Abstract
Transcription factors and epigenetic modulators are involved in the maintenance of self-renewal in embryonic stem (ES) cells. Here, we demonstrate the existence of a regulatory loop in ES cells between Sox2, an indispensable transcription factor for self-renewal, and embryonic ectoderm development (Eed), an epigenetic modulator regulating histone methylation. We found that Sox2 and Eed positively regulate each other's expression. Interestingly, Sox2 overexpression suppressed the induction of differentiation-associated genes in Eed-deficient ES cells without restoring histone methylation. This Sox2-mediated suppression was prevented by knockdown of the histone acetyltransferase (HAT), Tip60 or Elp3, and Sox2 stimulated expression of these HATs. Furthermore, forced expression of either HAT resulted in repression of differentiation-associated genes in Eed-deficient cells. These results suggest that Sox2 overcame the phenotype of Eed-deficient ES cells by promoting histone acetylation. We also found that knockout of Eed and knockdown of these HATs synergistically enhanced the upregulation of differentiation-associated genes in ES cells. Taken together, our results suggest that the Eed/Sox2 regulatory loop contributes to the maintenance of self-renewal in ES cells by controlling histone methylation and acetylation.
Keywords: Eed, embryonic stem cells, histone modification, self-renewal, Sox2
Introduction
Embryonic stem (ES) cells are derived from the inner cell mass (ICM) of the mammalian blastocyst (Evans and Kaufman, 1981; Martin, 1981). Mouse ES cells can self-renew and maintain their pluripotency when cultured in the presence of leukaemia inhibitory factor (LIF). Several transcription factors have important roles in the self-renewal capacity of mouse ES cells (Niwa, 2007). STAT3 is a downstream transcription factor activated by LIF and is essential and sufficient for the maintenance of self-renewal (Niwa et al, 1998; Matsuda et al, 1999; Ying et al, 2008). Nanog is a homeobox transcription factor whose overexpression can bypass the requirement of LIF for self-renewal, although it is dispensable for self-renewal (Chambers et al, 2003, 2007; Mitsui et al, 2003). The POU family transcription factor Oct3/4 has a central role in ES cell self-renewal and ICM production (Nichols et al, 1998; Niwa et al, 2000). Another indispensable transcription factor is the SRY-related HMG-box protein Sox2. Sox2-deficient mouse embryos die shortly after implantation (Avilion et al, 2003), and a study using inducible Sox2-deficient ES cells revealed that Sox2 stabilizes ES cells in a pluripotent state by maintaining the requisite level of Oct3/4 expression (Masui et al, 2007). It has been suggested that these transcription factors form networks and stimulate the expression of a set of self-renewal genes to maintain the ‘stemness’ of ES cells (Boyer et al, 2005; Loh et al, 2006; Chen et al, 2008).
In addition to transcription factors, histone modifiers also have an important role in ES cell self-renewal. The chromatin of self-renewing ES cells exhibits a characteristic structure of increased accessibility due to fewer and more loosely bound histones and architectural proteins (Meshorer and Misteli, 2006). When ES cells undergo differentiation, their chromatin structure changes dynamically in response to global histone modifications. Histone modifications have been shown to regulate gene activation and repression during development (Kouzarides, 2007). For example, acetylation of various residues of histone H3 (H3Ac) and histone H4 (H4Ac) is involved in transcriptional activation, whereas methylation of Lys-27 of histone H3 (H3K27me) is linked to transcriptional silencing.
Polycomb group (PcG) proteins are histone-modifying proteins that participate in transcriptional repression. Three PcG proteins, enhancer of zeste 2 (Ezh2), embryonic ectoderm development (Eed) and suppressor of zeste 12 homologue (Suz12), comprise the core of the Polycomb repressive complex 2 (PRC2), which mediates H3K27me (Cao et al, 2002; Czermin et al, 2002; Kuzmichev et al, 2002; Müller et al, 2002). Ezh2 is an SET domain-containing histone methyltransferase and functions as the catalytic subunit of PRC2. Eed exists in four isoforms (Eed1, Eed2, Eed3 and Eed4), which arise from alternate translation initiation sites in the same mRNA (Kuzmichev et al, 2004), and has a crucial role in boosting the enzymatic activity of Ezh2. Finally, Suz12 is involved in nucleosome binding of PRC2 (Nekrasov et al, 2005). Genome-wide location analysis in ES cells has revealed that many PcG target genes encode transcription factors important in development (Boyer et al, 2006). In fact, mouse embryos deficient for Suz12, Ezh2 or Eed displayed embryonic lethality with gastrulation arrest (Faust et al, 1998; O’Carroll et al, 2001; Pasini et al, 2004), underscoring the importance of these PcGs in early embryogenesis.
PcG proteins are also involved in the repression of differentiation-associated genes in self-renewing ES cells. Previously, we and others showed that Eed-deficient ES cells exhibit de-repression of multiple differentiation-associated genes and are prone to differentiation (Boyer et al, 2006; Chamberlain et al, 2008; Ura et al, 2008). In wild-type ES cells, H3K27me accumulated in the promoter regions of differentiation-associated genes. In contrast, the H3K27me accumulation disappeared and multiple differentiation-associated genes were upregulated in Eed-deficient ES cells. Moreover, some self-renewal genes were downregulated, although the cells still proliferated and expressed other self-renewal genes, including Oct3/4. These findings suggest that the molecular characteristics that define ‘stemness’ are slightly but significantly decreased in Eed-deficient ES cells. Similarly, Suz12-deficient ES cells exhibited decreased H3K27me with increased expression of differentiation-associated genes (Pasini et al, 2007). Ezh2-deficient ES cells also exhibited reduced H3K27me, although differentiation-associated genes were not upregulated due to compensation by Ezh1 (Shen et al, 2008).
During ES cell differentiation, the downregulation of self-renewal genes and upregulation of differentiation-associated genes should occur in a simultaneous and coordinated manner. It is expected, therefore, that highly regulated cross-talk(s) may exist between transcription factors that stimulate the expression of self-renewal genes and PcG proteins that suppress the expression of differentiation-associated genes. Here, we report the discovery of cross-talk between Sox2 and Eed. We determined that these molecules each positively regulate the other's expression through a regulatory feedback loop. We also found that Sox2 blocks the induction of differentiation-associated genes in Eed-deficient ES cells through the upregulation of self-renewal genes by binding to their promoters and/or triggering histone acetylation. These data suggest that the Eed/Sox2 regulatory loop regulates self-renewal and differentiation of ES cells by controlling histone methylation and acetylation.
Results
Eed is a downstream target of Sox2
It has been suggested that the expression of differentiation-associated genes is repressed by PRC2-mediated H3K27me in self-renewing ES cells, but de-repressed by the loss of PRC2 activity in differentiating ES cells (Boyer et al, 2006; Lee et al, 2006; Pasini et al, 2007; Shen et al, 2008; Ura et al, 2008). In fact, a reduction of trimethylation of Lys-27 of histone H3 (H3K27me3) was observed in differentiating ES cells (Figure 1A), suggesting that PRC2 activity is reduced during ES cell differentiation. Among the three major components of PRC2, a decrease in Eed mRNA occurs earlier than in Ezh2 or Suz12 (Ura et al, 2008). Disruption of the eed gene resulted in the loss of H3K27me3 (Montgomery et al, 2005; Chamberlain et al, 2008; Ura et al, 2008), and overexpression of Eed was able to suppress the reduction of H3K27me3 during differentiation (Figure 1A). Taken together, these results suggest that the expression level of Eed determines the level of H3K27me3 during ES cell differentiation, and raise the possibility that Eed expression is strictly regulated by self-renewal transcription factors.
Figure 1.
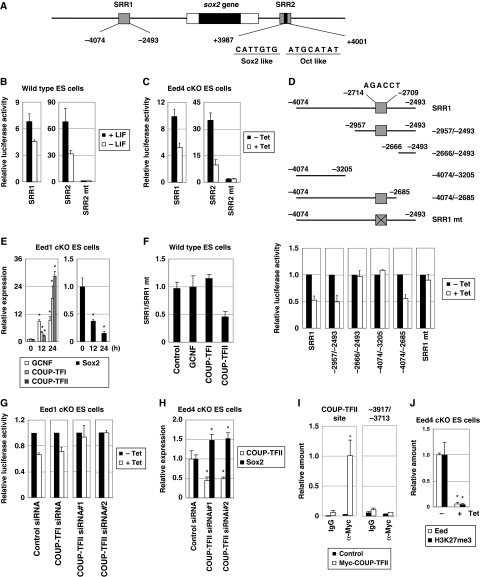
Eed is a downstream target of Sox2. (A) Eed overexpression restores H3K27me3 during differentiation. ES cells transfected with a control or Eed1 expression vector were cultured with or without LIF for 4 days. The total amount of H3K27me3 in cell lysates was examined by western blot analysis (lower panels). Signal intensity was presented as the fold change relative to the sample in the presence of LIF (upper panel). (B) Eed expression is reduced by repression of Sox2. 2TS22C ES cells were cultured with or without Tet for the indicated periods. Expression levels of Sox2 and Eed were quantified by quantitative RT–PCR (qRT–PCR) and presented as the fold change relative to an untreated control sample. Asterisk, significant difference from untreated control cells (P<0.05). Hash, significant difference from cells cultured with Tet for 24 h and then without Tet for 24 h (P<0.05). (C) Repression of Sox2 induces reduction of H3K27me3. 2TS22C ES cells were cultured with or without Tet for 4 days, and levels of H3K27me3 were detected by western blot analysis. (D) Sox2 is reduced following Eed depletion. Eed1 cKO ES cells were cultured with or without Tet for the indicated periods. Expression of Sox2 and Eed was examined by qRT–PCR. Asterisk, significant difference from untreated control cells (P<0.05). Hash, significant difference from cells cultured with Tet for 24 h and then without Tet for 24 h (P<0.05). (E) Schematic representation of the promoter region of the eed gene. (F–H) Regulation of the Eed promoter region by STAT3, Oct3/4 and Sox2. (F) The reporter plasmid pGL2-Eed(−2600/−13) was transfected into HEK293 cells together with a control empty vector or expression vectors for Sox2, Oct3/4, STAT3 or Nanog. After 2 days in culture, cells were harvested and subjected to luciferase assay. (G) ES cells were transfected with pGL2-Eed(−2600/−13) derivatives carrying mutations at Sox2-, Oct3/4- and/or STAT3-binding sites and cultured for 2 days. In each experiment, the data was normalized by setting the value of the wild-type promoter as 1.0. (H) 2TS22C ES cells were transfected with pGL2-Eed(−2600/−13) (wild-type) or pGL2-Eed(−2600/−13)Sox2mt (Sox2 mt) and cultured with or without Tet for 2 days. (I) Sox2 binds to the promoter region of the eed gene in vivo. ES cells transfected with a control or Flag-Sox2 expression vector were subjected to ChIP assay using an anti-Flag antibody, followed by qPCR using primers for the Sox2-binding site in the eed gene. The promoter region of the eed gene from −382 to −12 was used as a negative control. Asterisk, significant difference from sample precipitated with control IgG (P<0.05). In all experiments, error bars indicate s.d. (n=3).
Among the three indispensable transcription factors for ES cell self-renewal, STAT3 and Oct3/4 have already been shown to directly regulate Eed expression (Ura et al, 2008). Using 2TS22C ES cells, we examined whether Sox2 also regulates Eed expression. As reported previously by Masui et al (2007), Sox2 expression in this cell line can be regulated by addition of tetracycline (Tet). Upon Tet treatment, the expression level of Sox2 rapidly decreased, but recovered following Tet removal (Figure 1B). Similarly, expression of Eed was downregulated by Tet stimulation and recovered after Tet withdrawal. In agreement with Eed downregulation, the overall amount of H3K27me3 was diminished in Tet-treated 2TS22C ES cells (Figure 1C). These results suggest that Eed is downstream of Sox2. Interestingly, we found that the opposite is also true: the expression level of Sox2 was reduced when Eed expression was suppressed by Tet treatment in Eed conditional knockout (cKO) ES cells (Ura et al, 2008), but was restored by Tet removal with re-expression of Eed (Figure 1D). These findings suggest the intriguing possibility that the self-renewal promoting transcription factor Sox2 and the differentiation-suppressing, epigenetic regulator Eed engage in tightly regulated cross-talk and form a regulatory loop in ES cells. In contrast, expression of Stat3 and Oct3/4 was not affected by Eed downregulation (Supplementary Figure S1). Therefore, the primary focus of this study is the relationship between Eed and Sox2.
First, it was determined whether Eed is a direct target of Sox2. The 2.6-kb upstream region (−2600/−13) of the eed gene contains STAT3- and Oct3/4-binding sites (Ura et al, 2008). Since Sox2 often binds to a sequence adjacent to an Oct3/4-binding site, we searched for a putative Sox2 sequence near the Oct3/4-binding site (−2019/−2012) and found one such sequence (5′-AACAACAG-3′) at −2037/−2030 (Figure 1E). A luciferase assay was then performed using the 2.6-kb upstream region to determine if the identified putative site is an authentic Sox2-binding site. As shown in Figure 1F, promoter activity of the 2.6-kb region was stimulated by the presence of Sox2, Oct3/4 or STAT3, but not by Nanog, suggesting that this region contains an Sox2-responsive element in addition to Oct3/4- and STAT3-responsive elements. Disruption of the putative Sox2-binding site by mutagenesis reduced promoter activity, which was further decreased when combined with additional mutations at the Oct3/4- and/or STAT3-binding sites (Figure 1G). Similarly, when Sox2 was downregulated in 2TS22C ES cells, the promoter activity of the 2.6-kb region was reduced to a level comparable to that of the mutant lacking the Sox2-binding site (Figure 1H). Importantly, the promoter activity of the mutant 2.6-kb region was not reduced further by Sox2 downregulation. These results suggest that Sox2 stimulates Eed expression through the Sox2-binding site at −2037/−2030.
To confirm the in vivo binding of Sox2 to the promoter region of the eed gene, chromatin immunoprecipitation (ChIP) analysis was carried out. A DNA fragment (−2250/−1882) containing the identified Sox2-binding site in the eed gene was precipitated by anti-Flag antibody from Flag-Sox2-expressing ES cells, but not from control ES cells (Figure 1I). In contrast, another region (−382/−12) of the eed gene, which does not contain an Sox2-binding site, did not precipitate from Flag-Sox2-expressing ES cells. Taken together, these results indicate that Eed is directly regulated by Sox2 in ES cells.
Eed positively regulates Sox2 expression through repression of COUP-TFII
We next examined how Sox2 expression is positively regulated by Eed, an epigenetic regulator that usually suppresses expression of its target genes. The sox2 gene contains two enhancer regions, SRR1 and SRR2 (Tomioka et al, 2002) (Figure 2A). Withdrawal of LIF from the culture medium resulted in the rapid repression of Sox2 expression (Supplementary Figure S1), as well as a reduction in the enhancer activities of SRR1 and SRR2 (Figure 2B), suggesting that these regions act as regulatory elements of Sox2 expression in ES cells. Activity of these regions was also reduced in Eed cKO ES cells treated with Tet (herein referred to as Eed-deficient ES cells; Figure 2C). These results suggest that Eed regulates Sox2 expression through the SRR1 and SRR2 regions.
Figure 2.
Sox2 is regulated by Eed through COUP-TFII. (A) Schematic representation of SRR1 and SRR2 in the sox2 gene. (B) Enhancer activities of SRR regions decrease after LIF removal. After transfection with pGL4pro-SRR1, pGL4pro-SRR2 or pGL4pro-SRR2mt, ES cells were cultured with or without LIF for 2 days and subjected to a reporter assay to examine the enhancer activities of SRR1, SRR2 and SRR2mt. (C) The enhancer activities of the SRR regions decrease in Eed-deficient ES cells. Enhancer activities of SRR1, SRR2 and SRR2mt were examined in Eed4 cKO ES cells treated with or without Tet for 2 days. (D) Identification of the Eed-responsive element in SRR1. Luciferase assay was performed for SRR1 and its derivatives (shown in upper panel) in Eed4 cKO ES cells treated with or without Tet for 2 days. (E) Expression of GCNF, COUP-TFI, COUP-TFII (left panel) and Sox2 (right panel) in Eed-deficient ES cells. Eed1 cKO ES cells were cultured with or without Tet for the indicated times. Expression of each gene was examined by quantitative RT–PCR (qRT–PCR). Asterisk, significant difference from untreated cells (P<0.05). (F) COUP-TFII suppresses the enhancer activity of SRR1 through the Eed-responsive element. Enhancer activities of SRR1 and SRR1mt were examined in ES cells transfected with control, GCNF, COUP-TFI or COUP-TFII expression vectors. Note that all values are presented as the ratio of the enhancer activity of SRR1 to that of SRR1mt. (G) Decreased enhancer activity of SRR1 in Eed-deficient ES cells is restored by knockdown of COUP-TFII. The enhancer activity of SRR1 was examined in Eed1 cKO ES cells transfected with pFIV-control, pFIV-COUP-TFI, pFIV-COUP-TFII#1 or pFIV-COUP-TFII#2 in the presence or absence of Tet. (H) Knockdown of COUP-TFII induces Sox2 expression. After transfection with pFIV-COUP-TFII, Eed4 cKO ES cells were cultured in the presence of Tet, and mRNA expression of COUP-TFII and Sox2 was examined by qRT–PCR. Asterisk, significant difference from control cells (P<0.05). (I) In vivo binding of COUP-TFII to SRR1. ES cells transfected with either a control or a Myc-COUP-TFII expression vector were subjected to a ChIP assay using control IgG (IgG) or anti-Myc antibody (α-Myc), followed by qPCR using primers for the COUP-TFII-binding site of SRR1. The upstream region (−3917/−3713) of the COUP-TFII-binding site in SRR1 was used as a negative control. Asterisk, significant difference from sample precipitated with control IgG (P<0.05). (J) Eed regulates H3K27me3 at the promoter region of the coup-tfII gene. Eed4 cKO ES cells were cultured with or without Tet and subjected to a ChIP assay using anti-Myc (Eed) and anti-H3K27me3 (H3K27me3) antibodies, followed by qPCR using primers for the promoter region of the coup-tfII gene. Note that Eed4 cKO ES cells express Myc-tagged Eed4 in the absence of Tet. In all experiments, error bars represent s.d. (n=3).
SRR2 contains one Sox2/Oct3/4-binding element and this element has been identified as a core element of SRR2 (Tomioka et al, 2002). Consistent with this, SRR2 containing mutations at the Sox2/Oct3/4-binding site (SRR2mt) showed negligible enhancer activity (Figure 2B and C), suggesting that the reduced enhancer activity of SRR2 in Eed-deficient cells was likely due to the downregulation of Sox2.
Next, SRR1 was analysed for an Eed-responsive element. Deletion analysis revealed that the enhancer activities of the −2957/−2493 and −4074/−2685 regions of SRR1 were closely correlated with the expression level of Eed (Figure 2D), suggesting that an Eed-responsive element exists in the −2957/−2685 region of SRR1. This region contains a consensus-like sequence (5′-AGACCT-3′) found in the binding site for the transcriptional repressors GCNF, COUP-TFI and COUP-TFII. Interestingly, the enhancer activity of an SRR1mt with mutations in this sequence was not affected by Eed downregulation (Figure 2D), suggesting that this putative binding site is the Eed-responsive element in SRR1.
These findings prompted exploration of the possibility that expression of Sox2 is repressed by GCNF, COUP-TFI and/or COUP-TFII in Eed-deficient ES cells. We first examined whether Eed regulates expression of these repressors. Expression analysis using Eed cKO ES cells revealed that Eed downregulation resulted in the induction of each of the repressors, as well as the downregulation of Sox2 expression (Figure 2E), suggesting that the three repressors are downstream molecules of Eed. To identify the repressor(s) that binds to the Eed-responsive element, we compared the ratio of enhancer activity of SRR1 to that of SRR1mt (SRR1/SRR1mt) among the three repressors (Figure 2F). Forced expression of COUP-TFII significantly reduced SRR1/SRR1mt, suggesting that COUP-TFII represses the activity of SRR1 through the Eed-responsive element. On the other hand, neither GCNF nor COUP-TFI had an effect on this ratio. These results suggest that COUP-TFII binds to the Eed-responsive element in SRR1 to suppress Sox2 expression in Eed-deficient ES cells. This interpretation is supported by the additional observation that the enhancer activity of SRR1 was not reduced in Eed-deficient ES cells when COUP-TFII was knocked down (Figure 2G). Furthermore, the expression level of Sox2 in Eed-deficient ES cells was increased by knockdown of COUP-TFII (Figure 2H), and COUP-TFII was shown to bind to SRR1 in vivo (Figure 2I). We also observed that Eed, as well as K27-methylated histone H3, was associated with the promoter region of the coup-tfII gene, and both associations disappeared when Eed was repressed (Figure 2J). Taken together, these findings indicate that Eed positively regulates Sox2 expression through suppression of COUP-TFII, which binds to SRR1 and represses Sox2 expression.
Sox2 overcomes the phenotype of Eed-deficient ES cells
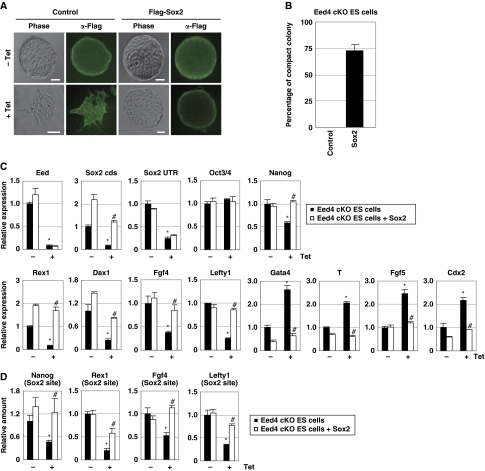
The downregulation of Sox2 in Eed-deficient ES cells (Figure 1D) prompted investigation of whether Sox2 can rescue the phenotype of Eed-deficient ES cells. When treated with Tet, Eed cKO ES cells underwent a morphological change and disruption of compact colony formation (Figure 3A and B). However, many Sox2-expressing, Eed-deficient ES cells formed compact colonies even in the presence of Tet. In addition, the expression of self-renewal genes (Nanog, Rex1, Dax1, Fgf4 and Lefty1) was downregulated by Tet treatment, but was restored by Sox2 overexpression (Figure 3C). Furthermore, induction of differentiation-associated genes (Gata4, T, Fgf5 and Cdx2) in Eed-deficient ES cells was suppressed by Sox2 overexpression. In contrast, Eed overexpression appeared to have negligible effect on the phenotype of Sox2-deficient ES cells, as determined by cellular morphology and gene expression (Supplementary Figure S2).
Figure 3.
Sox2 overcomes the phenotype of Eed-deficient ES cells. (A) The morphology of Sox2-expressing, Eed-deficient ES cells. Eed4 cKO ES cells were transfected with either a control or Flag-Sox2 expression vector and cultured in the presence or absence of Tet. Immunostaining with anti-Flag antibody confirmed the expression of transgenes. Bars, 50 μm. (B) Sox2-expressing, Eed-deficient ES cells form compact colonies. Eed4 cKO ES cells transfected with a control or Flag-Sox2 expression vector were cultured with Tet for 4 days, and the per cent that formed compact colonies was calculated by dividing the number of compact colonies by the total number of colonies. Results represent three independent experiments. (C) Sox2 suppresses downregulation of self-renewal genes and induction of differentiation-associated genes induced by Eed depletion. The indicated cells were cultured with or without Tet for 4 days. Expression of the indicated genes was examined by quantitative RT–PCR (qRT–PCR). Asterisk, significant difference from Eed4 cKO cells cultured in the absence of Tet (P<0.05). Hash, significant difference from Tet-treated Eed4 cKO cells (P<0.05). (D) Sox2 overexpression restores Sox2 binding to promoter regions of self-renewal genes in Eed-deficient ES cells. The indicated cells were cultured with or without Tet for 4 days and subjected to ChIP assay using an anti-Sox2 antibody, followed by qPCR using primers for the Sox2-binding site of the indicated genes. Asterisk, significant difference from Eed4 cKO cells cultured without Tet (P<0.05). Hash, significant difference from Tet-treated Eed4 cKO cells (P<0.05). In all experiments, error bars indicate s.d. (n=3).
Since H3K27me3 is absent in Eed-deficient ES cells, we next examined whether Sox2 expression can restore the loss of H3K27me3. Immunocytochemistry and western blot analysis revealed that the overall amount of H3K27me3 was reduced in Eed-deficient ES cells, and Sox2 overexpression had no effect on this reduction (Figure 4A and B; Supplementary Figure S3). Moreover, no increase in H3K27me3 in the promoter regions of differentiation-associated genes was detected in Sox2-expressing, Eed-deficient ES cells (Figure 4C). These results suggest that although Sox2 cannot restore H3K27me3, Sox2 can compensate for the absence of H3K27me3 in Eed-deficient ES cells.
Figure 4.
Sox2 promotes histone acetylation in Eed-deficient ES cells. Eed4 cKO ES cells transfected with either control or Flag-Sox2 expression vectors were cultured in the presence or absence of Tet for 4 days. (A, B) Global amounts of H3Ac, H4Ac and H3K56Ac are maintained in Sox2-expressing, Eed-deficient ES cells. Control or Sox2-expressing, Eed-deficient ES cells were subjected to immunostaining (A) or western blot analysis (B) using anti-H3K27me3, H3Ac, H4Ac and H3K56Ac antibodies. Bars, 50 μm. (C) Sox2 cannot restore H3K27me3 in the promoter regions of differentiation-associated genes in Eed-deficient ES cells. After culturing with or without Tet, the indicated cells were subjected to ChIP assay using an anti-H3K27me3 antibody, followed by qPCR using primers for the promoter region of the indicated genes. Asterisk, significant difference from Eed4 cKO cells cultured without Tet (P<0.05). (D) The relative amounts of H3Ac and H4Ac were quantitatively determined by ELISA. (E–G) Sox2 restores histone acetylation in the promoter regions of self-renewal genes in Eed-deficient ES cells. The indicated cells were subjected to ChIP assay using anti-H3Ac (E), H4Ac (F) and H3K56Ac (G) antibodies, followed by qPCR using primers for the promoter regions of the indicated genes. Asterisk, significant difference from Eed4 cKO cells cultured without Tet (P<0.05). Hash, significant difference from Eed4 cKO cells cultured with Tet (P<0.05). In all experiments, error bars indicate s.d. (n=3).
Sox2 promotes histone acetylation in Eed-deficient ES cells
Next, we explored the molecular mechanism of how Sox2 suppresses the downregulation of self-renewal genes and the induction of differentiation-associated genes in Eed-deficient ES cells in an H3K27me3-independent manner. Genome-scale ChIP-chip analyses revealed previously that Sox2 binds to the promoter regions of many self-renewal genes, probably to stimulate their expression (Boyer et al, 2005; Chen et al, 2008). Since Eed deficiency leads to the downregulation of Sox2 expression, the effect of Eed deficiency on Sox2 binding to the promoter regions of target genes was examined. We performed a ChIP assay for several known Sox2 target genes and found that the binding of Sox2 to the promoter regions was indeed reduced in Eed-deficient cells (Figure 3D). On the other hand, overexpression of Sox2 restored the amount of promoter-bound Sox2.
The involvement of histone acetyltransferases (HATs) in the self-renewal of ES cells has been suggested previously (Fazzio et al, 2008). We, therefore, examined the effect of Sox2 on histone acetylation, including H3Ac, H4Ac and acetylation at Lys-56 of histone H3 (H3K56Ac). Interestingly, immunostaining suggested that the overall amounts of H3Ac, H4Ac and H3K56Ac were reduced in Eed-deficient ES cells, while levels were maintained in Sox2-expressing, Eed-deficient ES cells (Figure 4A). Multiple quantitative analyses confirmed that levels of H3Ac, H4Ac and H3K56Ac were increased by Sox2 overexpression (Figure 4B and D). Furthermore, ChIP analysis demonstrated that reduced levels of H3Ac, H4Ac and H3K56Ac at the promoter regions of self-renewal genes in Eed-deficient ES cells were restored by Sox2 overexpression (Figure 4E–G). These results suggest that Sox2 promotes histone acetylation in the promoter regions of self-renewal genes in Eed-deficient ES cells.
Sox2 positively regulates expression of HATs
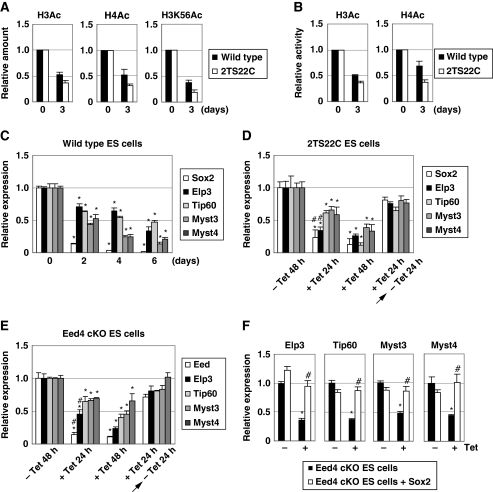
As Sox2 overexpression maintained histone acetylation levels in Eed-deficient ES cells, the relationship between Sox2 and histone acetylation was further investigated. As shown in Figure 5A, western blot analysis revealed that overall levels of H3Ac, H4Ac and H3K56Ac decreased in Sox2-deficient ES cells (i.e. Tet-treated 2TS22C ES cells). ELISA also confirmed the reduction of H3Ac and H4Ac in Sox2-deficient ES cells (Figure 5B). These results suggest that Sox2 regulates histone acetylation in ES cells.
Figure 5.
HATs are downstream effectors of Sox2. (A, B) Global levels of H3Ac, H4Ac and H3K56Ac are reduced during ES cell differentiation and by Sox2 downregulation. Wild-type and 2TS22C ES cells were cultured for 0 or 3 days without LIF and with Tet, respectively. The total amounts of modified histones were determined by western blot analysis (A) and ELISA (B). (C) Expression levels of HATs are reduced after the removal of LIF. Wild-type ES cells were cultured without LIF for the indicated number of days, and the expression of each HAT gene was examined by quantitative RT–PCR (qRT–PCR). Asterisk, significant difference from wild-type cells at day 0 (P<0.05). (D) Expression of HATs is regulated by Sox2. 2TS22C ES cells were cultured with or without Tet for the indicated period, and the expression levels of HATs were determined. Asterisk, significant difference from untreated control cells (P<0.05). Hash, significant difference from cells cultured with Tet for 24 h and then without Tet for 24 h (P<0.05). (E) Expression of HATs is regulated by Eed. Eed4 cKO ES cells were cultured with or without Tet for the indicated period and the expression of HATs was examined. Asterisk, significant difference from untreated control cells (P<0.05). Hash, significant difference from cells cultured with Tet for 24 h and then without Tet for 24 h (P<0.05). (F) Reduced expression of HATs in Eed-deficient ES cells is restored by Sox2 expression. The indicated cells were cultured with or without Tet for 4 days, and expression of the indicated HATs was measured. Asterisk, significant difference from Eed4 cKO cells cultured without Tet (P<0.05). Hash, significant difference from Tet-treated Eed4 cKO cells (P<0.05). In all experiments, error bars indicate s.d. (n=3).
Analysis using GEO Profiles (http://www.ncbi.nlm.nih.gov/sites/entrez?db=geo) revealed that several HATs, including Elp3, Tip60, Myst3 and Myst4, are expressed in undifferentiated ES cells. We, therefore, examined whether these HATs are downstream molecules of Sox2 in ES cells. When LIF was withdrawn from the culture medium, Sox2 expression was downregulated in ES cells, followed by the downregulation of Elp3, Tip60, Myst3 and Myst4 (Figure 5C), as well as a reduction in H3Ac, H4Ac and H3K56Ac (Figure 5A and B). When Sox2 expression was downregulated by Tet treatment in 2TS22C ES cells, expression of these HATs was also downregulated, and this effect was reversed by the removal of Tet (Figure 5D). These results suggest that Elp3, Tip60, Myst3 and Myst4 are downstream targets of Sox2. Similarly, expression levels of these HATs decreased when Eed expression was suppressed and were restored after re-expression of Eed (Figure 5E). Moreover, reduced expression of HATs in Eed-deficient ES cells was restored by Sox2 overexpression (Figure 5F). These data suggest that Eed regulates the expression of Elp3, Tip60, Myst3 and Myst4 by controlling Sox2 expression.
Role of histone acetylation in Sox2 activity in Eed-deficient ES cells
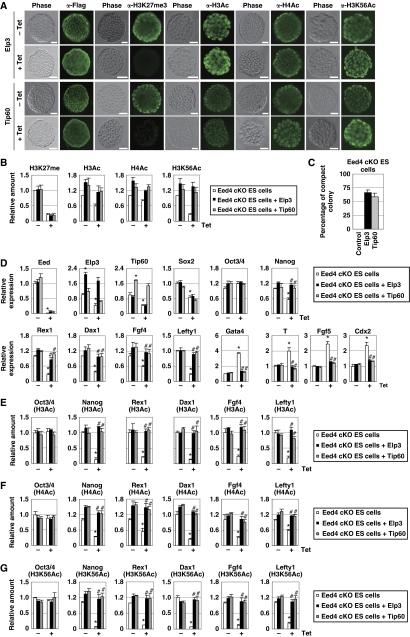
To evaluate the importance of histone acetylation in Sox2-mediated compensation for Eed deficiency, we first examined whether overexpression of HATs had an effect similar to that of Sox2 on Eed-deficient ES cells. First, Elp3- or Tip60-expressing Eed cKO ES cells were generated. An overall reduction in the amount of H3K27me3 in Eed-deficient ES cells was not reversed by Elp3 or Tip60 overexpression (Figure 6A and B). However, global levels of H3Ac, H4Ac and H3K56Ac were maintained in Elp3- or Tip60-expressing, Eed-deficient ES cells, and were comparable to those in undifferentiated control cells (Figure 6A and B; Supplementary Figure S4A and B).
Figure 6.
Elp3 and Tip60 overcome the phenotype of Eed-deficient ES cells. Eed4 cKO ES cells transfected with control, Flag-Elp3 or Flag-Tip60 expression vectors were cultured in the presence or absence of Tet for 4 days. (A) Immunostaining of Elp3- or Tip60-expressing Eed4 cKO ES cells using anti-Flag, H3K27me3, H3Ac, H4Ac and H3K56Ac antibodies. Bars, 50 μm. (B) Global amounts of H3Ac, H4Ac and H3K56Ac are restored by Elp3 or Tip60 expression in Eed-deficient ES cells. Overall amounts of the modified histones were examined by western blot analysis. (C) Elp3- or Tip60-expressing, Eed-deficient ES cells form compact colonies. The ratio of compact colonies was calculated by dividing the number of compact colonies by the total number of colonies. Results are representative of three independent experiments. (D) Elp3 or Tip60 suppresses downregulation of self-renewal genes and induction of differentiation-associated genes induced by Eed depletion. Expression of the indicated genes was examined by quantitative RT–PCR (qRT–PCR). Asterisk, significant difference from Eed4 cKO cells cultured without Tet (P<0.05). Hash, significant difference from Eed4 cKO cells cultured with Tet (P<0.05). (E–G) Elp3 or Tip60 restores histone acetylation in the promoter regions of self-renewal genes in Eed-deficient ES cells. The indicated cells were subjected to ChIP assay with anti-H3Ac (E), anti-H4Ac (F) and anti-H3K56Ac (G) antibodies, followed by qPCR using primers for the promoter regions of the indicated genes. Asterisk, significant difference from Eed4 cKO cells cultured without Tet (P<0.05). Hash, significant difference from Eed4 cKO cells cultured with Tet (P<0.05). In all experiments, error bars indicate s.d. (n=3).
When the effect of HAT expression on ES cell self-renewal was examined, more than half of the Elp3- or Tip60-expressing, Eed-deficient ES cells formed compact colonies (Figure 6A and C). Elp3 and Tip60 restored decreased expression of self-renewal genes and repressed the induction of differentiation-associated genes in Eed-deficient ES cells (Figure 6D). Importantly, expression of Sox2 was still repressed in either Elp3- or Tip60-expressing, Eed-deficient ES cells, suggesting that the Elp3- or Tip60-mediated effect is independent of Sox2. As expected, loss of H3K27me3 at the locus of differentiation-associated genes was not restored in Elp3- or Tip60-expressing, Eed-deficient ES cells (Supplementary Figure S4C), while a reduction in histone acetylation at the promoter regions of self-renewal genes in Eed-deficient ES cells was reversed by either Elp3 or Tip60 expression (Figure 6E–G). These results suggest that, similar to Sox2, Elp3 and Tip60 can compensate for the phenotype of Eed-deficient ES cells.
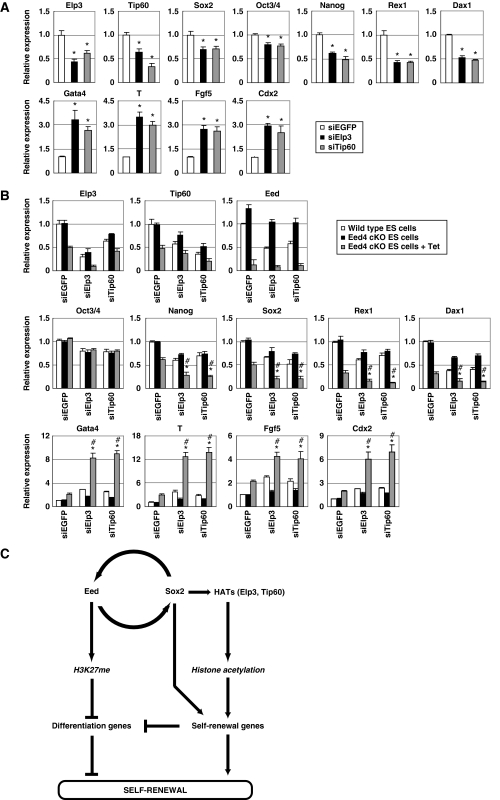
It was next determined whether the expression of HATs is required for Sox2 activity in Eed-deficient ES cells. When either Elp3 or Tip60 was knocked down, the expression of self-renewal genes was repressed in Sox2-expressing, Eed-deficient ES cells (Figure 7A). In addition, expression of differentiation-associated genes was increased in Elp3- or Tip60-knockdown cells, suggesting that expression of HATs is required for Sox2-mediated compensation for the loss of H3K27me3 in Eed-deficient ES cells.
Figure 7.
H3K27me and histone acetylation cooperatively regulate stemness in ES cells. (A) Knockdown of Elp3 or Tip60 reverses the effect of Sox2 on Eed-deficient ES cells. Sox2-expressing Eed4 cKO ES cells were transfected with EGFP siRNA (siEGFP), Elp3 siRNA (siElp3) or Tip60 siRNA (siTip60) and cultured with Tet for 2 days. Expression of the indicated genes was examined by quantitative RT–PCR (qRT–PCR). Asterisk, significant difference from EGFP siRNA-transfected cells (P<0.05). (B) Synergistic effect of Eed depletion and HAT knockdown on ES cell differentiation. Wild-type ES cells and Eed4 cKO ES cells were transfected with EGFP siRNA, Elp3 siRNA or Tip60 siRNA, and cultured with or without Tet for 2 days. The expression of the indicated genes was examined by qRT–PCR. Asterisk, significant difference from EGFP siRNA-transfected Eed4 cKO cells cultured with Tet (P<0.05). Hash, significant difference from Elp3 or Tip60 siRNA-transfected Eed4 cKO cells cultured without Tet (P<0.05). In all experiments, error bars represent s.d. (n=3). (C) The proposed role of the Eed/Sox2 regulatory loop in ES cell self-renewal.
H3K27me and histone acetylation are important for ES cell stemness
Finally, the relationship between histone acetylation and H3K27me in the regulation of self-renewal and differentiation-associated genes in ES cells was investigated (Figure 7B). As described above, either knockout of Eed or knockdown of Elp3 or Tip60 reduced expression of self-renewal genes and increased expression of the differentiation-associated genes. When the two were combined, downregulation of self-renewal genes and upregulation of differentiation-associated genes were further enhanced, suggesting that both H3K27me and histone acetylation are required for maintenance of self-renewal genes and repression of differentiation-associated genes.
Discussion
STAT3, Oct3/4 and Sox2 have been shown to be indispensable for self-renewal of mouse ES cells. We demonstrated previously that STAT3 and Oct3/4 directly regulate the expression of Eed (Ura et al, 2008). In the present study, we showed that Sox2 also directly binds to the promoter region of the eed gene and positively regulates Eed expression. Therefore, it is likely that Eed expression in ES cells is strictly regulated by these three crucial factors. On the other hand, Sox2 expression is regulated through SRR1 and SRR2 (Tomioka et al, 2002). Although it has been shown that SRR2 contains Oct3/4- and Sox2-binding sites and is regulated by these factors, the regulatory mechanism of SRR1 has not been elucidated yet. Our current study demonstrated that SRR1 contains a COUP-TFII-binding site, through which COUP-TFII can repress the enhancer activity of SRR1. We also found that Eed negatively controls COUP-TFII expression. These results indicate that SRR1 is positively regulated by Eed through suppression of COUP-TFII.
SRR2 has been shown to possess higher enhancer activity than SRR1 in undifferentiated embryonal carcinoma cells (Tomioka et al, 2002). Similarly, Figure 2B shows that SRR2 has much higher activity than SRR1 in ES cells. In addition, SRR2 is positively regulated by two key transcription factors, Oct3/4 and Sox2. It is likely, therefore, that SRR2 has a dominant role in induction of Sox2 expression in ES cells. On the other hand, SRR1 contains a suppressive site that negatively controls the enhancer activity, such as the COUP-TFII-binding site, suggesting that SRR1 is involved in regulation of Sox2 expression. For example, we speculate that SRR1 region has an important role in initiation of Sox2 downregulation during ES cell differentiation. Although it is well established that Oct3/4 regulates Sox2 expression, reduction in Sox2 expression occurs earlier than that of Oct3/4 during ES cell differentiation (Supplementary Figure S1), suggesting that Sox2 downregulation during differentiation is not triggered by Oct3/4 repression. Considering that Eed is downregulated during the early phase of differentiation (Supplementary Figure S1), suppression of SRR1 by COUP-TFII, which is caused by Eed downregulation, appears to be the initial step of Sox2 downregulation.
Several studies have suggested the importance of histone acetylation in ES cell self-renewal. Global reduction of acetylated histones is observed during differentiation of ES cells, and inhibition of histone deacetylase activity prevents differentiation (Lee et al, 2004). In human ES cells, approximately 1% of histone H3 is acetylated at Lys-56, and H3K56Ac is frequently observed in the promoter regions of self-renewal genes (Xie et al, 2009). In the present study, we demonstrated that total levels of H3Ac, H4Ac and H3K56Ac were restored by Sox2 overexpression in Eed-deficient ES cells. Sox2 also restored histone acetylation in the promoter regions of self-renewal genes. Furthermore, knockdown of HATs cancelled the effect of Sox2 without significant reduction of Sox2 expression, and expression of HATs exhibited an effect similar to that of Sox2 on the phenotype of Eed-deficient cells. Taken together, these results indicate that Sox2 overcomes the loss of H3K27me by stimulating HAT activity.
As candidate downstream targets of Sox2, we identified the HATs Elp3 and Tip60. Elp3 is the catalytic subunit of an HAT elongator complex (Svejstrup, 2007), and Tip60 is a subunit of a chromatin remodelling complex, the Tip60-p400 complex, which is involved in DNA damage response and cell-cycle regulation (Squatrito et al, 2006). Both HATs were downregulated when Sox2 expression was reduced, and their expression was recovered when Sox2 expression was restored. In addition, reduced expression levels of these HATs in Eed-deficient cells were restored by Sox2 expression. These results suggest that Sox2 promotes HAT activity in Eed-deficient ES cells by upregulating Tip60 and Elp3. This hypothesis is supported by recent reports suggesting that Tip60 is involved in ES cell self-renewal (Fazzio et al, 2008; Hu et al, 2009).
Analyses using Eed- or Suz12-deficient ES cells have revealed that although H3K27me is important for repression of differentiation-associated genes, loss of H3K27me is not sufficient for complete differentiation of ES cells (Montgomery et al, 2005; Azuara et al, 2006; Bernstein et al, 2006; Chamberlain et al, 2008; Ura et al, 2008). One possible explanation for the incomplete differentiation of H3K27me-deficient ES cells is that H3K27me has an important role in ES cell differentiation (Pasini et al, 2007). However, the data presented here show that knockdown of Elp3 or Tip60 further enhanced repression of self-renewal genes and induction of differentiation-associated genes in Eed-deficient ES cells. These observations suggest the possibility that ES cell differentiation requires the reduction of histone acetylation in addition to the loss of H3K27me.
Several reports have described the phenotype of Eed-deficient ES cells (Morin-Kensicki et al, 2001; Montgomery et al, 2005; Azuara et al, 2006; Boyer et al, 2006; Schoeftner et al, 2006; Chamberlain et al, 2008; Shen et al, 2008; Ura et al, 2008). Basically, all the reports including ours have reached the same conclusion. For example, H3K27me is lost by Eed deficiency. Although some differentiation-associated genes are upregulated and some self-renewal genes are downregulated, Eed-deficient ES cells can be maintained in culture. The phenotype of Eed deficiency is reversible (Ura et al, 2008). Eed-deficient ES cells contribute to all lineages in chimeric embryos (Morin-Kensicki et al, 2001; Chamberlain et al, 2008). Taken together, these observations indicate that Eed is dispensable for ES cell self-renewal and pluripotency. The observed discrepancies in the level of some marker genes among reports are probably due to difference in culture condition, clonal variation and/or adaptation. Despite dispensability, the fact that several differentiation-associated genes are upregulated by Eed deficiency indicates the importance of Eed in maintenance of ‘complete’ self-renewal.
Recently, it was reported that ES cells comprise a heterogeneous population (Toyooka et al, 2008), suggesting the possibility that the observed change of gene expression may occur in not all but a portion of Eed-deficient ES cells. Although immunostaining has suggested that an endodermal marker Gata4 is upregulated in most of Eed-null ES cells (Boyer et al, 2006), further detailed analyses, such as a single-cell PCR analysis, should be done to clarify this point.
In this study, we discovered a regulatory loop between Eed and Sox2 in ES cells. The existence of this loop allows us to hypothesize the following model for the molecular mechanism of ES cell self-renewal (Figure 7C). In self-renewing ES cells, Eed represses the expression of differentiation-associated genes through H3K27me, and Sox2 positively regulates self-renewal genes through direct binding and histone acetylation. Eed and Sox2 positively feed back to each other and maintain both H3K27me and histone acetylation at high levels. The finding that Sox2 can inhibit the induction of differentiation-associated genes in Eed-deficient ES cells without increasing H3K27me3 suggests that self-renewal gene products can somehow suppress the induction of differentiation-associated genes, even in the absence of H3K27me. Upon differentiation, the Eed/Sox2 loop is inactivated and both H3K27me and histone acetylation are downregulated simultaneously, leading to upregulation of differentiation-associated genes and downregulation of self-renewal genes. This mechanism allows the expression of self-renewal and differentiation-associated genes to be coordinately regulated in ES cells.
The discovery of the Eed/Sox2 regulatory loop also raises the intriguing possibility that the differentiation process in ES cells may consist of two stages. In the first stage, the Eed/Sox2 regulatory loop is inactivated even though Oct3/4 expression is maintained, and ES cells may begin to lose stemness and stay in an ‘incomplete’ differentiated state. In the second stage, ‘complete’ differentiation is accomplished by Oct3/4 downregulation. We will explore these possibilities in future studies.
Materials and methods
Cell culture and plasmid transfection
ES cell lines, A3-1 (Azuma and Toyoda, 1991), 2TS22C (Masui et al, 2007) and Eed cKO, were cultured on gelatin-coated dishes with Dulbecco’s modified Eagle’s medium (DMEM), as described previously (Ura et al, 2008). Eed1 cKO and Eed4 cKO ES cells are Eed-null ES cells that express Myc-tagged Eed1 and Eed4 isoforms, respectively, under the control of ‘Tet-Off’ system (Ura et al, 2008). Sox2- or HATs-expressing Eed cKO ES cells were established by introducing pCAGIHisDR-Flag-Sox2, -Elp3 or -Tip60 into Eed cKO ES cells, and cultured in the presence of 3 mM histidinol (Sigma, St Louis, MO). Human embryonal kidney (HEK) 293 cells were cultured in DMEM containing 10% fetal bovine serum. Plasmids were introduced into cultured cells by lipofection with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) or calcium phosphate-mediated transfection. The medium was replaced with fresh medium 1 day after transfection and samples were analysed 2 days after transfection.
Plasmid construction
Construction of the expression vectors pCAG-IP, pCAGIP-STAT3, pCAGIP-mycOct3/4, pCAGIP-mycNanog, pCAGIP-mycGCNF and pCAGIP-mycCOUP-TFI was described previously (Yoshida-Koide et al, 2004; Akagi et al, 2005; Kinoshita et al, 2007; Takao et al, 2007). The expression vector pCAG-IHisDR was constructed by replacing the IRES-puromycin resistance-poly A cassette of pCAG-IP with the IRES-histidiol resistance-DsRedT4-poly A cassette of pBRCAG-cHA-IRES-HisDsRedT4. The coding regions of mouse Sox2, COUP-TFII, Elp3 and Tip60 were amplified by PCR using a cDNA library of A3-1 cells as a template with the following primers: 5′-GAA TTC ATG TAT AAC ATG ATG GAG ACG-3′ and 5′-GCG GCC GCT CAC ATG TGC GAC AGG GGC AG-3′ for Sox2, 5′-GCG GCC GCT TAT TGA ATT GCC ATA TAT GGC CAG TTA AAA CTG CTG CCG-3′ and 5′-GCG GCC GCT TAT TGA ATT GCC ATA TAT GGC CAG TTA AAA CTG CTG CCG-3′ for COUP-TFII, 5′-GAA TTC ATG AGG CAA AAG AGG AAA GG-3′ and 5′-GCG GCC GCT TAT TTT AGC ATC TTT ACC A-3′ for Elp3 and 5′-GAA TTC ATG GCG GAG GTG GGG GAG AT-3′ and 5′-GCG GCC GCT CAC CAC TTT CCT CTC TTG C-3′ for Tip60. The plasmids pCAGIHisDR-Flag-Sox2, pCAGIHisDR-Flag-Elp3 and pCAGIHisDR-Flag-Tip60 were constructed by inserting the corresponding cDNAs into pCAG-IHisDR. pCAGIP-Flag-Sox2, pCAGIP-mycSox2, pCAGIP-mycEed1, pCAGIP-mycEed4 and pCAGIP-mycCOUP-TFII were generated by inserting the corresponding cDNAs into pCAG-IP. The small-interfering RNA (siRNA) expression vectors pFIV-control, pFIV-COUP-TFI, pFIV-COUP-TFII#1 and pFIV-COUP-TFII#2 were constructed by inserting 5′-TGC GTT GCT AGT ACC AAC T-3′, 5′-GCA GTT TCA ACT GGC CTT A-3′, 5′-CCA CAT ACG GAT CTT CCA A-3′ and 5′-CCG AGT ATA GCT GCC TCA A-3′ into pFIV-H1/U6-puro (System Biosciences, Mountain View, CA), respectively.
Construction of the three reporter plasmids, pGL2-Eed(−2600/−13), pGL2-Eed(−2600/−13)STAT3mt and pGL2-Eed(−2600/−13)Oct3/4mt, was described previously (Ura et al, 2008). To produce pGL2-Eed(−2600/−13)Sox2mt, mutations (AACAACAG to AACCCCAG) at the Sox2-binding site were introduced into pGL2-Eed(−2600/−13) by PCR. To generate pGL4 promoter, the SV40 promoter sequence of pGL2 promoter (Promega, Madison, WI) was transferred into the Bgl II and Hind III sites of pGL4.10 (Promega). SRR1 and SRR2 were amplified by PCR and subcloned into pGL4 promoter to obtain pGL4pro-SRR1 and pGL4pro-SRR2, respectively. The SRR2 mutant (SRR2mt) containing mutations (CATTGTGATGCATAT to CCTGGGGCTTCCTCT) at Sox2- and Oct3/4-binding sites, the deletion mutants of SRR1 (−2957/−2493, −2666/−2493, −4074/−3205 and −4074/−2685) and the SRR1 mutant (SRR1mt) carrying mutations (AGACCT to CGCCAT) at COUP-TFII-binding site were generated by PCR and inserted into pGL4 promoter.
Luciferase reporter assay
ES cells in a 6-cm dish were transfected with various combinations of plasmids by calcium phosphate-mediated transfection. Two days after transfection, ES cells were harvested and lysed in cell lysis buffer (20 mM Hepes-NaOH (pH 7.2), 10 mM MgCl2, 1 mM EDTA, 10 mM sodium fluoride, 25 mM β-glycerophosphate, 1 mM sodium orthovanadate, 20 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml pepstatin A, 1% nonidet P-40 and 10% glycerol). Luciferase activity was measured using a luciferase assay system (Promega) in a luminometer (Luminescencer AB-2200, ATTO, Tokyo, Japan).
Western blot analysis, immunostaining and ELISA
ES cells were harvested, lysed in 1 × sample buffer (6% glycerol, 10 mM Tris–HCl, 2% SDS, 50 mM DTT, 2 mM EDTA, 0.002% Coomassie Brilliant Blue R250) and heat denatured. Samples were subjected to SDS–PAGE and electrophoretically transferred to a nitrocellulose membrane. The membranes were then probed with antibodies against H3K27me3, H3Ac, H4Ac, H3K56Ac (Millipore, Billerica, MA) and lamin B1 (Santa Cruz Biotechnology, Santa Cruz, CA). The signals were visualized using enhanced chemiluminescence reagents (Perkin-Elmer, Norwalk, CT) with an LAS-1000 image analyser (Fuji Film, Tokyo, Japan), and signal intensity was normalized to lamin B1.
For immunostaining, ES cells were fixed with 4% paraformaldehyde at 4°C for 30 min. After permeabilization with 0.5% Triton X-100 in phosphate-buffered saline (PBS) (−), the cells were pre-incubated with 1% BSA in PBS (−). The cells were then incubated with 1 μg/ml monoclonal anti-Flag-antibody (Sigma), or 300-fold diluted rabbit anti-H3K27me3, anti-H3Ac, anti-H4Ac or anti-H3K56Ac antibodies at 4°C overnight, followed by incubation with 1000-fold diluted goat anti-rabbit or anti-mouse IgG FITC conjugate (Santa Cruz Biotechnology). Signal intensity was determined with Image J software. ELISA was performed using an EpiQuik global histone acetylation kit (Epigentek Group Inc, Brooklyn, NY) according to the manufacturer's protocol.
Knockdown of target genes
Double-stranded siRNAs were purchased from Operon Biotechnologies (Huntsville, AL). The target sequences used were 5′-CUA UCC GUG CUA GAU AUG ACC-3′ for Elp3, 5′-CUA CGU AAU GAC GGA GUA UGA-3′ for Tip60 and 5′-GCC ACA ACG UCU AUA UCA UGG-3′ for EGFP. ES cells (1 × 105 cells) in a 6-cm dish were transfected with siRNA or the siRNA expression vector using Lipofectamine 2000. Two days after transfection, ES cells were harvested and subjected to gene expression analysis.
Quantitative RT–PCR analysis
Total RNAs were isolated from ES cells with Sepasol reagent (Nacalai tesque, Kyoto, Japan) and converted to cDNAs using Revertra Ace (Toyobo, Osaka, Japan) with oligo(dT)12–18 primers (Nippon Gene, Tokyo, Japan). The amount of each cDNA was evaluated by quantitative PCR using MxPro Mx3005P (Stratagene, La Jolla, CA). All samples were tested in triplicate, and the results from each sample were normalized relative to Gapdh expression. The sequences of the primer sets are shown in Supplementary Table S1.
ChIP assay
ChIP assay was performed using antibodies against the Myc epitope, Sox2 (sc-17319) (Santa Cruz Biotechnology), Flag epitope (Sigma), Lys-27-methylated histone H3, acetylated histone H3, acetylated histone H4 or Lys-56-acetylated histone H3 (Millipore), as described previously (Ura et al, 2008). For the detection of precipitated genomic DNA, quantitative PCR was performed. All samples were tested in triplicate and the results from each sample were normalized relative to input DNA. The sequences of the primer sets are shown in Supplementary Table S2.
Supplementary Material
Acknowledgments
This work was supported in part by a Research Fellowship from the Japan Society for the Promotion of Science for Young Scientists (HU), the Naito Foundation (HU and HK), Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (TA, HK and TY) and the Mochida Memorial Foundation for Medical and Pharmaceutical Research (TA).
Footnotes
The authors declare that they have no conflict of interest.
References
- Akagi T, Usuda M, Matsuda T, Ko MSH, Niwa H, Asano M, Koide H, Yokota T (2005) Identification of Zfp-57 as a downstream molecule of STAT3 and Oct-3/4 in embryonic stem cells. Biochem Biophys Res Commun 331: 23–30 [DOI] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R (2003) Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17: 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG (2006) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 [DOI] [PubMed] [Google Scholar]
- Azuma S, Toyoda Y (1991) Production of a germ-line chimeric mouse derived from newly established embryonic stem cells. Jpn J Anim Reprod 37: 37–43 [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T (2008) Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 26: 1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A (2003) Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113: 643–655 [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A (2007) Nanog safeguards pluripotency and mediates germline development. Nature 450: 1230–1234 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK et al. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292: 154–156 [DOI] [PubMed] [Google Scholar]
- Faust C, Lawson KA, Schork NJ, Thiel B, Magnuson T (1998) The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development 125: 4495–4506 [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B (2008) An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134: 162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Fisher JB, Koprowski S, McAllister D, Kim MS, Lough J (2009) Homozygous disruption of the Tip60 gene causes early embryonic lethality. Dev Dyn 238: 2912–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Ura H, Akagi T, Usuda M, Koide H, Yokota T (2007) GABPα regulates Oct-3/4 expression in mouse embryonic stem cells. Biochem Biophys Res Commun 353: 686–691 [DOI] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Jenuwein T, Tempst P, Reinberg D (2004) Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell 14: 183–193 [DOI] [PubMed] [Google Scholar]
- Lee JH, Hart SR, Skalnik DG (2004) Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis 38: 32–38 [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K et al. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL et al. (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440 [DOI] [PubMed] [Google Scholar]
- Martin GR (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA 78: 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H (2007) Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol 9: 625–635 [DOI] [PubMed] [Google Scholar]
- Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, Yokota T (1999) STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J 18: 4261–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Misteli T (2006) Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol 7: 540–546 [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642 [DOI] [PubMed] [Google Scholar]
- Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T (2005) The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol 15: 942–947 [DOI] [PubMed] [Google Scholar]
- Morin-Kensicki EM, Faust C, LaMantia C, Magnuson T (2001) Cell and tissue requirements for the gene eed during mouse gastrulation and organogenesis. Genesis 31: 142–146 [DOI] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Nekrasov M, Wild B, Müller J (2005) Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep 6: 348–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A (1998) Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95: 379–391 [DOI] [PubMed] [Google Scholar]
- Niwa H (2007) How is pluripotency determined and maintained? Development 134: 635–646 [DOI] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A (1998) Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev 12: 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24: 372–376 [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T (2001) The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 21: 4330–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K (2007) The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 27: 3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K (2004) Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23: 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A (2006) Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J 25: 3110–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH (2008) EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M, Gorrini C, Amati B (2006) Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ (2007) Elongator complex: how many roles does it play? Curr Opin Cell Biol 19: 331–336 [DOI] [PubMed] [Google Scholar]
- Takao Y, Yokota T, Koide H (2007) β-Catenin up-regulates Nanog expression through interaction with Oct-3/4 in embryonic stem cells. Biochem Biophys Res Commun 353: 699–705 [DOI] [PubMed] [Google Scholar]
- Tomioka M, Nishimoto M, Miyagi S, Katayanagi T, Fukui N, Niwa H, Muramatsu M, Okuda A (2002) Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res 30: 3202–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H (2008) Identification and characterization of subpopulations in undifferentiated ES cell culture. Development 135: 909–918 [DOI] [PubMed] [Google Scholar]
- Ura H, Usuda M, Kinoshita K, Sun C, Mori K, Akagi T, Matsuda T, Koide H, Yokota T (2008) STAT3 and Oct-3/4 control histone modification through induction of Eed in embryonic stem cells. J Biol Chem 283: 9713–9723 [DOI] [PubMed] [Google Scholar]
- Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, Conway AE, Garcia BA, Plath K, Clark AT, Grunstein M (2009) Histone H3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell 33: 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A (2008) The ground state of embryonic stem cell self-renewal. Nature 453: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida-Koide U, Matsuda T, Saikawa K, Nakanuma Y, Yokota T, Asashima M, Koide H (2004) Involvement of Ras in extraembryonic endoderm differentiation of embryonic stem cells. Biochem Biophys Res Commun 313: 475–481 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.