Abstract
EMBO J 30 11, 2153–2166 (2011); published online April 15 2011
Actin and actin-related proteins (Arps) are critical components of chromatin remodelling and modifying complexes, where they are commonly found in pairs. Yet, little is known about their role in these complexes. In this issue of The EMBO Journal, Fenn et al (2011) report the first structural investigation of two nuclear Arps, the INO80 complex subunits Arp4 and Arp8. Importantly, their results also shed light on the critical role that the nucleotide state of actin may have on its interactions with the Arps.
Actin forms part of a large family of structurally related proteins that include sugar kinases, glycerol kinases, Hsp70 chaperones, as well as the bacterial actin homologues MreB and ParM. Sequence identity among these proteins is negligible, but they all share a common fold, consisting of two structurally similar α/β domains connected by a hinge and by a molecule of ATP binding in a large cleft at the interdomain interface. As a result of this architecture, these proteins undergo nucleotide-dependent conformational changes, and while their core functions are extremely diverse, they can be generally viewed as ATP-dependent molecular switches. In the case of actin, the ATP switch is used to control the transition between its monomeric (G-actin) and filamentous (F-actin) forms in the cytoplasm. More closely related to actin are the Arps (Arp1–Arp11), sharing 17–52% sequence identity with actin, with larger numbers in this nomenclature, indicating increasing divergence from actin (Muller et al, 2005). The Arps can be subdivided into cytosolic (Arps1–3, Arps10–11) and nuclear (Arps4–9). Regardless of their cellular location, Arps are frequently found as hetero-pairs or higher oligomers, forming part of large multiprotein complexes that frequently associate with actin. Thus, in the cytoplasm, Arp2 and Arp3 form part of the Arp2/3 complex that mediates actin filament nucleation and branching, whereas Arp1 and Arp11 form part of the dynactin complex that targets the dynein motor to specific subcellular locations and cargoes. In the nucleus, Arps are found exclusively as components of large chromatin-modifying complexes, including nucleosome remodelling and certain histone acetyltransferase (HAT) complexes, usually involving at least two of the Arps and actin itself. Of particular interest is Arp4 (Baf53 in humans), which is the nuclear Arp most frequently found in such complexes, including INO80, SWI/SNF, SWR1, PBAP, and NuA4. Despite their diverse roles and structures, a unifying feature among Arp-containing chromatin-modifying complexes is that they all include a protein with a helicase-SANT-associated (HSA) domain (Szerlong et al, 2008). Variations in the sequence of the HSA domain allow for the selective recruitment of specific pairs of Arps or actin–Arp. The HSA domain thus emerges as the basic scaffold upon which nuclear Arps and actin are recruited to chromatin-remodelling complexes.
The role of the Arps and specifically Arp4 in chromatin-remodelling complexes is only beginning to emerge, but by analogy with other members of the actin superfamily, nuclear Arps would be expected to modulate the activities of their host complexes through nucleotide-dependent conformational transitions. Evidence is accumulating in support of this view. Thus, for instance, actin and Arp4 are both required for association of the yeast HAT complex NuA4 with histones (Harata et al, 1999; Galarneau et al, 2000), with the binding of ATP to Arp4, breaking it apart from other subunits of the complex and inducing dissociation of the complex from nucleosomes (Sunada et al, 2005).
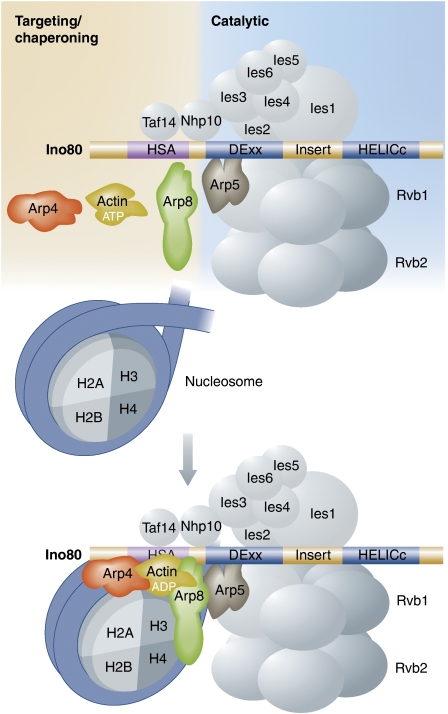
A similar situation may occur in INO80 (Figure 1), a chromatin-remodelling complex with roles in transcription, DNA repair, and DNA replication (Conaway and Conaway, 2009; Morrison and Shen, 2009). This large complex contains stoichiometric amounts of Arp4, Arp5, Arp8, and actin (Shen et al, 2000). Genetic depletion of Arp5 leaves the rest of the complex intact, whereas removal of Arp8 results in concomitant loss of Arp4 and actin from the complex (Shen et al, 2003). This result suggests that the latter three subunits, which are known to interact with the N-terminal HSA domain of the Ino80 core ATPase subunit (Shen et al, 2003; Szerlong et al, 2008), bind to the complex as a unit. Like Arp4, Arp8 has been shown to bind histones in vitro (Harata et al, 1999; Galarneau et al, 2000; Shen et al, 2003), and accordingly the complex lacking Arp8–Arp4–actin is also defective for DNA binding and nucleosome mobilization, suggesting that these subunits have a nucleosome-targeting or histone chaperone function for chromatin remodelling by the INO80 complex (Shen et al, 2003; Kashiwaba et al, 2010; Chen et al, 2011). How the nucleotide state of actin and the Arps may regulate this process is still unknown, but the new results of Fenn et al offer some interesting clues. For the first time, these authors demonstrate direct and preferential interactions of both Arp4 and Arp8 with ADP-bound actin. Specifically, Arp4 was found to bind at the barbed end of actin, and possibly bind simultaneously with Arp8. Their results also suggest that Arp4 could have a role in maintaining the pool of monomeric actin in the nucleus, analogous to the role of profilin and thymosin-β4 in the cytoplasm.
Figure 1.
Model of the INO80 chromatin-remodelling complex. (Top) Untargeted INO80 complex. The core ATPase, Ino80, contains an N-terminal HSA domain that binds Arp4, Arp8, and actin (Szerlong et al, 2008), and a C-terminal ATPase domain that is split into two parts, DExx box domain (DExx) and helicase domain (HELICc). Other subunits of the complex not discussed here are shown in grey. (Bottom) Targeted INO80 complex. The results of Fenn et al suggest that Arp4 and Arp8 may form a complex with actin in the ADP-bound state. Actin–Arp4–Arp8 are probably recruited as a unit to the HSA domain of Ino80 (Shen et al, 2003; Szerlong et al, 2008). The acidic loop insertions of the Arps as compared with actin mediate specific interactions with histones. These interactions bring the INO80 complex to nucleosomes. This complex might be ultimately recycled by a conformational change due to ATP binding to actin, somewhat analogous to the dissociation of NuA4 from histones upon ATP binding to Arp4 (Sunada et al, 2005).
Fenn et al also determined the atomic structure of Arp4 by X-ray crystallography, and solution structures of both Arp4 and Arp8 using small angle X-ray scattering (SAXS). As expected, Arp4 shares the basic fold of actin, but its nucleotide-binding pocket is significantly less accessible to the solvent than that of actin, which the authors suggest correlates with the strong ATP-binding affinity and weak ATPase activity of Arp4 as compared with actin. The solution SAXS structures of Arp4 and Arp8 reveal large loop insertions compared with actin. The SAXS low-resolution envelopes show these insertions towards the pointed and barbed ends of the actin monomer (see cartoons in Figure 1), which in actin are involved in subunit–subunit contacts in the filament, explaining the inability of these Arps to polymerize. Of note, the amino acids in the insertions of Arp4 and Arp8 are predominantly acidic, and thus are thought to mediate specific interactions with histones, which are markedly basic, with Arp8 binding preferentially to histones H3 and H4 and Arp4 to histones H3, H2B, and H2A (Harata et al, 1999; Shen et al, 2003).
While this study represents a significant advance, our understanding of nuclear Arp structure function is still in its infancy. It will be important to perform similar studies on other Arps, and particularly pairs of Arps in association with their specific HSA motifs or even entire chromatin-modifying complexes. It is also becoming clear that nuclear actin is predominantly monomeric, contrary to the main actin functions in the cytoskeleton that are mediated by F-actin. It would be important to understand which factors regulate actin's monomeric state in the nucleus, specifically by structure-function studies of the Arp4–actin complex established by Fenn et al in their work.
Acknowledgments
Supported by NIH National Institute of Mental Health grant MH087950.
Footnotes
The authors declare that they have no conflict of interest.
References
- Chen L, Cai Y, Jin J, Florens L, Swanson SK, Washburn MP, Conaway JW, Conaway RC (2011) Subunit organization of the human INO80 chromatin remodeling complex: an evolutionarily conserved core complex catalyzes atp-dependent nucleosome remodeling. J Biol Chem 286: 11283–11289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW (2009) The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem Sci 34: 71–77 [DOI] [PubMed] [Google Scholar]
- Fenn S, Breitsprecher D, Gerhold CB, Witte G, Faix J, Hopfner KP (2011) Structural biochemistry of nuclear actin-related proteins 4 and 8 reveals their interaction with actin. EMBO J 30: 2153–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau L, Nourani A, Boudreault AA, Zhang Y, Heliot L, Allard S, Savard J, Lane WS, Stillman DJ, Cote J (2000) Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell 5: 927–937 [DOI] [PubMed] [Google Scholar]
- Harata M, Oma Y, Mizuno S, Jiang YW, Stillman DJ, Wintersberger U (1999) The nuclear actin-related protein of Saccharomyces cerevisiae, Act3p/Arp4, interacts with core histones. Mol Biol Cell 10: 2595–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwaba S, Kitahashi K, Watanabe T, Onoda F, Ohtsu M, Murakami Y (2010) The mammalian INO80 complex is recruited to DNA damage sites in an ARP8 dependent manner. Biochem Biophys Res Commun 402: 619–625 [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Shen X (2009) Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol 10: 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Oma Y, Vallar L, Friederich E, Poch O, Winsor B (2005) Sequence and comparative genomic analysis of actin-related proteins. Mol Biol Cell 16: 5736–5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C (2000) A chromatin remodelling complex involved in transcription and DNA processing. Nature 406: 541–544 [DOI] [PubMed] [Google Scholar]
- Shen X, Ranallo R, Choi E, Wu C (2003) Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol Cell 12: 147–155 [DOI] [PubMed] [Google Scholar]
- Sunada R, Gorzer I, Oma Y, Yoshida T, Suka N, Wintersberger U, Harata M (2005) The nuclear actin-related protein Act3p/Arp4p is involved in the dynamics of chromatin-modulating complexes. Yeast 22: 753–768 [DOI] [PubMed] [Google Scholar]
- Szerlong H, Hinata K, Viswanathan R, Erdjument-Bromage H, Tempst P, Cairns BR (2008) The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol 15: 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]