Abstract
Nuclear actin and actin-related proteins (Arps) are integral components of various chromatin-remodelling complexes. Actin in such nuclear assemblies does not form filaments but associates in defined complexes, for instance with Arp4 and Arp8 in the INO80 remodeller. To understand the relationship between nuclear actin and its associated Arps and to test the possibility that Arp4 and Arp8 help maintain actin in defined states, we structurally analysed Arp4 and Arp8 from Saccharomyces cerevisiae and tested their biochemical effects on actin assembly and disassembly. The solution structures of isolated Arp4 and Arp8 indicate them to be monomeric and the crystal structure of ATP–Arp4 reveals several differences to actin that explain why Arp4 does not form filaments itself. Remarkably, Arp4, assisted by Arp8, influences actin polymerization in vitro and is able to depolymerize actin filaments. Arp4 likely forms a complex with monomeric actin via the barbed end. Our data thus help explaining how nuclear actin is held in a discrete complex within the INO80 chromatin remodeller.
Keywords: actin-related proteins, chromatin-remodelling, INO80 complex, nuclear actin, structural biology
Introduction
Nuclear actin has emerging functions in a variety of DNA associated processes. It is an integral part of several chromatin-remodelling complexes, for example, the yeast INO80 complex (Shen et al, 2000), the yeast SWR1 complex (Krogan et al, 2003) or the mammalian BAF complex (Zhao et al, 1998), suggesting an important role in chromatin dynamics. Moreover, actin binds transcription factors and ribonucleoprotein complexes (Kukalev et al, 2005), is required for optimal transcriptional activity of all three RNA polymerases (Franke, 2004), and has been implicated in the movement of nuclear loci (Chuang et al, 2006).
In the cytoplasm, actin is found predominantly in two forms: monomeric G-actin and filamentous F-actin with a dynamic interchange between both, regulated by a host of different actin-binding proteins (Campellone and Welch, 2010). Although canonical F-actin has not been observed within the nucleus by conventional methods, there is growing evidence that distinct filamentous forms of actin do indeed have important roles in the nucleus (Hofmann et al, 2001; Krauss et al, 2003; Pendleton et al, 2003; Bohnsack et al, 2006; McDonald et al, 2006). The major form of nuclear actin, however, seems to be as a monomeric component of different nuclear complexes. How actin is integrated into those complexes in a discrete monomeric state is poorly understood.
One class of proteins that regulates actin dynamics in the cytoplasm and is also found in the nucleus are actin-related proteins (Arps; Dion et al, 2010). Arps are structurally similar to actin and comprise 10 family members in S. cerevisiae (Poch and Winsor, 1997), with Arp1 to Arp3 and Arp10 being mainly cytoplasmic and Arp4 to Arp9 mainly nuclear. The Arp2/3 complex for instance organizes cytoplasmic actin filaments into branched networks (Pollard, 2007), but also has effects on transcription by RNA polymerase II (Wu et al, 2006; Yoo et al, 2007). The role of classical nuclear Arps, however, is less clear. Arp4 is the most conserved ‘primordial’ Arp and found in the largest number of nuclear complexes (Dion et al, 2010). Arp4 occurs together with actin in human BAF, PBAF and Tip60 complexes (Clapier and Cairns, 2009), is an essential gene in yeast (Harata et al, 1994) and has been implicated in histone binding (Galarneau et al, 2000). In addition to being associated with higher molecular weight assemblies, a species of uncomplexed Arp4 was found in yeast cell extracts as well, indicating a role of Arp4 independently of chromatin-remodelling machines (Sunada et al, 2005).
Within the INO80 chromatin remodeller, Arp4 forms a discrete and stable subcomplex with Arp8, actin and the helicase-SANT-associated (HSA) domain of Ino80p (Szerlong et al, 2008). INO80 is a multifunctional complex that evicts and/or slides histones at sites of DNA double-strand breaks, transcription and replication forks (Morrison et al, 2004; van Attikum et al, 2004; Papamichos-Chronakis and Peterson, 2008). Similarly, a subcomplex containing Arp4, actin and the HSA domain has been identified in the yeast histone variant exchange factor SWR1 a remodeller highly related to INO80 and human SWI–SNF (the mammalian Arp4 homologue BAF53-actin-HSA) (Szerlong et al, 2008). Altogether, this suggests that the complex of Arp4, actin and the HSA domain is a conserved functional module. In INO80, this module additionally contains Arp8. Although Arp8 has been mainly characterized as an INO80-specific subunit (Shen et al, 2000), it also functions independently of INO80. Specifically, human Arp8 persists on chromatin during mitosis and its depletion increases chromosome misalignment independent from other components of the INO80 complex (Aoyama et al, 2008).
To shed light on the enigmatic structures and biochemical functions of nuclear Arps and their interactions with actin, we determined the crystal structure of S. cerevisiae Arp4, as well as the X-ray scattering solution structures of Arp4 and Arp8. The crystal structure can explain why Arp4, in contrast to highly related conventional actin, does not form filaments, and reveals that Arp4 binds to ATP. To uncover the biochemical relationship of Arp4, Arp8 and actin, we biochemically analysed the effects of Arp4 and Arp8 on actin filament dynamics. We observe that both proteins affect the polymerization and depolymerization of actin albeit in different ways. While Arp4 appears to interact with the barbed end of monomeric actin, thereby preventing the nucleation of actin filaments, Arp8 efficiently sequesters actin monomers with slow kinetics by interaction with a region of the monomer distinct from the barbed end. Most notably, Arp4 and Arp8 cooperate in monomer sequestering, consistent with formation of a discrete Arp4–Arp8–actin module within the INO80 complex. In summary, we show that Arp4 and Arp8 evidently stabilize monomeric actin within chromatin-modifying complexes. Potential mechanistic roles in the context of chromatin remodellers are discussed.
Results
Crystal structure of Arp4 reveals the actin fold with characteristic loop insertions and deletions
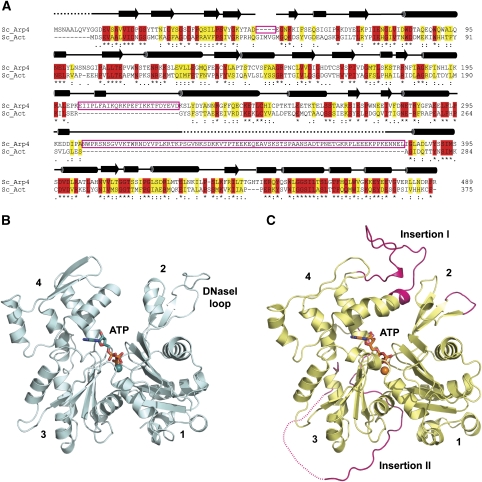
To provide a first framework for nuclear Arps and to characterize similarities and key differences between S. cerevisiae Arp4 and actin (Figure 1A), we crystallized S. cerevisiae Arp4 and determined its crystal structure to 3.4 Å resolution. Crystals belonged to space group P61 and contained four molecules per asymmetric unit. We derived phases by molecular replacement using yeast actin (pdb code: 1YAG) as search model and obtained interpretable electron density for the entire Arp4 polypeptide except the nine N-terminal amino acids and 49 amino acids within Insertion II of Arp4 (Ser328–Lys377). The model was refined to an R-value of 19.2% (R-free 22.1%) with good stereochemistry (Table I). Additional density was clearly visible in the centre of the four subdomains and could be interpreted as a bound ATP molecule with one coordinated metal ion.
Figure 1.
Structure and sequence comparisons between Arp4 and actin. (A) Sequence alignment between S. cerevisiae Arp4 (top) and actin (bottom). Identical residues are shaded in red and similar residues in yellow. The main differences between the two proteins: shortening of the DNaseI loop, Insertion I (25 residues) and Insertion II (80 residues) are highlighted by pink boxes. The secondary structure of Arp4 is displayed on the top of the alignment; residues missing in the electron density are depicted as dashed lines. (B) Crystal structure of S. cerevisiae actin (pdb: 1YAG). The four subdomains are numbered and the DNaseI loop is labelled. ATP is represented in sticks and the metal ion is displayed as a sphere. (C) Crystal structure of S. cerevisiae Arp4 (pdb: 3QB0). The two insertions are labelled and shown in pink, as is the shortened DNaseI loop. The 49 amino acids within Insertion II that are not visible in the electron density are indicated by the dashed line. ATP is represented in sticks and the metal ion is displayed as a sphere.
Table 1. Data collection and refinement statistics.
| Wt S. cerevisiae Arp4 | |
|---|---|
| Data collection | |
| Space group | P61 |
| Cell dimensions | |
| a, b, c (Å) | 119.3, 119.3, 396.7 |
| α, β, γ (deg) | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50–3.4 (3.61–3.40)a |
| Rmerge (%) | 16.5 (38.6) |
| I/σI | 11.6 (4.4) |
| Completeness (%) | 99.5 (99.0) |
| Redundancy | 5.7 (5.3) |
| Refinement | |
| Resolution (Å) | 50.0–3.4 |
| No. of reflections | 43 013 |
| Rwork/Rfree | 19.2/22.1 |
| No. of atoms | |
| Protein | 13657 |
| Ligand/ion | 124 |
| Water | 0 |
| B-factors | |
| Protein | 63.3 |
| Ligand/ion | 52.6 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (deg) | 0.805 |
| aValues in parentheses are for highest resolution shell. | |
Arp4 displays the typical actin fold with a central nucleotide-binding cleft between subdomains one and two on one side and subdomains three and four on the other side (Figure 1B). Despite its high overall structural similarity to conventional actin there are important differences on the level of primary structure, namely loop insertions and deletions that lead to consequences in the tertiary structure and molecular surface of Arp4 compared with actin. Two large insertions can be identified with a length of 25 and 80 amino acids, respectively. In a structural alignment Arp4's smaller Insertion I expands amino acids 196–203 in subdomain four of actin and its larger Insertion II between actins' amino acids 268 and 272 bridges subdomains three and four (Figure 1A). Insertion I lies mainly atop the pointed end of Arp4 whereas Insertion II winds itself around domain three on the backside of the Arp4 molecule (Figure 1C). Additionally, the DNaseI-binding loop is substantially shorter in Arp4 compared with actin.
Solution structures of Arp4 and Arp8
To verify the crystal structure in solution and to confirm lack of polymerization, we determined the solution structures of Arp4 and Arp8 by SAXS measurements. The Kratky-plot of Arp4 shows a bell-shaped curve, indicating that the protein is properly folded (Supplementary Figure S1). Ab initio shape reconstructions reveal a compact protein with a small protrusion. The reconstructed envelopes display the characteristic flatness typical for actin. The crystal structure can be nicely docked into the envelope with the additional protrusion allowing for the accommodation of the 49 unstructured amino acids of Insertion II, which could not be located in the electron density (Figure 2A). The theoretical SAXS curve of the Arp4 structure calculated with CRYSOL is in good agreement with the measured SAXS data further indicating that the crystal structure is similar to the structure in solution (Supplementary Figure S1). The apparent molecular weight determined by SAXS is about 51 kDa, which corresponds well to the actual mass of an Arp4 monomer (56 kDa). The protein concentrations in the SAXS measurements (up to 130 μM) lie well above the critical concentration, at which free actin monomers start to polymerize to actin filaments, which is about 0.1 μM at the barbed end and 0.7 μM at the pointed end of a filament (Bonder et al, 1983). This indicates that ATP–Arp4 does not form actin-like filaments.
Figure 2.
Solution structures of Arp4 and Arp8. (A) Overlay of the final averaged ab initio shape reconstruction of Arp4 derived by SAXS experiments (blue envelope) with the docked crystal structure of Arp4 (yellow). Front and side views indicate a good fit between the crystal and the solution structure. The likely position of the disordered 49 amino acids of Insertion II within the solution structure is indicated by the dashed line. (B) Overlay of the final averaged ab initio shape reconstruction of full-length Arp8 (blue envelope) and N-terminally truncated Arp8 lacking the first 244 amino acids (red envelope) derived from SAXS experiments. The crystal structure of yeast actin (pdb: 1YAG, cyan) is docked into the envelopes for comparison. Front and side views indicate that the core actin fold fits into both solution structures of Arp8. Additional density can be attributed to insertions present in Arp8 compared with actin. The large N-terminus of Arp8 (amino acids 1–244) seems to form an extended protrusion consistent with the secondary structure prediction for it to be mainly unstructured.
We next analysed the solution structure and oligomerization state of S. cerevisiae Arp8, which resides in a complex with Arp4 and actin (Supplementary Figure S2). No aggregation effects or changes in oligomerization at higher concentrations (up to 80 μM) were detected. The apparent molecular weight determined by SAXS was about 105 kDa, which corresponds to the mass of an Arp8 monomer (101 kDa). Ab initio shape reconstruction reveals Arp8 to be more elongated than Arp4. The thicker part of the structure, however, is perfectly able to accommodate the actin fold present in Arp8 (Figure 2B). SAXS measurements on an N-terminally truncated Arp8 construct lacking the first 244 amino acids reveal a much more compact shape. This is consistent with the prediction that those 244 amino acids are highly unstructured and confirms that the truncated version of Arp8 is still properly folded (Figure 2B). For a sequence alignment of Arp8 with actin, see Supplementary Figure S3.
In summary, SAXS data indicate that both Arp4 and Arp8 are monomeric under the conditions used, consistent with their approximately equal stoichiometric complex formation with actin and the HSA domain within the INO80 complex.
ATP is tightly bound to Arp4
ATP as well as a metal ion are clearly defined in the electron density in the nucleotide-binding cleft of Arp4 (Figure 3A), consistent with previous biochemical observations (Sunada et al, 2005). The presence of the nucleotide in the Arp4 structure is still somewhat surprising since neither ATP nor divalent metal ions were present during the purification process or the crystallization set-ups and must have been co-purified from the expression host. This argues not only for strong ATP binding but also for weak ATPase activity of Arp4 alone.
Figure 3.
ATP binding by Arp4. (A) Stereo image of the simulated annealing difference omit map calculated for the ATP molecule and the metal ion using CNS. Electron density is clearly present between the four domains of Arp4 (yellow), indicating binding of the ATP and metal ligands (orange sticks and sphere). The map is displayed as blue mesh and contoured at 1.0 σ. (B) Detailed view of the ATP nucleotide bound by actin (pdb: 2HF4). The protein is displayed in cyan ribbon and important residues are represented as sticks and labelled. (C) Detailed view of the ATP nucleotide bound by Arp4 (in yellow). Important residues are represented as sticks and labelled. Note that Tyr 24 stacks on the ribose of ATP and Asp163 forms a hydrogen bond with Ser23 leading to tighter closure of the two nucleotide-binding loops P1 and P2. Together with His162 and helix 227–246 (in pink) the nucleotide is more strongly shielded from the solvent as compared with actin.
The divalent cation is coordinated by the conserved Asp159Arp4 (corresponding to Asp154actin) (Figure 3B and C). The side chain of Ser23Arp4 (corresponding to Ser14actin) is rotated outward to accommodate the third phosphate moiety of ATP, resulting in a conformation of the sensor loop (residues 75–77 in Arp4) similar to that observed in the structure of non-polymerizable ATP-actin (Rould et al, 2006). Interestingly, Gly15actin in the P1 nucleotide-clamping loop of actin is substituted by Tyr24Arp4. This tyrosine stacks on the ribose moiety of the bound ATP leading to an even tighter closure of the P1 loop around ATP in Arp4 as compared with actin. Accordingly, the P2 nucleotide-clamping loop has Gly158actin and Asp157actin substituted by Asp163Arp4 and His162Arp4, respectively. Asp163Arp4 forms a hydrogen bond with Ser23Arp4 leading to a tighter closure of the P2 loop around the phosphate moieties in Arp4 (Figure 3B and C). Together with His162Arp4, the P1 and P2 nucleotide-binding loops in Arp4 seem to be shielding the phosphate residues of the nucleotide more tightly from the environment than in actin. Furthermore, an α-helix of Arp4 (residues 227–246 in Arp4) is longer by one and a half turns leading to a positional shift of this helix further closing the nucleotide-binding cleft from the top. A surface representation of the two proteins nicely illustrates that the nucleotide is more accessible to the environment in actin compared with Arp4 (Supplementary Figure S4).
Sequence comparison shows that His161actin and Gln137actin are substituted by Ser166Arp4 and Thr142Arp4, respectively. According to mutational and structural studies, His161actin takes part in the ATP hydrolysis cycle of actin by positioning a nucleophilic water (Vorobiev et al, 2003; Martin et al, 2006) and the mutation of Gln137actin to alanine reduces actin's rate of ATP hydrolysis (Iwasa et al, 2008). Taken together, the absence of a catalytic histidine and glutamine and the tighter closure of the P1 and P2 nucleotide-binding loops of Arp4 around the phosphate moieties might lead to strong ATP binding and the apparent absence of ATPase activity of Arp4.
The intimate binding of ATP to Arp4 is further emphasized by our observation that ATP-binding mutants (S23DArp4, D159GArp4 and G161DArp4) were found to be insoluble upon expression in insect cells. This is analogous to the instability of actin upon removal of the divalent cation and the nucleotide by EDTA (Altschuler et al, 2005) and indicates that ATP (or ADP) is required for proper folding of Arp4.
The structure of Arp4 explains why it is unable to form actin-like filaments
Our observation that Arp4 is apparently polymerization incompetent can be understood by analysing its structure in the context of current models of F-actin (Oda et al, 2009). In the filament, actin residues 283–294 at the barbed end of one monomer are enclosed by residues 61–65, 200–208 and 241–247 at the pointed end of the preceding monomer. Additionally, the DNaseI-binding loop (residues 38–49 in actin) at the pointed end is extended to contact the hydrophobic groove between subdomains one and three at the barbed end of a following monomer (Figure 4A). On top of these intra-strand contacts of the two-stranded filament additional inter-strand contacts are formed. Specifically, the C-terminus of the actin α-helix 191–199 contacts the amino terminus of the actin α-helix 110–115 in the neighbouring strand and the hydrophobic plug (residues 265–271 in actin) contacts four regions in the opposite strand, including actin residues 201–203 and 39–42 of one subunit and actin residues 170–174 and 285–286 of an adjacent subunit (Figure 4B).
Figure 4.
Arp4 is not capable of forming actin-like filaments. (A) Model of two intra-strand actin monomers (n and n+2) preceding each other within the actin filament (Oda et al, 2009) (two shades of cyan). Important residues for forming intra-strand contacts are highlighted in dark green and labelled. Note the extended DNaseI loop of the bottom monomer reaching into the hydrophobic groove between subdomains one and three of the following monomer. (B) Model of inter-strand interactions between two preceding actin monomers (n and n+2) with a third monomer from the second strand (n+1) of the double-stranded actin filament (Oda et al, 2009) (three shades of cyan). Important residues for forming inter-strand contacts are highlighted in dark green and labelled. Note the hydrophobic plug region extending from the n+1 monomer to contact both opposing actin monomers. (C) Overlay between actin (cyan) and Arp4 (yellow) detailing differences at the pointed end. Highlighted in dark green and labelled are actin residues important for forming contacts within the actin filament (see Figure 4A). In pink are changes present in Arp4 that render those contacts largely impossible (Insertion I and shortened DNaseI loop). (D) View of the ‘backside’ of actin (cyan) and Arp4 (yellow). The hydrophobic plug region of actin is coloured in dark green and labelled. Insertion II (in pink; dashed line represents the unstructured 49 residues of Insertion II) expands this region in Arp4 and Trp315 (represented in sticks and labelled) caps α-helix 110–115 in actin, rendering important inter-strand contacts within the actin filament impossible (see Figure 4B).
Taking a closer look at the corresponding regions in Arp4 readily reveals that most of the mentioned contacts are not possible in Arp4 due to the presence of the Insertions I and II, as well as the shortening of the DNaseI loop. Insertion I (residues 199–226 in Arp4) forms a mainly unstructured loop on top of the pointed end of Arp4. In addition to the inserted residues, α-helices at both ends of this loop differ in length in Arp4 as compared with actin (see Figure 3C). Intriguingly, this insertion coincides with several stretches of amino acids which are important for forming contacts within the actin filament (Oda et al, 2009). Specifically, it masks region 200–208 of actin by positioning part of the loop (residues 201–209 in Arp4) above of this region and by elongating the actin helix 202–216. Moreover, residues 241–247 of actin are also masked by a stretch of this loop (residues 210–218 in Arp4), which positions itself above this interaction surface prohibiting it to form contacts with an adjacent actin monomer (Figure 4C).
The other substantial difference at the pointed end of Arp4 compared with actin is the shortened DNaseI-binding loop. This unstructured loop reaches into the hydrophobic pocket present at the barbed end of an adjacent actin monomer. It is shorter by about five residues in Arp4 (residues 47–54 in Arp4) impairing the contact to the barbed end of the adjacent monomer (Figure 4C).
Insertion II (amino acids 300–382 in Arp4) was partially disordered. Whereas residues 300–327 and 378–382 were ordered in the crystal, the position of residues 328–377 can be inferred from the SAXS envelope (see above). The loop emanates from domain four and packs on the ‘backside’ of Arp4, mainly contacting domains three and four before it enters the actin fold again in domain three (Figure 4D). It coincides with the hydrophobic plug region of actin (residues 265–274 in actin) which is predicted by the filament model (Oda et al, 2009) and mutational studies (Shvetsov et al, 2002) to be involved in inter-strand contacts. Very likely the expansion of this region in Arp4 disturbs the ability to form the proper inter-strand interactions within actin filaments. In addition, Insertion II caps the actin α-helix 110–115, another intra-stand interacting element, by stacking of Trp315Arp4 on top of that helix (Figure 4D).
Taken together, the surface properties of Arp4 are very different compared with actin. As a consequence, key interactions which are important for intra-strand filament contacts in actin are no longer possible. Additionally, crucial inter-strand interactions, especially through the hydrophobic plug region of actin are disturbed, nicely explaining why Arp4 itself is monomeric.
Arp4 inhibits actin polymerization by binding to monomers
Given that Arp4, Arp8 and actin form a complex together with the HSA domain of the Ino80 subunit of the INO80 chromatin remodeller (Szerlong et al, 2008) we addressed the question if Arp4 and/or Arp8 can directly interact with actin. Since actin exists in a dynamic equilibrium between F- and G-actin, we tested the effect of Arp4 and Arp8 on actin polymerization and depolymerization.
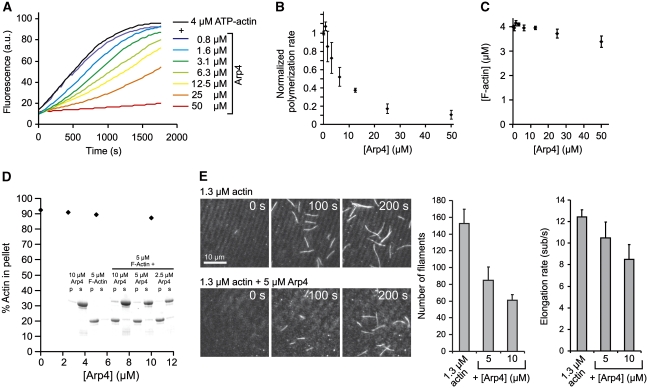
First, we analysed the spontaneous polymerization of monomeric, pyrene-labelled Mg2+-ATP-actin in the presence of Arp4 by measuring the increase in pyrene fluorescence. Arp4 decreased the polymerization rate of actin in a concentration-dependent manner (Figure 5A and B). However, it did not markedly sequester actin monomers at steady state, resulting in nearly constant levels of F-actin after 16 h (Figure 5C). Co-sedimentation analyses with F-actin and Arp4 confirmed that Arp4 does not reduce the amount of actin filaments, and moreover showed that Arp4 does not bind to actin filaments (Figure 5D). This suggests that it interacts with monomers but without efficiently sequestering them, reminiscent of the G-actin-binding protein profilin (Korenbaum et al, 1998).
Figure 5.
Arp4 inhibits F-actin assembly. (A) Pyrene assays of actin polymerization. In all, 4 μM Mg2+-ATP-actin was polymerized by addition of KMEI buffer and the Arp4 concentrations indicated, and the increase in pyrene fluorescence was detected over time. (B) Normalized polymerization rates derived from three independent experiments equivalent to (A) show a dose-dependent inhibition of polymerization. (C) Steady-state F-actin fluorescence from experiments equivalent to (A) indicate that Arp4 does not sequester monomers. (D) Co-sedimentation assays of 5 μM Mg2+-ATP-actin polymerized in the presence of the Arp4 concentrations indicated shows that Arp4 neither sequesters monomers nor binds to F-actin. (E) In vitro TIRF microscopy of single actin filaments growing in the presence and absence of Arp4 at time points 0, 100 and 200 s. In all, 1.3 μM Mg2+-ATP-G-actin was polymerized in TIRF buffer and the elongation rates and numbers of filaments were analysed. Addition of Arp4 markedly decreases the number of growing filaments and has a slight effect on the elongation rate as well.
To verify the direct interaction of Arp4 with actin, we used surface plasmon resonance (SPR) to test the binding of Arp4 to actin immobilized on the surface of a sensor chip (Supplementary Figure S5). SPR clearly indicates that Arp4 specifically binds to actin and the KD of the interaction can be estimated to be around 2 μM.
In order to gain a more detailed picture of the inhibitory effects of Arp4 on actin assembly, we employed in vitro total internal reflection fluorescence (TIRF) microscopy on Oregon-Green (OG)-labelled, polymerizing actin filaments. We found that Arp4 decreased both the elongation rate of actin filaments as well as the total number of spontaneously formed filaments (Figure 5E). The effect on filament nucleation was more pronounced, since 5 μM Arp4 were already sufficient to reduce the number of growing filaments by 50%, while filament elongation rates were only slightly decreased. Thus, we propose that Arp4, like profilin, interacts with monomeric actin and that this interaction inhibits primarily the formation of F-actin nuclei.
Effects of Arp4 on the equilibrium of G- and F-actin depends on nucleotides
To further analyse the effects of Arp4 on the G-/F-actin equilibrium, we determined the critical concentration of F-actin with free (CCrit(free)) or CapZ-capped barbed ends (CCrit(capped)). CCrit(free) of F-actin alone was 0.1 μM, while it was shifted to 0.6 μM when the capping protein CapZ was present (Figure 6A). In all, 20 μM Arp4 did not change CCrit(free), which is in line with the observation that Arp4 did not sequester monomers at steady state (Figure 5C and D). However, it shifted CCrit(capped) to ∼2 μM in the presence of CapZ, suggesting that it interferes with monomer addition to the pointed end of filaments by binding to the barbed end of monomers (Figure 6A). Moreover, these assays served to calculate the KD for the Arp4/G-actin interaction to be ∼8 μM, which is in the same range as the results obtained from SPR. Most notably, analogous effects on the critical concentration of free and capped filaments were again previously observed for the small G-actin-binding protein profilin, which binds the barbed end of the monomer and thus prevents monomer addition to the pointed end of the filament when barbed ends are capped (Korenbaum et al, 1998).
Figure 6.
Effects of Arp4 on the G-/F-actin equilibrium. (A) Critical concentration plot of F-actin in the presence and absence of Arp4 and CapZ. F-actin at different concentrations, either with free barbed ends or with CapZ-capped barbed ends was incubated in the presence and absence of 20 μM Arp4. Arp4 only increases the critical concentration of capped filaments. (B) Time course of the effects of Arp4 on the G-/F-actin equilibrium. Pyrene fluorescence of F-actin was detected for 200 s. Subsequently, KMEI buffer alone or supplemented with Arp4 or LatA was added, and the decrease in fluorescence was followed for 10 000 s. The smaller timescale highlights fast, instant depolymerization upon addition of Arp4 (right). (C) Effect of Arp4 on the G-/F-actin equilibrium when barbed ends are capped. The experiments were performed as in (B) with the exception that CapZ was added. The smaller timescale illustrates that rapid Arp4-mediated F-actin disassembly is abolished when barbed ends were capped (right). (D) Effect of Arp4 on the F-/G-actin equilibrium in the absence of ATP. The experiments were performed as in (B). Buffers contained ADP instead of ATP, residual ATP was removed by adding hexokinase and glucose before the experiments. The smaller timescale highlights immediate and complete depolymerization of ADP-actin by Arp4 (right). (E) ADP-actin assembly assay. In all, 10 μM Mg2+-ADP-actin was polymerized in the presence of the Arp4 concentrations indicated. (F) Normalized polymerization rates derived from (E) and Figure 5A are shown for comparison. The inhibition of actin assembly by Arp4 is much more efficient for ADP-actin. (G) Addition of ATP to Arp4-inhibited ADP-Mg2+-actin polymerization restored actin assembly.
To further investigate the effect of Arp4 on filament elongation, we performed pyrene assays with CapZ-capped filaments where only pointed end elongation of the filaments is possible. In this case, we could monitor a concentration-dependent reduction of pointed end elongation through the addition of Arp4 (Supplementary Figure S6), supporting the results of the critical concentration assay. Therefore, these results suggest that Arp4 interacts with the barbed end of the monomer.
Next, we analysed the time course of Arp4-mediated sequestering on F-actin in more detail by employing pyrene assays with pre-formed F-actin. Surprisingly, we found that addition of 10 μM Arp4 to 2 μM F-actin with free barbed ends triggered an immediate disassembly of actin filaments indicated by the rapid loss of pyrene fluorescence. The initial rate of this disassembly reaction was comparable to that of LatrunculinA (LatA)-mediated F-actin disassembly, which was used as a control (Figure 6B). LatA was shown to depolymerize F-actin completely by binding to the pointed end of monomers with nM affinities preventing both re-incorporation into the filament, and nucleotide exchange (Coue et al, 1987; Yarmola et al, 2000). In contrast to LatA, Arp4-mediated F-actin disassembly was not complete, but stopped after ∼100 s. Subsequently, a new G-/F-actin equilibrium was slowly restored, suggesting that Arp4 binding to the barbed end of the monomer does not inhibit its re-incorporation into the filament, consistent with the findings above that Arp4 does not efficiently sequester monomers under these conditions (Figure 5C and D). When using F-actin with CapZ-capped barbed ends, we found that rapid Arp4-mediated disassembly was inhibited, but now F-actin was depolymerized more slowly, corroborating the results from the critical concentration analysis (Figure 6C).
The remaining question was why Arp4 rapidly depolymerized uncapped F-actin only to some extend? Since F-actin is composed of ADP-actin, we hypothesized that Arp4 efficiently removes ADP-actin from the barbed end, but that it still allows exchange of ADP to ATP of the bound monomer. In order to test this hypothesis, we performed equivalent experiments in KMEI/Mg-exchange buffer containing ADP instead of ATP and removed ATP entirely from the mixture by adding hexokinase and glucose before the experiment. We found that Arp4 now completely sequestered monomers, resulting in a depolymerization rate comparable to LatA-mediated filament disassembly (Figure 6D). To further verify different binding of Arp4 to ADP- and ATP-actin, we performed polymerization assays with monomeric Mg2+-ADP-actin, and found that the inhibitory effect of Arp4 on the polymerization of ADP-actin was markedly enhanced when compared with ATP-actin (Figure 6E and F). Most strikingly, addition of ATP during the polymerization process released the strong inhibition of ADP-actin polymerization by Arp4 and led to enhanced polymerization, supporting that Arp4 binding to the monomer does not interfere with nucleotide exchange and suggesting that the affinity of Arp4 for ADP-actin is higher than for ATP-actin (Figure 6G). These data indicate that Arp4 primarily binds and sequesters ADP-actin via a barbed end interaction, and that this interaction does not interfere with nucleotide exchange.
Arp8 does not inhibit actin polymerization but sequesters ADP-actin
As Arp8 is part of the complex containing Arp4, actin and the HSA domain of Ino80p, we tested whether Arp8 also affects actin dynamics. In contrast to Arp4, Arp8 did not affect the polymerization kinetics of ATP-actin in pyrene assays (Figure 7A and B). TIRF assays on single filaments confirmed that Arp8 does neither change filament elongation nor nucleation (Figure 7C). Unexpectedly, and in contrast to Arp4, however, we found that Arp8 efficiently sequestered actin monomers at steady state, as assessed by fluorescence measurements and high-speed sedimentation analyses (Figure 7D and E). Consistently, 20 μM Arp8 shifted CCrit of both capped and free filaments from 0.1 and 0.6 μM, respectively, to about 1.3 μM, which corresponds to a KD of ∼16.5 μM (Figure 7F). These results indicate that Arp8 does not interact with the barbed end of actin monomers, and that it might exclusively bind and sequester actin monomers in a manner that prevents their re-incorporation into either end of the filament.
Figure 7.
Arp8 does not inhibit actin assembly but sequesters actin monomers. (A) Pyrene assays of actin polymerization. In all, 4 μM Mg2+-ATP-actin was polymerized by addition of KMEI buffer and the Arp8 concentrations indicated, and the increase in pyrene fluorescence was detected over time. (B) Normalized polymerization rates derived from three independent measurements equivalent to (A) show no inhibition of polymerization by Arp8. (C) Number of filaments and barbed end elongation rates in the presence and absence of Arp8 determined by TIRF microscopy as described in Figure 5E are not influenced by Arp8. (D) Steady-state F-actin fluorescence measurements indicate that Arp8 sequesters monomers. (E) Co-sedimentation assays of 5 μM actin polymerized in the presence of the Arp8 concentrations indicated corroborate that Arp8 sequesters monomers. (F) Critical concentration plot of F-actin in the presence and absence of Arp8 and CapZ. Conditions are as in Figure 6A. Note that the change in CCrit does not depend on the presence of CapZ. CCrit(free) and CCrit(capped) are both shifted to 1.3 μM corresponding to a KD of the Arp8–G-actin interaction of ∼16.5 μM. (G) Arp8 depolymerizes F-actin with very slow kinetics. Conditions are as in Figure 6B and D. Note that removal of ATP from the reaction mixture led to a loss of the pronounced lag-phase of Arp8-mediated F-actin disassembly. (H) ADP-actin assembly assay. In all, 10 μM Mg2+-ADP-actin was polymerized in the presence of the Arp8 concentrations indicated. The amount of F-actin was reduced with increasing amounts of Arp8.
SPR experiments to verify the direct interaction of Arp8 with actin indicate that Arp8 indeed binds to actin (Supplementary Figure S7). Due to problems with some unspecific binding to the reference channels of the chips, however, we were not able to determine an accurate KD.
Analysis of the kinetics of Arp8-mediated sequestration of F-actin revealed that while no obvious sequestration of monomers was observed immediately after Arp8 addition, F-actin depolymerized after a lag-phase of ∼2500 s with very slow kinetics. Conversion of ATP to ADP in the reaction mixture by adding hexokinase and glucose before the experiment resulted in loss of the lag-phase of this reaction, leading to instant depolymerization of F-actin (Figure 7G). However, the kinetics of the Arp8-mediated depolymerization were significantly slower compared with Arp4-mediated sequestration of ADP-actin monomers in the absence of ATP (Figure 6D), and the depolymerization reaction was not complete, in line with a lower affinity of Arp8 to ADP-actin monomers. Consistent with very slow binding kinetics of Arp8 to ADP-actin, we detected no reduction in the initial polymerization rate of ADP-actin in the presence of Arp8 using pyrene assays, but instead found that the amount of F-actin was reduced with increasing amounts of Arp8 (Figure 7H). Thus, we suggest that Arp8 binds ADP-actin with slow kinetics in a region of the monomer that is distinct from the barbed end and prevents its re-incorporation into either end of the filament, possibly the side of an actin molecule.
Arp4 and Arp8 synergistically inhibit actin polymerization
Our hypothesis that Arp8 and Arp4 might interact with distinct regions of the actin monomer and the findings that both proteins affect the G-/F-actin equilibrium differently prompted us to investigate whether they act in concert during actin monomer binding. Therefore, we performed pyrene assays with ATP-actin in presence of a constant concentration of 6 μM Arp4, which only slightly inhibited actin polymerization, and added different amounts of Arp8. Most notably, Arp8 enhanced the inhibitory effect of Arp4 on actin polymerization in a concentration-dependent manner by as much as three-fold (Figure 8A and B). Analysis of the polymerization reaction by TIRF microscopy showed that excess amounts of Arp8 enhanced the inhibitory effect of Arp4 on both filament nucleation and elongation, resulting in a reduction of growing filaments by ∼75% and a reduction in the filament elongation rate by ∼25% (Figure 8C). Moreover, Arp4 enhanced actin monomer sequestration by Arp8, leading to a reduction of F-actin by ∼75% after 16 h incubation as assessed by pyrene-fluorescence measurements and to a ∼50% reduction after 5 h incubation as quantified by F-actin sedimentation analyses, respectively (Figure 8D and E). A more detailed analysis of the combined effects of Arp4 and Arp8 on monomer sequestration following the drop of pyrene fluorescence of F-actin after Arp4 and Arp8 addition revealed that the effects of both proteins, namely instantaneous, fast depolymerization by Arp4 and slow depolymerization by Arp8, seem to add up to achieve an enhanced sequestration activity (Figure 8F). Thus, we propose that both proteins efficiently sequester monomeric ADP-actin in a synergistic manner. If this happens by simultaneous co-binding of actin by both proteins remains to be demonstrated.
Figure 8.
Arp4 and Arp8 synergistically sequester G-actin. (A) Pyrene assays of actin polymerization. In all, 4 μM Mg2+-ATP-actin was polymerized by addition of KMEI buffer in the presence of 6 μM Arp4 and the Arp8 concentrations indicated. (B) Arp8 enhances Arp4-mediated inhibition of polymerization. Polymerization rates were obtained from (A) and normalized to the slope of the assembly of 4 μM Mg2+-ATP-actin in presence of 6 μM Arp4. (C) Single filament elongation rates and numbers of filaments in the presence and absence of 5 μM Arp4 and 20 μM Arp8 obtained by TIRF microscopy as described in Figure 5E indicate that Arp8 increases the effect of Arp4. (D) Steady-state F-actin fluorescence experiments indicate that Arp4 increases Arp8-mediated sequestering of monomers. (E) Co-sedimentation assays of 5 μM actin polymerized in the presence of the Arp4 and Arp8 concentrations indicated. The sequestration effect of Arp8 is more pronounced in the presence of Arp4 (compare with Figure 7E). (F) Combined effects of Arp4 and Arp8 on the F-/G-actin equilibrium suggest an additive and sequential effect of fast depolymerization by Arp4 and slow depolymerization by Arp8. Experiments were performed as in Figure 6B.
Discussion
The specific presence and functional role of actin and nuclear Arps, especially the actin–Arp4 pair, in a set of chromatin-modifying complexes has been a big puzzle over the years (Dion et al, 2010). Arp4 is essential in yeast (Harata et al, 1994) and the multiple interactions of actin and its prominent role in the cytoskeleton make the analysis of actin and associated Arps in nuclear complexes so difficult.
To help reveal the structure and function of nuclear actin-related protein complexes in chromatin-remodelling enzymes, we report the structure of the nuclear actin-related protein Arp4 and studied its functional interaction with Arp8 and actin. Although Arp4 has a structure highly related to actin, it contains several loop insertions and deletions at sites where actin protomers form contacts according to the current model of F-actin (Oda et al, 2009). The different surface properties of Arp4 likely have consequences in terms of protein–protein interactions between Arp4 and actin, or classical actin-binding proteins. While our data suggest that Arp4 displays a high affinity for ADP-G-actin, it does not form filaments on its own or with actin, and the position of the insertion loops and deletions make it unlikely that Arp4 forms complexes with regulatory actin-binding proteins like profilin. Arp4 also has a more shielded ATP-binding site compared with actin, with several divergent residues that lead to formation of a stable ATP state with little if not lacking ATP hydrolysis by isolated Arp4 in solution.
We found that Arp4 rapidly depolymerizes actin filaments in biochemical assays and our combined data suggest that it does so by interacting with ADP-G-actin via the barbed end without interfering with the exchange of the actin-bound nucleotide. The inhibition of actin filament nucleation by Arp4 is therefore reminiscent of profilin, a protein that helps to regulate the structure and dynamics of actin filaments and promotes ADP to ATP exchange in G-actin (Witke, 2004).
Interestingly, we found that Arp8 acts synergistically with Arp4 in depolymerizing actin filaments. As Arp4, Arp8 and actin are found within a stable subcomplex in the INO80 remodeller, Arp4 and Arp8 might specifically stabilize monomeric actin in the INO80 complex. While our SPR measurements indicate that in the absence of the INO80 HSA domain, Arp4 and Arp8 can interact with actin only the co-expression of the three proteins together with the HSA domain leads to formation of a stable complex (Szerlong et al, 2008). Thus, the interaction between Arp4, Arp8 and actin could be stabilized by the HSA domain. In any case, the interaction of Arp4 and Arp8 with actin and their effect on actin polymerization properties suggest that nuclear Arps might have functions in regulating the properties of nuclear actin and such a role might explain why nuclear forms of actin are prominently found in monomeric states within chromatin-modifying complexes. As Arp4 and Arp8 are also implicated in histone binding (Galarneau et al, 2000; Shen et al, 2003), they might link chromatin structure and dynamics to nuclear actin regulation.
It will require further experiments to test, whether Arp4 and Arp8 not only stabilize monomeric actin within nuclear complexes but also regulate actin dynamics in the nucleus on a more general level. Especially, once the defined Arp–actin complexes are assembled within chromatin remodellers they could gain additional functions in actin regulation. In support of this, the BAF complex binds to pointed ends in vitro in the presence of phosphatidyl inositol 4/5 bisphosphate and stabilizes filaments (Rando et al, 2002). Alternatively, the apparently preferential interaction of ATP bound Arp4 with ADP-G-actin could hint that ATP/ADP could help control the interactions of proteins within the remodelling complex and be part of its regulation or functional cycle, consistent with data that show that the ATP binding and possibly hydrolysis of Arp4 can regulate interaction with the other remodeller subunits in vitro (Sunada et al, 2005).
A role of nuclear Arps in regulating nuclear actin in the context of chromatin, however, is not completely unexpected. It has recently been proposed, based on a variety of circumstantial evidence, that nuclear actin, nuclear myosin and perhaps nuclear Arps are involved in long-range organization of nuclei (Dion et al, 2010). The functionally best-understood Arps are those of the cytoplasmic Arp2/3 complex that is most prominently involved in branching of cytoplasmic actin filaments (Pollard, 2007). Like Arp2 and Arp3, Arp4 might require additional partners for its full activity. As mentioned, Arp8 does act synergistically with Arp4 and another possible factor to stimulate its function could be the INO80 HSA domain. Thus, it will be important to address whether Arp4, Arp8 and the INO80 HSA domain could form a complex to specifically regulate nuclear actin at the sites of INO80 chromatin-remodelling activity, such as Arp2/3 regulates cytoplasmic actin nucleation at barbed end branch points. In support of this, the knockdown of mammalian Arp4 (BAF53) increases chromosome territory size and deletion of Arp8 in yeast leads to an abnormally increased cell volume and irregular, elongated cell morphology compared with wild type together with unusually elongated buds (Hibbs et al, 2007; Watanabe et al, 2009). Although one cannot rule out more indirect effects at this point, these observations are consistent with a more general function of Arp4 and Arp8 in actin metabolism.
In summary, our structural and biochemical results for Arp4 and Arp8 now open a biochemical and structure–function analysis path to specifically decipher the role of nuclear Arps and—more generally—nuclear actin in chromatin structure and dynamics.
Materials and methods
Protein expression and purification
Arp4 and Arp8 genes were amplified from yeast genomic DNA and cloned via SalI–NotI into pFBDM vector (Berger et al, 2004). An N-terminal hexa-His tag cleavable by Tagzyme (Qiagen) was introduced by the primers. Baculovirus was produced in Sf21 cells (Invitrogen) using standard methods. The proteins were expressed in High Five Insect cells (Invitrogen) following 72 h of infection. The truncated version of Arp8, lacking the first 244 amino acids was expressed in E. coli and used for the described assays. Protein purification was carried out in 20 mM EPPS 8.0, 200 mM NaCl, 5% v/v glycerol and 5 mM β-ME by subsequent Ni-NTA (Qiagen) affinity chromatography, ion exchange on HiTrapQ HP (GE Healthcare) and gel filtration on Superdex-200 (GE Healthcare). Pure protein was concentrated in 10 kDa cutoff centrifugal devices (Amicon) to final concentrations of 8–10 mg/ml, flash frozen in liquid nitrogen and stored at −80°C.
Protein crystallization
For protein crystallization, Arp4 was reductively methylated (Kim et al, 2008). After complete methylation, the protein was subjected to gel filtration on Superdex-200 (GE Healthcare) and freshly concentrated to about 8 mg/ml. Crystallization was performed by hanging drop vapour diffusion against 27% PEG2000MME, 0.1 M HEPES-NaOH pH 7.0, 6% w/v D+-Trehalose, 50 mM glycine at 20°C. Rod-shaped crystals in space group P61 grew to useful sizes in about 2–3 weeks and were frozen in liquid nitrogen with 20% v/v glycerol as cryoprotectant.
Crystal structure determination
Diffraction data to a limiting resolution of 3.4 Å were collected on a single crystal at 100 K with a wavelength of 1.0 Å at X06SA (Swiss Light Source, Villigen, Switzerland). The data set was processed and scaled using XDS (Kabsch, 1993). A molecular replacement model was generated from the structure of yeast actin (pdb: 1YAG) using CHAINSAW. Molecular replacement was carried out with PHASER (McCoy et al, 2007). The model was optimized by cyclic rounds of manual model building using COOT (Emsley and Cowtan, 2004) and refinement with PHENIX (Adams et al, 2010) applying NCS restraints for all chains. The stereochemistry of the refined structure was analysed by PROCHECK (Laskowski et al, 1993) with 86.8, 12.4, 0.8 and 0% of residues in the most favoured, additionally allowed, generously allowed and disallowed regions of the Ramachandran plot, respectively. Coordinates have been deposited in the protein data bank (accession number: 3QB0).
Small-angle X-ray scattering
Protein samples for SAXS were freshly purified by preparative size-exclusion chromatography (20 mM EPPS pH 8.0, 100 mM NaCl, 2% v/v glycerol and 5 mM β-ME) using a Superdex-200 (GE Healthcare). The flow through of the concentration step was used as a buffer reference. Data were collected at beamline X33, EMBL/DESY (Hamburg, Germany) at a cell temperature of 20°C. Molecular weight estimation of Arp4 and Arp8 samples was done by comparison of the corresponding I(o)-values from Guinier approximation with BSA (66 kDa) and Porod-volume analysis (Putnam et al, 2007). Arp4 and Arp8 were measured at protein concentrations of 2.1, 4.7 and 7.2 mg/ml and 1.4, 4.3 and 8.1 mg/ml, respectively. Data were processed using the ATSAS package (Konarev et al, 2006). A set of 10 independent ab initio structures was calculated using GASBOR, without any prior symmetry information. Models were aligned and averaged using DAMAVER (Volkov and Svergun, 2003). The bead models were transformed to an electron density using the SITUS package (Wriggers and Chacon, 2001). Docking of structures into these densities was done using UCSF CHIMERA (Pettersen et al, 2004). Comparison between measured scattering data and theoretical scattering data was done using CRYSOL (Svergun et al, 1995).
Pyrene actin assays
Before the experiments, Ca2+-ATP-actin was converted to Mg2+-ATP-actin by addition of 10 × Mg-exchange buffer (20 mM Tris, pH 8.0, 2 mM ATP, 1 mM MgCl2 and 0.5 mM DTT). For assembly assays, protein was diluted in storage buffer (20 mM EPPS, pH 8.0, 100 mM KCl, 5 mM β-ME, 2% v/v glycerol,) and 10 × low salt KMEI buffer was added (250 mM KCl, 10 mM MgCl2, 10 mM EGTA and 100 mM imidazole, pH 7.3). Anti-foam 204 (Sigma) and KCl were added at final concentrations of 0.005% and 50 mM, respectively. In all, 18 μl of a 40 μM solution of 10% pyrene-labelled Mg2+-ATP-G-actin (in 2 mM Tris/HCl, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2 and 0.5 mM DTT) was placed in an 8-well microtiter assembly strip (Thermo Scientific). The assembly reaction was initiated by transferring 162 μl of the protein solution to the pyrene-labelled actin. Polymerization of actin was followed by measuring the fluorescence of pyrene actin (excitation at 364 nm and emission at 407 nm) in a Fluoroskan II plate reader (Thermo Scientific) for at least 1500 s. For polymerization experiments with Mg2+-ADP-actin monomers, ATP was replaced by ADP in all buffers. The relative polymerization rate was derived by measuring the slopes from the points where 10–50% of the actin had assembled. The amount of F-actin at steady state was obtained by measuring pyrene fluorescence after 16 h incubation of the reaction mixtures at 4°C.
For analysis of the effects of Arp4 and Arp8 on F-actin, 40 μM Mg2+-ATP-actin (15% pyrene labelled) was polymerized overnight in KMEI buffer. For depolymerization experiments in ADP-containing buffer, ATP was hydrolysed by adding 10 U/ml hexokinase (Sigma) and 20 mM glucose to the mixture. The assays were performed by rapidly mixing 10 μl of the protein in KMEI buffer with 90 μl of a solution of 2.2 μM F-actin. Pyrene-fluorescence decrease was monitored with a Jasco FP-6500 fluorimeter.
In vitro TIRF microscopy
Time-lapse TIRF microscopy (TIRFM) on OG-labelled actin was performed as described (Breitsprecher et al, 2008). Images from an Olympus IX-81 inverted microscope were captured every 5 s with exposures of 200 ms with a Hamamatsu Orca-R2 CCD camera (Hamamatsu Corp., Bridgewater, NJ). The pixel size corresponded to 0.11 μm.
Reactions in TIRF assays contained 1.3 μM actin (30% OG labelled). Before the experiments, Ca2+-ATP-actin was converted to Mg2+-ATP-actin as described. Polymerization experiments were performed in TIRF buffer (10 mM imidazole, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.2 mM ATP, 10 mM DTT, 15 mM glucose, 20 μg/ml catalase, 100 μg/ml glucose oxidase and 0.5% w/v methylcellulose (4000 cP), pH 7.4). The elongation rates of filaments were calculated with ImageJ software using the plugins MtrackJ and Manual Tracking. Each experiment was repeated at least three times. For each polymerization measurement, at least 15 barbed ends of individual filaments were manually tracked. The average number of filaments was obtained by counting actin filaments in an area of 100 × 140 μm 200 s after initiation of the polymerization reaction.
Critical concentration assay
For determination of the critical concentration (CCrit) of F-actin in the presence of Arp4 and Arp8, 20 μM of the respective protein was mixed with pre-polymerized F-actin (20% pyrene labelled) in a concentration range from 10 μM to 5 nM in KMEI- and Mg-exchange buffer and incubated at room temperature in the dark. After 16 h, the pyrene fluorescence was detected in a Fluoroskan II plate reader (Thermo Scientific). CCrit was derived from plots of fluorescence versus actin concentration and corresponds to the intercept of the linear regressions of the signals for filamentous and monomeric actin. Dissociation constants were derived using the equation
 |
where [Arp] is the total concentration of Arp4 or Arp8, respectively, [CCrit(Arp)] is the concentration of unpolymerized actin given by the intercept of the linear regression lines of the signals for filamentous and monomeric actin in the presence of Arp and capping protein, and [CCrit(capped)] is the critical concentration for polymerization determined as the intercept of the regression lines for filamentous and monomeric actin in the absence of Arp and presence of capping protein.
Sedimentation assay
In all, 100 μl of a solution of 5 μM Mg2+-ATP-actin was polymerized either alone or in presence of Arp4 and Arp8 by addition of 1 × KMEI buffer and incubated for 5 h at room temperature. F-actin was sedimented by centrifugation for 30 min at 120 000 g. Subsequently, pellets and supernatants were analysed by SDS–PAGE, and the band intensities were quantified using the profile plotting tool implemented in the ImageJ software.
Supplementary Material
Acknowledgments
We thank Dr Kristina Lakomek for help with calculating the omit electron density map used in Figure 3, Dr Maria Lucas for help with the Biacore experiments and Brigitte Keßler for help with cloning. We thank Professor Michael Schleicher and Dr Annette Müller-Taubenberger from the LMU for providing us with acetone powder of rabbit skeletal muscle. We thank the staffs of beamline PXI at the Swiss Light Source (Villingen), ID23-1 at the European Synchrotron Radiation Facility (Grenoble) and X33 at EMBL/DESY (Hamburg) for help with data collection. This work was supported by a scholarship from the Boehringer Ingelheim Fonds to SF and grants from the DFG (SFB/TR5, SFB646 and SFB684) and German Excellence initiative (Center for Integrated Protein Science and Munich Center for Advanced Photonics) to KPH as well as grant FA 330/4-2 from the DFG to JF. CBG was supported by the Elite Network Bavaria in Macromolecular Science.
Author contributions: SF cloned, expressed and purified Arp4 and Arp8; SF crystallized Arp4 and solved the structure; SF performed the Biacore measurements; SAXS measurements and data analysis were performed by SF and GW with the help of CBG; CBG also helped with protein purification; DB performed the experiments concerning actin biochemistry; experiments were devised by SF, DB, JF and KPH; the paper was written by SF and KPH with the help of DB.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66(Part 2): 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschuler GM, Klug DR, Willison KR (2005) Unfolding energetics of G-alpha-actin: a discrete intermediate can be re-folded to the native state by CCT. J Mol Biol 353: 385–396 [DOI] [PubMed] [Google Scholar]
- Aoyama N, Oka A, Kitayama K, Kurumizaka H, Harata M (2008) The actin-related protein hArp8 accumulates on the mitotic chromosomes and functions in chromosome alignment. Exp Cell Res 314: 859–868 [DOI] [PubMed] [Google Scholar]
- Berger I, Fitzgerald DJ, Richmond TJ (2004) Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol 22: 1583–1587 [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Stuven T, Kuhn C, Cordes VC, Gorlich D (2006) A selective block of nuclear actin export stabilizes the giant nuclei of Xenopus oocytes. Nat Cell Biol 8: 257–263 [DOI] [PubMed] [Google Scholar]
- Bonder EM, Fishkind DJ, Mooseker MS (1983) Direct measurement of critical concentrations and assembly rate constants at the two ends of an actin filament. Cell 34: 491–501 [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Kiesewetter AK, Linkner J, Urbanke C, Resch GP, Small JV, Faix J (2008) Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J 27: 2943–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD (2010) A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol 11: 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS (2006) Long-range directional movement of an interphase chromosome site. Curr Biol 16: 825–831 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Coue M, Brenner SL, Spector I, Korn ED (1987) Inhibition of actin polymerization by latrunculin A. FEBS Lett 213: 316–318 [DOI] [PubMed] [Google Scholar]
- Dion V, Shimada K, Gasser SM (2010) Actin-related proteins in the nucleus: life beyond chromatin remodelers. Curr Opin Cell Biol 22: 1–9 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 (Part 12 Part 1): 2126–2132 [DOI] [PubMed] [Google Scholar]
- Franke WW (2004) Actin′s many actions start at the genes. Nat Cell Biol 6: 1013–1014 [DOI] [PubMed] [Google Scholar]
- Galarneau L, Nourani A, Boudreault AA, Zhang Y, Heliot L, Allard S, Savard J, Lane WS, Stillman DJ, Cote J (2000) Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell 5: 927–937 [DOI] [PubMed] [Google Scholar]
- Harata M, Karwan A, Wintersberger U (1994) An essential gene of Saccharomyces cerevisiae coding for an actin-related protein. Proc Natl Acad Sci USA 91: 8258–8262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbs MA, Hess DC, Myers CL, Huttenhower C, Li K, Troyanskaya OG (2007) Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics 23: 2692–2699 [DOI] [PubMed] [Google Scholar]
- Hofmann W, Reichart B, Ewald A, Muller E, Schmitt I, Stauber RH, Lottspeich F, Jockusch BM, Scheer U, Hauber J, Dabauvalle MC (2001) Cofactor requirements for nuclear export of Rev response element (RRE)- and constitutive transport element (CTE)-containing retroviral RNAs. An unexpected role for actin. J Cell Biol 152: 895–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa M, Maeda K, Narita A, Maeda Y, Oda T (2008) Dual roles of Gln137 of actin revealed by recombinant human cardiac muscle alpha-actin mutants. J Biol Chem 283: 21045–21053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Kim Y, Quartey P, Li H, Volkart L, Hatzos C, Chang C, Nocek B, Cuff M, Osipiuk J, Tan K, Fan Y, Bigelow L, Maltseva N, Wu R, Borovilos M, Duggan E, Zhou M, Binkowski TA, Zhang RG, Joachimiak A (2008) Large-scale evaluation of protein reductive methylation for improving protein crystallization. Nat Methods 5: 853–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev PV, Petoukhov MV, Volkov VV, Svergun DI (2006) ATSAS 2.1, a program package for small-angle scattering data analysis. J Appl Crystallogr 39: 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenbaum E, Nordberg P, Bjorkegren-Sjogren C, Schutt CE, Lindberg U, Karlsson R (1998) The role of profilin in actin polymerization and nucleotide exchange. Biochemistry 37: 9274–9283 [DOI] [PubMed] [Google Scholar]
- Krauss SW, Chen C, Penman S, Heald R (2003) Nuclear actin and protein 4.1: essential interactions during nuclear assembly in vitro. Proc Natl Acad Sci USA 100: 10752–10757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, Richards DP, Wu X, Emili A, Hughes TR, Buratowski S, Greenblatt JF (2003) A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell 12: 1565–1576 [DOI] [PubMed] [Google Scholar]
- Kukalev A, Nord Y, Palmberg C, Bergman T, Percipalle P (2005) Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat Struct Mol Biol 12: 238–244 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Martin AC, Welch MD, Drubin DG (2006) Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat Cell Biol 8: 826–833 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40(Part 4): 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ (2006) Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol 172: 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X (2004) INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119: 767–775 [DOI] [PubMed] [Google Scholar]
- Oda T, Iwasa M, Aihara T, Maeda Y, Narita A (2009) The nature of the globular- to fibrous-actin transition. Nature 457: 441–445 [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Peterson CL (2008) The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol 15: 338–345 [DOI] [PubMed] [Google Scholar]
- Pendleton A, Pope B, Weeds A, Koffer A (2003) Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J Biol Chem 278: 14394–14400 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Poch O, Winsor B (1997) Who′s who among the Saccharomyces cerevisiae actin-related proteins? A classification and nomenclature proposal for a large family. Yeast 13: 1053–1058 [DOI] [PubMed] [Google Scholar]
- Pollard TD (2007) Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct 36: 451–477 [DOI] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA (2007) X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys 40: 191–285 [DOI] [PubMed] [Google Scholar]
- Rando OJ, Zhao K, Janmey P, Crabtree GR (2002) Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc Natl Acad Sci USA 99: 2824–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rould MA, Wan Q, Joel PB, Lowey S, Trybus KM (2006) Crystal structures of expressed non-polymerizable monomeric actin in the ADP and ATP states. J Biol Chem 281: 31909–31919 [DOI] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C (2000) A chromatin remodelling complex involved in transcription and DNA processing. Nature 406: 541–544 [DOI] [PubMed] [Google Scholar]
- Shen X, Ranallo R, Choi E, Wu C (2003) Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell 12: 147–155 [DOI] [PubMed] [Google Scholar]
- Shvetsov A, Musib R, Phillips M, Rubenstein PA, Reisler E (2002) Locking the hydrophobic loop 262-274 to G-actin surface by a disulfide bridge prevents filament formation. Biochemistry 41: 10787–10793 [DOI] [PubMed] [Google Scholar]
- Sunada R, Gorzer I, Oma Y, Yoshida T, Suka N, Wintersberger U, Harata M (2005) The nuclear actin-related protein Act3p/Arp4p is involved in the dynamics of chromatin-modulating complexes. Yeast 22: 753–768 [DOI] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ (1995) CRYSOL—a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr 28: 768–773 [Google Scholar]
- Szerlong H, Hinata K, Viswanathan R, Erdjument-Bromage H, Tempst P, Cairns BR (2008) The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol 15: 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Hohn B, Gasser SM (2004) Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119: 777–788 [DOI] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI (2003) Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr 36: 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobiev S, Strokopytov B, Drubin DG, Frieden C, Ono S, Condeelis J, Rubenstein PA, Almo SC (2003) The structure of nonvertebrate actin: implications for the ATP hydrolytic mechanism. Proc Natl Acad Sci USA 100: 5760–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Watanabe D, Nogami S, Morishita S, Ohya Y (2009) Comprehensive and quantitative analysis of yeast deletion mutants defective in apical and isotropic bud growth. Curr Genet 55: 365–380 [DOI] [PubMed] [Google Scholar]
- Witke W (2004) The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol 14: 461–469 [DOI] [PubMed] [Google Scholar]
- Wriggers W, Chacon P (2001) Using Situs for the registration of protein structures with low-resolution bead models from X-ray solution scattering. J Appl Crystallogr 34: 773–776 [Google Scholar]
- Wu X, Yoo Y, Okuhama NN, Tucker PW, Liu G, Guan JL (2006) Regulation of RNA-polymerase-II-dependent transcription by N-WASP and its nuclear-binding partners. Nat Cell Biol 8: 756–763 [DOI] [PubMed] [Google Scholar]
- Yarmola EG, Somasundaram T, Boring TA, Spector I, Bubb MR (2000) Actin-latrunculin A structure and function. Differential modulation of actin-binding protein function by latrunculin A. J Biol Chem 275: 28120–28127 [DOI] [PubMed] [Google Scholar]
- Yoo Y, Wu X, Guan JL (2007) A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J Biol Chem 282: 7616–7623 [DOI] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR (1998) Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95: 625–636 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.