s0960-8966(97)00057-6. [DOI] [PubMed] [Google Scholar]
- 115.Nonaka I. Animal models of muscular dystrophies. Lab Anim Sci. 1998;48(1):8–17. [PubMed] [Google Scholar]
- 116.Collins CA, Morgan JE. Duchenne’s muscular dystrophy: animal models used to investigate pathogenesis and develop therapeutic strategies. Int J Exp Pathol. 2003;84(4):165–172. doi: 10.1046/j.1365-2613.2003.00354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shelton GD, Engvall E. Canine and feline models of human inherited muscle diseases. Neuromuscul Disord. 2005;15(2):127–138. doi: 10.1016/j.nmd.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 118.Wells DJ, Wells KE. What do animal models have to tell us regarding Duchenne muscular dystrophy? Acta Myol. 2005;24(3):172–180. [PubMed] [Google Scholar]
- 119.Nakamura K, Ariga T, Yahagi T, Miyatake T, Suzuki A, Yamakawa T. Interspecies comparison of muscle gangliosides by two-dimensional thin-layer chromatography. J Biochem. 1983;94(5):1359–1365. doi: 10.1093/oxfordjournals.jbchem.a134482. [DOI] [PubMed] [Google Scholar]
- 120.Shimatsu Y, Yoshimura M, Yuasa K, et al. Major clinical and histopathological characteristics of canine X-linked muscular dystrophy in Japan, CXMDJ. Acta Myol. 2005;24(2):145–154. [PubMed] [Google Scholar]
- 121.Valentine BA, Cooper BJ, de Lahunta A, O’Quinn R, Blue JT. Canine X-linked muscular dystrophy. An animal model of Duchenne muscular dystrophy: clinical studies. J Neurol Sci. 1988;88(1–3):69–81. doi: 10.1016/0022-510x(88)90206-7. [DOI] [PubMed] [Google Scholar]
- 122.Van Ham LML, Desmidt M, Tshamala M, Hoorens JK, Mattheeuws DRG. Canine X-linked muscular dystrophy in Belgian Groenendaeler shepherds. J Am Anim Hosp Assoc. 1993;29(6):570–574. [Google Scholar]
- 123.Van Ham LML, Roels SLMF, Hoorens JK. Congenital dystrophy-like myopathy in a Brittany Spaniel puppy. Prog Vet Neurol. 1995;6(4):135–138. [Google Scholar]
- 124.Walmsley GL, Arechavala-Gomeza V, Fernandez-Fuente M, et al. A duchenne muscular dystrophy gene hot spot mutation in dystrophin-deficient cavalier king charles spaniels is amenable to exon 51 skipping. PLoS One. 2010;5(1):e8647. doi: 10.1371/journal.pone.0008647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schatzberg SJ, Olby NJ, Breen M, et al. Molecular analysis of a spontaneous dystrophin ‘knockout’ dog. Neuromuscul Disord. 1999;9(5):289–295. doi: 10.1016/s0960-8966(99)00011-5. [DOI] [PubMed] [Google Scholar]
- 126.Cooper BJ, Winand NJ, Stedman H, et al. The homologue of the Duchenne locus is defective in X-linked muscular dystrophy of dogs. Nature. 1988;334(6178):154–156. doi: 10.1038/334154a0. [DOI] [PubMed] [Google Scholar]
- 127.Kornegay JN, Tuler SM, Miller DM, Levesque DC. Muscular dystrophy in a litter of golden retriever dogs. Muscle Nerve. 1988;11(10):1056–1064. doi: 10.1002/mus.880111008. [DOI] [PubMed] [Google Scholar]
- 128.Klarenbeek S, Gerritzen-Bruning MJ, Rozemuller AJ, van der Lugt JJ. Canine X-linked muscular dystrophy in a family of Grand Basset Griffon Vendéen dogs. J Comp Pathol. 2007;137(4):249–252. doi: 10.1016/j.jcpa.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 129.Wentink GH, van der Linde-Sipman JS, Meijer AEFH, et al. Myopathy with a possible recessive X-linked inheritance in a litter of Irish Terriers. Vet Pathol. 1972;9(5):328–349. doi: 10.1177/030098587200900504. [DOI] [PubMed] [Google Scholar]
- 130.Jones BR, Brennan S, Mooney CT, et al. Muscular dystrophy with truncated dystrophin in a family of Japanese Spitz dogs. J Neurol Sci. 2004;217(2):143–149. doi: 10.1016/j.jns.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 131.Bergman RL, Inzana KD, Monroe WE, et al. Dystrophin-deficient muscular dystrophy in a Labrador retriever. J Am Anim Hosp Assoc. 2002;38(3):255–261. doi: 10.5326/0380255. [DOI] [PubMed] [Google Scholar]
- 132.Paola JP, Podell M, Shelton GD. Muscular dystrophy in a miniature Schnauzer. Prog Vet Neurol. 1993;4(1):14–18. [Google Scholar]
- 133.Wieczorek LA, Garosi LS, Shelton GD. Dystrophin-deficient muscular dystrophy in an old English sheepdog. Vet Rec. 2006;158(8):270–273. doi: 10.1136/vr.158.8.270. [DOI] [PubMed] [Google Scholar]
- 134.Wetterman CA, Harkin KR, Cash WC, Nietfield JC, Shelton GD. Hypertrophic muscular dystrophy in a young dog. J Am Vet Med Assoc. 2000;216(6):878–881. doi: 10.2460/javma.2000.216.878. [DOI] [PubMed] [Google Scholar]
- 135.Presthus J, Nordstoga K. Congenital myopathy in a litter of Samoyed dogs. Prog Vet Neurol. 1993;4(2):37–40. [Google Scholar]
- 136.Baltzer WI, Calise DV, Levine JM, Shelton GD, Edwards JF, Steiner JM. Dystrophin-deficient muscular dystrophy in a Weimaraner. J Am Anim Hosp Assoc. 2007;43(4):227–232. doi: 10.5326/0430227. [DOI] [PubMed] [Google Scholar]
- 137.Smith BF, Kornegay JN, Duan D. Independent canine models of Duchenne muscular dystrophy due to intronic insertions of repetitive DNA; Seattle, WA. 10th Annual Meeting of the American Society of Gene Therapy; 2007. May 30, Jun 3. [Google Scholar]
- 138.Winand N, Pradham D, Cooper B. Molecular characterization of severe Duchennetype dystrophy in a family of Rottweiler dogs; Tucson, AZ. Proceedings of the Muscular Dystrophin Association: Molecular Mechanisms of Neuromuscular Disease.1994. Jan 24–25, [Google Scholar]
- 139.Smith BF, Yue Y, Woods PR, et al. An intronic LINE-1 element insertion in the dystrophin gene aborts dystrophin expression and results in Duchenne-like muscular dystrophy in the corgi breed. Lab Invest. 2011;91(2):216–231. doi: 10.1038/labinvest.2010.146. 2011:91. Doi: 10.1038/labinvest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Thioudellet C, Blot S, Squiban P, Fardeau M, Braun S. Current protocol of a research phase I clinical trial of full-length dystrophin plasmid DNA in Duchenne/Becker muscular dystrophies. Part I: rationale. Neuromuscul Disord. 2002;12 Suppl 1:S49–S51. doi: 10.1016/s0960-8966(02)00082-2. [DOI] [PubMed] [Google Scholar]
- 141.Braun S. Naked plasmid DNA for the treatment of muscular dystrophy. Curr Opin Mol Ther. 2004;6(5):499–505. [PubMed] [Google Scholar]
- 142.McClorey G, Moulton HM, Iversen PL, Fletcher S, Wilton SD. Antisense oligonucleotide-induced exon skipping restores dystrophin expression in vitro in a canine model of DMD. Gene Ther. 2006;13(19):1373–1381. doi: 10.1038/sj.gt.3302800. [DOI] [PubMed] [Google Scholar]
- 143.Spitali P, Rimessi P, Fabris M, et al. Exon skipping-mediated dystrophin reading frame restoration for small mutations. Hum Mutat. 2009;30(11):1527–1534. doi: 10.1002/humu.21092. [DOI] [PubMed] [Google Scholar]
- 144.Yokota T, Lu QL, Partridge T, et al. Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs. Ann Neurol. 2009;65(6):667–676. doi: 10.1002/ana.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yue Y, Ghosh A, Long C, et al. A single intravenous injection of adeno-associated virus serotype-9 leads to whole body skeletal muscle transduction in dogs. Mol Ther. 2008;16(12):1944–1952. doi: 10.1038/mt.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kornegay JN, Li J, Bogan JR, et al. Widespread muscle expression of an AAV9 human mini-dystrophin vector after intravenous injection in neonatal dystrophin-deficient dogs. Mol Ther. 2010;18(8):1501–1508. doi: 10.1038/mt.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang Z, Kuhr CS, Allen JM, et al. Sustained AAV-mediated dystrophin expression in a canine model of Duchenne muscular dystrophy with a brief course of immunosuppression. Mol Ther. 2007;15(6):1160–1166. doi: 10.1038/sj.mt.6300161. [DOI] [PubMed] [Google Scholar]
- 148.Wang Z, Allen JM, Riddell SR, et al. Immunity to adeno-associated virus-mediated gene transfer in a random-bred canine model of Duchenne muscular dystrophy. Hum Gene Ther. 2007;18(1):18–26. doi: 10.1089/hum.2006.093. [DOI] [PubMed] [Google Scholar]
- 149.Yue Y, Shin JH, Duan D. Whole body skeletal muscle transduction in neonatal dogs with AAV-9. Methods Mol Biol. 2011;709:313–329. doi: 10.1007/978-1-61737-982-6_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zucconi E, Valadares MC, Vieira NM, et al. Ringo: discordance between the molecular and clinical manifestation in a golden retriever muscular dystrophy dog. Neuromuscul Disord. 2010;20(1):64–70. doi: 10.1016/j.nmd.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 151.Ambrósio CE, Valadares MC, Zucconi E, et al. Ringo, a golden retriever muscular dystrophy (GRMD) dog with absent dystrophin but normal strength. Neuromuscul Disord. 2008;18(11):892–893. doi: 10.1016/j.nmd.2008.06.385. [DOI] [PubMed] [Google Scholar]
- 152.Bretag A. Too much hype, not enough hope: are balanced reporting and proper controls too much to expect from therapeutic studies in animal models of neuromuscular diseases that presage clinical trials in humans? Neuromuscul Disord. 2007;17(3):203–205. doi: 10.1016/j.nmd.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 153.Morris G, Man N, Sewry CA. Monitoring duchenne muscular dystrophy gene therapy with epitope-specific monoclonal antibodies. Methods Mol Biol. 2011;709:39–61. doi: 10.1007/978-1-61737-982-6_3. [DOI] [PubMed] [Google Scholar]
- 154.Bostick B, Yue Y, Duan D. Phenotyping cardiac gene therapy in mice. Methods Mol Biol. 2011;709:91–104. doi: 10.1007/978-1-61737-982-6_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hakim CH, Li D, Duan D. Monitoring murine skeletal muscle function for muscle gene therapy. Methods Mol Biol. 2011;709:75–89. doi: 10.1007/978-1-61737-982-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dellorusso C, Crawford RW, Chamberlain JS, Brooks SV. Tibialis anterior muscles in mdx mice are highly susceptible to contraction-induced injury. J Muscle Res Cell Motil. 2001;22(5):467–475. doi: 10.1023/a:1014587918367. [DOI] [PubMed] [Google Scholar]
- 157.Yokota T, Takeda S, Lu QL, Partridge TA, Nakamura A, Hoffman EP. A renaissance for antisense oligonucleotide drugs in neurology: exon skipping breaks new ground. Arch Neurol. 2009;66(1):32–38. doi: 10.1001/archneurol.2008.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Palmucci L, Doriguzzi C, Mongini T, Restagno G, Chiadò-Piat L, Maniscalco M. Unusual expression and very mild course of Xp21 muscular dystrophy (Becker type) in a 60-year-old man with 26 percent deletion of the dystrophin gene. Neurology. 1994;44(3 Pt 1):541–543. doi: 10.1212/wnl.44.3_part_1.541. [DOI] [PubMed] [Google Scholar]
- 159.Tuffery-Giraud S, Saquet C, Thorel D, et al. Mutation spectrum leading to an attenuated phenotype in dystrophinopathies. Eur J Hum Genet. 2005;13(12):1254–1260. doi: 10.1038/sj.ejhg.5201478. [DOI] [PubMed] [Google Scholar]
- 160.Fanin M, Freda MP, Vitiello L, Danieli GA, Pegoraro E, Angelini C. Duchenne phenotype with in-frame deletion removing major portion of dystrophin rod: threshold effect for deletion size? Muscle Nerve. 1996;19(9):1154–1160. doi: 10.1002/mus.880190902. [DOI] [PubMed] [Google Scholar]
- 161.Duan D. Challenges and opportunities in dystrophin-deficient car- diomyopathy gene therapy. Hum Mol Genet. 2006;15(Spec No 2):R253–R261. doi: 10.1093/hmg/ddl180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Anderson JL, Head SI, Rae C, Morley JW. Brain function in Duchenne muscular dystrophy. Brain. 2002;125(Pt 1):4–13. doi: 10.1093/brain/awf012. [DOI] [PubMed] [Google Scholar]
- 163.Bostick B, Yue Y, Long C, et al. Cardiac expression of a minidystrophin that normalizes skeletal muscle force only partially restores heart function in aged Mdx mice. Mol Ther. 2009;17(2):253–261. doi: 10.1038/mt.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bostick B, Yue Y, Duan D. Gender influences cardiac function in the mdx model of Duchenne cardiomyopathy. Muscle Nerve. 2010;42(4):600–603. doi: 10.1002/mus.21763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Desguerre I, Christov C, Mayer M, et al. Clinical heterogeneity of duchenne muscular dystrophy (DMD): defnition of sub-phenotypes and predictive criteria by long-term follow-up. PLoS One. 2009;4(2):e4347. doi: 10.1371/journal.pone.0004347. [DOI] [PMC free article] [PubMed] [Google Scholar]