Abstract
Background
Huntington disease (HD) is an inherited neurodegenerative disease caused by an abnormal expansion of a CAG repeat in the huntingtin HTT (HD) gene. The primary genetic determinant of the age at onset (AO) is the length of the HTT CAG repeat; however, the remaining genetic contribution to the AO of HD has largely not been elucidated. Recent studies showed that impaired functioning of the peroxisome proliferator-activated receptor gamma coactivator 1a (PGC-1alpha) contributes to mitochondrial dysfunction and appears to play an important role in HD pathogenesis. Further genetic evidence for involvement of PGC-1alpha in HD pathogenesis was generated by the findings that sequence variations in the PPARGC1A gene encoding PGC-1alpha exert modifying effects on the AO in HD. In this study, we hypothesised that polymorphisms in PGC-1alpha downstream targets might also contribute to the variation in the AO.
Results
In over 400 German HD patients, polymorphisms in the nuclear respiratory factor 1 gene, NRF-1, and the mitochondrial transcription factor A, encoded by TFAM showed nominally significant association with AO of HD. When combining these results with the previously described modifiers rs7665116 in PPARGC1A and C7028T in the cytochrome c oxidase subunit I (CO1, mt haplogroup H) in a multivariable model, a substantial proportion of the variation in AO can be explained by the joint effect of significant modifiers and their interactions, respectively.
Conclusions
These results underscore that impairment of mitochondrial function plays a critical role in the pathogenesis of HD and that upstream transcriptional activators of PGC-1alpha may be useful targets in the treatment of HD.
Background
Huntington disease (HD) is an autosomal dominantly transmitted, progressive neurodegenerative disease associated with a polymorphic CAG trinucleotide repeat in the 5' part of the HTT (HD) gene, which is expanded and translated into an elongated polyglutamine tract in the huntingtin protein [1].
The length of the expanded CAG tract is inversely related to the age at clinical onset of HD, accounting for more than half of the overall variance in age at onset (AO) [2-4]. Yet, despite this strong correlation, there remains considerable variation in AO (of more than 40 years) in individuals with identical repeat lengths. Evidence has been provided for genetic as well as for environmental factors that affect the AO [5]. Identifying these modifiers in human HD and defining their precise role in the causal pathogenesis of HD could help to develop more effective treatment regimen for HD. To date, several candidate modifier genes of HD have been described in independent studies, all of them implicating a variety of processes apparently contributing to HD pathogenesis [6-10]. Recently, mitochondrial DNA (mtDNA) haplogroup H (7028C) and variations in the peroxisome proliferator-activated receptor gamma coactivator 1a (PPARGC1A) gene encoding PGC-1alpha were shown to exert modifying effects on the AO in HD, thus providing genetic evidence that complex interrelations of mitochondrial dysfunction have effects on the pathogenic process in HD [11-14]. A number of studies suggested that PGC-1alpha dysfunction may be central to HD pathogenesis [15]. PGC-1alpha-deficient mice show hyperkinetic movement disorder and striatal degeneration [16,17]. Gene expression analyses in cell lines, transgenic mouse models of HD and in different tissues from HD patients revealed a disruption of the PGC-1alpha regulatory pathway [18-20]. Recently, investigations on the ability of AMP-activated protein kinase (AMPK) to activate PGC-1alpha in brain, liver, brown adipose tissue (BAT) and muscle of HD transgenic mice strengthened the theory that impaired activation of PGC-1alpha plays an important role in the metabolic disturbances involved in the pathology of HD [20,21]. Accordingly, modulation of PGC-1alpha levels and activity has been proposed as a therapeutic option for HD pathology [15]. Indeed, activation of the PGC-1alpha signaling pathway via resveratrol-induced activation of the silent information regulator T1 (SIRT1), a mammalian sirtuin, in transgenic mice achieved positive effects in BAT [22]. Yet, PGC-1alpha as a coactivator protein responds to environmental influences and subsequently regulates various pathways in a tissue-specific and highly coordinated manner. Therefore, such pharmacological interventions aimed at PGC-1alpha may suffer from lack of specificity. Targeting key factors of the wide PGC-1alpha transcriptional network could, therefore, represent another approach for a specific modulation of PGC-1alpha activity. PGC-1alpha controls many aspects of oxidative metabolism, including respiration and mitochondrial biogenesis through coactivation and enhancing the expression and activity of several transcription factors including the nuclear respiratory factors (NRF)-1 and NRF-2 (GABP) and the estrogen related receptor alpha (ERRalpha) [23,24]. PGC-1alpha is also indirectly involved in regulating the expression of mtDNA transcription via increased expression of mitochondrial transcription factor A (TFAM) which is coactivated by NRF-1 [23,25]. In the present study, we addressed the question of the role of a diverse set of PGC-1alpha related factors in modifying the AO of HD. We investigated polymorphisms in the genes encoding ERRalpha (ESRRA), Mitofusin 2 (MFN2), NRF-1 and NRF-2 (NRF-1, GABPA and GABPB1), PGC-1beta (PPARGC1B), peroxisome proliferator-activated receptor-gamma (PPARG), SIRT1 (SIRT1) and TFAM (TFAM, see Table 1). A more comprehensive understanding of the pleiotropic effects of the PGC-1alpha family regulatory network in mitochondrial biogenesis and HD pathogensis could help to identify and fine tune pharmacological interventions targeting PGC-1alpha or alternatively its transcriptional complexes.
Table 1.
Candidate gene and SNP characteristics
| Gene | Chromosome | SNPs | Selection Criteria | |
|---|---|---|---|---|
| ESRRA | 11q13.1 | rs3217060 | 23 bp microsatellite repeat located in promoter region | |
| MFN2 | 1p36.22 | rs3753579 | located in promoter region | |
| NRF-1 | 7q32.2 | rs7781972, rs6949152 | previously reported association [29] | |
| rs1882094, rs3735006, | non-synonymous SNPs | |||
| rs10275661, rs10225103, rs10268267, rs6962005, rs10231985, rs11487138, rs11761434, rs1962039, rs2402970, rs6948697, rs10500120 | tag SNPs using public databases (dbSNP) | |||
| NRF-2 | GABPA | 21q21.3 | rs2829897 (A291V), rs2829898 (W323X), rs2829900 (E345K) | non-synonymous SNPs |
| GABPB1 | 15q21.2 | rs12594956, rs8031031 | previously reported association [39] | |
| PPARGC1B | 5q33.1 | rs7732671 (A164P), rs11959820 (R253S) | non-synonymous SNPs | |
| PPARG | 3p25.2 | rs1801282 (P12A) | non-synonymous SNP | |
| rs2938392, rs3856806 | tag SNPs using public databases (dbSNP) | |||
| SIRT1 | 10q21.3 | rs3758391, rs7069102 | previously reported association [40] | |
| rs10997860, rs2273773, rs35461348 | tag SNPs using public databases (dbSNP) | |||
| TFAM | 10q21.1 | rs1937 (S12T) | non-synonymous SNP | |
| rs4390300, rs10826178, rs1049432, rs11006132 | tag SNPs using public databases (dbSNP) | |||
Results
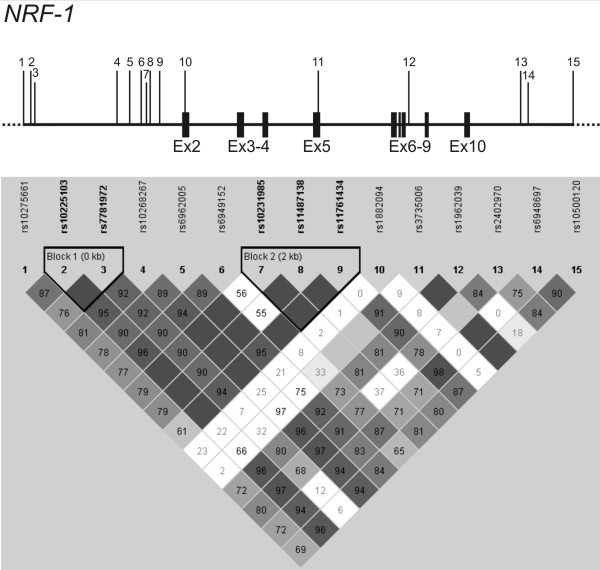
In our cohort of 401 HD patients, the expanded HTT allele accounts for nearly 73% of the variance in motor AO (R2 = 0.729) and shows a highly significant influence on the AO (p < 0.0001). Multiple regression models were used to test all SNPs for association with the AO. Of these, four showed a nominal p-value < 0.05 (Table 2). For NRF-1, addition of the intronic variations rs7781972 and rs6949152 showed an association with the motor AO. Inclusion of the rs7781972 genotypes in the model increased the R2 statistic from 0.729 to 0.733 in both the dominant and the additive model (p = 0.017 and p = 0.011, Table 2). Inclusion of the rs6949152 genotypes increased the R2 from 0.729 to 0.734 (p = 0.004) according to the dominant model and to 0.733 (p = 0.013) in the additive model (Table 2). Examining linkage disequilibrium (LD) among the 15 NRF-1 variations revealed, that the variations rs10275661, rs10225103, rs7781972, rs10268267, rs6962005 and rs6949152 in IVS1 were in high LD in the cohort (D' = 1.0, r2 = 0.87-0.92). In 3' direction the LD breaks down, and a second block of very strong LD (D' = 1.0, r2 ≥ 0.98) is observed for rs10231985, rs11487138 and rs11761434. The remaining SNPs covering exon 2 to IVS10 (rs1882094, rs3735006, rs1962039, rs2402970, rs6948697 and rs10500120) showed lower LD coefficients (D') and r2 values (Figure 1).
Table 2.
Details of SNPs included in the multivariable model
| Gene | SNP | Chromosome | Allele Freq. | Type | Biological Processes | ΔR 2 | Standardized beta | ||
|---|---|---|---|---|---|---|---|---|---|
| Coefficients | t | P * | |||||||
| NRF-1 | rs7781972 | 7q32.2 | 189/170/42 | IVS1 | control of nuclear genes required for respiration, heme biosynthesis, and mtDNA transcription and replication | .004 | -.066 | -2.56 | .011 |
| rs6949152 | 289/101/11 | IVS1 | .005 | -.076 | -2.94 | .004 | |||
| TFAM | rs1049432 | 10q21.1 | 239/145/17 | 3' near gene | mtDNA transcription and maintenance factor | .002 | -.053 | -2.04 | .042 |
| rs11006132 | 208/170/23 | 3' near gene | .004 | -.063 | -2.45 | .015 | |||
| PPARGC1A | rs7665116 | 4p15.2 | 309/82/10 | IVS2 | transcriptional coactivator, regulation of key mitochondrial genes | .004 | -.065 | -2.52 | .012 |
| CO1 | C7028T | mt7028 | 207/194 | mitochondrial | component of the respiratory chain, catalyses the reduction of oxygen to | .003 | -.057 | -2.22 | .027 |
The variability in motoric AO attributable to the CAG repeat length was assessed by linear regression using the logarithmically transformed onset age as the dependent variable and genotypes as independent variables. Delta (Δ) R2 quantifies the relative improvement of the regression model when the genotypes are considered in addition to the CAG repeats. * nominal P-values (not adjusted for multiple testing)
Figure 1.
Graphical representation of single-nucleotide polymorphisms (SNPs) in relation to the exon-intron structure (top) and the Haploview pairwise linkage disequilibrium (LD) structure of part of NRF-1 (bottom). Exons are indicated by solid black boxes, and the numbered vertical lines indicate positions of the SNPs analysed in NRF-1. Haploview plot showing pairwise LD (D' values) for all 15 SNPs based on genotypes of 401 HD patients of the study. Each square plots the level of LD between a pair of SNPs, comparisons between neighboring SNPs are arranged along the first line under the names of the SNPs. Dark grey coloring indicates strong LD, medium grey shading indicates less strong LD, light grey indicates intermediate LD, and white indicates weak LD. LD blocks are framed in black and were classified according to Gabriel et al. (2002).
In TFAM two SNPs showed association with motor AO among which, rs11006132 in the 3' region of the gene was most strongly associated (0.729 to 0.733; p = 0.015, Table 2). Including the genotypes of the coding Ser12Thr polymorphism (rs1937) in the model for AO did not increase the R2 statistic. The TFAM variations were in moderate LD with one another (D' < 1.0, r2 = 0.34-0.47), only the associated variations rs1049432 and rs11006132 in the 3' region are highly correlated with each other (pairwise r2 values >0.97). All other selected polymorphisms in NRF2, SIRT1, PGC1beta, MFN2 and PPARgamma showed no significant influence on the AO.
After consideration of SNP genotypes individually, a multivariable model was built in order to determine if a significant proportion of the variation in AO could be explained by the joint effect of mitochondrion-related modifier variations and their interactions. NRF-1 and TFAM SNPs showing significant main effects (nominal P < 0.05) together with the previously analysed modifier variations in PPARGC1A (rs7665116) and CO1 (C7028T, defining mt haplogroup H) were included in a multivariable model (Table 2). Here, the main effects together with all possible pairwise interactions of the SNPs were included in a forward selection process. The final multivariable model increased the R2 statistic from 0.729 to 0.747 and explained 4.8% additional residual variance in the motor AO of HD (Table 3).
Table 3.
SNP-SNP interaction included in final multivariable model
| Interaction | R 2 | Adjusted R 2 | ΔR 2 | Standardized beta | ||
|---|---|---|---|---|---|---|
| Coefficients | t | P | ||||
| CAG | .730 | .729 | -.864 | -34.23 | <0.0001 | |
| NRF-1 rs6949152 * CO1 C7028T | .739 | .738 | .009 | -.090 | -3.55 | <0.0001 |
| NRF-1 rs7781972 * TFAM rs11006132 | .745 | .743 | .014 | -.077 | -3.03 | <0.003 |
| PPARGC1A rs7665116 | .749 | .747 | .018 | -.069 | -2.72 | <0.007 |
A multivariable model including a total of four interactions was built in the total HD cohort using forward selection. With the addition of each SNP-SNP interaction, along with each SNP's respective main effect, the variability in adjusted log motor AO increased (assessed by adjusted R2). The final multivariable model explains 4.8% additional residual variance in HD motor AO.
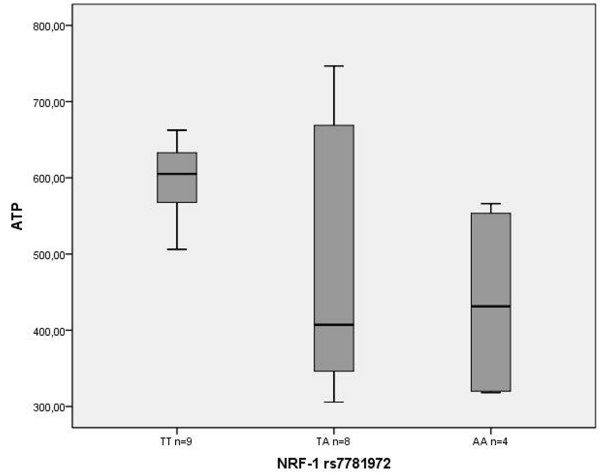
When correlating the ATP concentrations with the NRF-1 and TFAM genotypes, HD patients carrying at least one rare NRF-1 rs7781972 allele showed significantly lower ATP concentrations (487.1 ± 179 ng, n = 8; 436.7 ± 135.1 ng, n = 4) than homozygous individuals carrying the frequent allele (600.6 ± 48.7 ng, n = 9, p = 0.03; Figure 2). Considering an additive allele effect the ATP levels were negatively correlated with the rare NRF-1 rs7781972 allele (Pearson coefficient -0.478, p < 0.029). Yet, this effect was not obvious in a group of 38 healthy controls (529 ± 175.5 ng, n = 14; 487.4 ± 148 ng, n = 22 vs. 492 ± 120.5 ng, n = 2, Pearson coefficient -0.130, p < 0.437). In both groups the ATP levels were not significantly correlated with the mtDNA:nDNA ratios (HD: Spearman coefficient -0.383, p < 0.095, controls: 0.134, p < 0.417). Regarding the entire patient cohort (n = 401), the mtDNA content was also not associated with age, sex, AO, disease duration, CAG repeat lengths or any other genotype.
Figure 2.
ATP concentration in HD patients. Box plot shows medians, quartiles and extreme values. The mean ATP levels of HD patients with the TT genotype (600.6 ± 48.7 ng, n = 9) and those carrying at least one rare NRF-1 rs7781972 allele (TA and AA, 487.1 ± 179 ng, n = 8; 436.7 ± 135.1 ng, n = 4) were significantly different (p = 0.03 two-sample t test).
Discussion
Here, we performed an association study for AO modifiers of HD, based on a candidate gene approach including variations in PGC-1alpha target genes involved in the regulatory network that controls mitochondrial function. We found that SNPs in NRF-1 and TFAM showed nominally significant association with AO of HD.
The NRFs and - most importantly NRF-1 - are potent stimulators of the expression of nuclear genes required for mitochondrial respiratory function [23-25]. NRF-1 directly regulates the expression of several nuclear encoded genes involved in the expression, assembly and functions of the respiratory chain or indirectly regulates the mitochondrion-encoded cytochrome c oxidase (COX) subunit genes by activating TFAM [23-25]. Beyond the transcriptional expression of the respiratory chain NRF-1 is also supposed to control the key components of the protein import and assembly machinery, thus suggesting a broader meaning for NRF-1 in orchestrating events in the mitochondrial biogenesis [26]. Yet, a direct functional effect of the associated polymorphisms remains to be determined. Comparing the mtDNA content with various clinical and genetic parameters of the HD patients did not yield statistically significant results. Yet, the interpretation of these results has to be considered with caution, since expressing the mitochondrial DNA concentration as a ratio to nuclear DNA varies dependent on blood-processing protocols [27].
Given the relatively small sample sizes, the additive allele effect of NRF-1 rs7781972 on the ATP levels in HD patients should be regarded as preliminary. Nevertheless, these data could be indicative of genotype-dependent variation in the response to chronic energy stress conditions. Since high-intensity exercise also causes metabolic stress, physical activity on a very high level for prolonged time implies, amongst other effects, increased oxidative stress and the consequences of cellular dysfunction due to insufficient supply of ATP [28]. In this context, it is interesting to note that NFR-1 genotypes - particularly regarding rs6949152 - significantly influence phenotype traits indicative of endurance capacity in humans and their trainability [29].
Deviation in the response of certain NRF-1 genotypes on chronic energy stress conditions (endurance exercise training, but also neurodegeneration) could therefore explain the association with the variation in the onset age of HD. The same could apply to the other genotypes, since TFAM, PPARGC1A and mt haplogroups have also been described to be associated with differences in physical capabilities and training-induced effects [30-34]. Recently, Chaturvedi et al. [21] demonstrated that chronic energy deprivation in mice by administering the catabolic stressor β-guanidinopropionic acid increased expression of PGC-1alpha, NRF-1 and TFAM. Yet, this pathway, leading to mitochondrial biogenesis, increased mtDNA and numbers of mitochondria in response to energetic stress, was blocked in HD transgenic mice [21]. Furthermore, when combining the newly detected modifier variations in NRF-1 and TFAM with the previously described modifier variations in PPARGC1A and mt haplogroup H, in our study much more variability in AO can be explained than in seperate analyses. The combination of the polymorphisms defines nearly 5% of the unexplained variance in residual AO in our sample, thus highlighting their coordinately regulated metabolic interplay and the possible involvement in pathogenic HD conditions.
The important role of PGC-1alpha in the regulation of mitochondrial function together with the association between mitochondrial dysfunction and HD pathogenesis implies that activation of PGC-1alpha could have critical potential in the treatment of HD. Yet, pharmacological interventions directly aimed at PGC-1alpha have to overcome inherent limitations of targeting a coactivator protein [35]. Therefore, targeting the regulators of PGC-1alpha, as already demonstrated for its down-stream target ERRalpha and the NAD+-dependent deacetylase SIRT1, may represent an alternative approach [36].
Conclusions
Polymorphisms in NRF-1 and TFAM influence the AO of HD. Furthermore, we have demonstrated evidence for gene-gene (SNP-SNP) interactions among these SNPs and the modifier variations in PPARGC1A and CO1, thus providing further genetic evidence that impaired mitochondrial biogenesis in response to energetic stress plays a critical role in the pathogenesis of HD. These data also support the idea that upstream transcriptional activators of PGC-1alpha may be useful in the treatment of HD. However, since no multiple testing correction was applied, caution is necessary in interpreting. Further studies will be necessary to replicate these associations and to elucidate the pathways through which the modifier variations exert their effects on metabolic deficits underlying HD pathogenesis and potentially other late-onset diseases.
Methods
Study population
The study population has been described before [12] and consisted of 401 unrelated German patients (208 men and 193 women) with the clinical and genetic diagnosis of HD, recruited from the Huntington Center NRW, Bochum (Germany). AO was defined as the age at which, according to the experienced neurologists of the Center, the first motor signs of HD appeared (motor AO). The expanded trinucleotide repeats ranged from 40 to 66 with a mean (±SD) of 44.48 ± 3.8 CAGs, motor AO ranged from 16 to 76 years, with an onset (mean ± SD) of 44.9 ± 11.6 years. The normal CAG blocks ranged from from 10 to 32 with a mean (±SD) of 18.24 ± 2.8 CAGs. HD CAG repeat sizes were determined by polymerase chain reaction using an assay counting the perfectly repeated (CAG) n units. The study was performed in a manner that fully complies with the Code of Ethics of the World Medical Association (Declaration of Helsinki) and was approved by the ethics review board of the Ruhr-University Bochum (Germany).
Candiate gene and SNP selection strategy
In order to estimate a possible modifier effect conferred by individual SNPs, as well SNP-SNP interactions, we studied SNPs from 9 candidate genes contributing to the PGC-1alpha family regulatory network in mitochondrial biogenesis (Table 1). The candidate genes and SNPs were selected using the available published evidence at the beginning of the project. SNPs were chosen based on a number of different criteria including the published data, non-synonymous SNPs and tag SNPs from public databases such as dbSNP [http://www.ncbi.nlm.nih.gov/SNP].
Genotyping
We studied a total of 15 SNPs in NRF-1 (rs10275661, rs10225103, rs7781972, rs10268267, rs6962005, rs6949152, rs10231985, rs11487138, rs11761434, rs1882094, rs3735006, rs1962039, rs2402970, rs10500120, rs6948697), three in GABPA (NRF-2a, rs2829897, rs2829898, rs2829900), two in GABPB1 (NRF-2b, rs12594956, rs8031031), five in TFAM (rs4390300, rs1937, rs10826178, rs1049432, rs11006132), five SNPs in SIRT1 (rs3758391, rs10997860, rs7069102, rs2273773, rs35461348), two in PPARGC1B (rs11959820, rs7732671), one in MFN2 (rs3753579), three in PPARγ (rs1801282, rs2938392, rs2938392) and one in ESRRA (rs3217060). Genotyping was performed by PCR-RFLP techniques and commercially available TaqMan genotyping assay (Applied Biosystems). For fragment analysis of the 23 bp microsatellite repeat (ESRRA23, rs3217060) in the promoter of ESRRA and the insertion-deletion (INDEL) polymorphism (rs35461348) in SIRT1 we used fluorescence 5'FAM labelled, tailed oligonucleotide added to the 5'-part of the sequence specific primer as described before [37]. All primers were designed with the Primer Express 2.0 Software (Applied Biosystems, Foster City, USA). All other details of the methodology and primer sequences are available upon request.
mtDNA Quantification
Quantitative real-time PCR (qPCR) was used for mitochondrial DNA content measurement using an Applied Biosystem StepOne cycler (Applied Biosystems, Foster City, CA). Correction for mtDNA quantity was performed by simultaneous measurement of a single copy nuclear RNAseP gene. Quantification of nuclear (n) DNA was done with a commercial kit (RNAseP, Control Reagents, Applied Biosystems P/N 4316844) together with nDNA-specific fluorescent probe which was labelled internally using VIC fluorescent dye. Two primers and one probe used for mtDNA 12S ribosomal RNA quantification which were as here: mtF805 (5'CCACGGGAAACAGCAGTGATT3'), mtR927 (5'CTATTGACTTGGGTTAATCGTGTGA3') and TaqMan probe (Applied Biosystems) (6FAM-5'TGCCAGCCACCGCG3'-MGB) (labelled at the 5' end with a fluorescent reporter, 6FAM). The 10-μL PCR reaction contains 1 × TaqMan Universal PCR Master Mix (Applied Biosystems P/N 4304437), 0.5 μL of PDARs RNAseP and 112 nM of each mtDNA primer, 125 nM of mtDNA TaqMan probe, and 25 ng of total genomic DNA extract. PCR conditions were 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 sec of denaturation at 95°C and 60 sec of annealing/extension at 60°C.
In order to determine the quantities of mtDNA and nDNA, the average threshold cycle number (Ct) values of the nDNA and mtDNA were obtained from each case. Measurements were performed in triplicates and presented as means. The level of mtDNA was calculated using the delta Ct (ΔCt) of average Ct of mtDNA and nDNA (ΔCt = CtmtDNA-CtnDNA) in the same well as an exponent of 2 (2ΔCt). To test reproducibility a constant reference sample was analysed in each run.
Assessment of intracellular ATP concentrations in peripheral leukocytes
Sodium heparin blood was obtained from 21 HD patients and 38 age-/sex matched healthy controls devoid of acute infections (clinical aspect, white blood cell count, C-reactive protein). Assessment of intracellular ATP concentrations was performed as described before [13].
Statistical analysis
Variability in AO attributable to the CAG repeat number was controlled by linear regression using the logarithmically transformed AO as the dependent variable SNP genotypes as independent variables. All analyses were performed assuming a dominant or an additive effect for each polymorphism. In the dominant model, both, the heterozygous and the rarely observed homozygous variation were combined. In the additive model, both, rare homozygous and heterozygous variation effects were estimated using two dummy variables. We used a two-stage approach in order to identify both main and interactive genetic effects associated with the motor AO. In the first stage we conducted association analyses for SNP main effects. After this, we performed multivariable SNP modelling with associations passing the first stage of analysis as well as two previously published modifier SNPs in PPARGC1A (rs7665116) and CO1 (C7028T) [12,13]. The results were not adjusted for multiple testing as a Bonferroni adjustment would have been very conservative when taking into account that the SNPs were not independent, rather they were in tight LD. Hardy-Weinberg equilibrium (HWE) was tested for each SNP. Relationships between variables were determined by Pearson's correlation coefficient. The strength of LD between pairs of SNPs was measured as D' by using HAPLOVIEW [http://www.broad.mit.edu/mpg/haploview/]. LD blocks were inferred from the definition proposed by Gabriel et al. [38] as implemented in HAPLOVIEW with D' confidence bounds of 0.7-0.92. Comparison of dependent variables was performed using unpaired t tests with nominal significance assigned when p ≤ 0.05. SPSS Ver.18.0 (SPSS Inc.) was used for all statistical analyses.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
ETF carried out the molecular genetic experiments. CS ascertained the clinical status of the patients. DAA, SW, AH and AC contributed to the experimental design. JTE supervised the overall project, and LA designed the study including statistical analysis and drafted the manuscript. All authors read and approved the final version of the manuscript.
Contributor Information
Elahe Taherzadeh-Fard, Email: elahe.taherzadeh@yahoo.com.
Carsten Saft, Email: carsten.saft@rub.de.
Denis A Akkad, Email: amer.Akkad@rub.de.
Stefan Wieczorek, Email: stefan.wieczorek@rub.de.
Aiden Haghikia, Email: aiden.haghikia@rub.de.
Andrew Chan, Email: andrew.chan@rub.de.
Jörg T Epplen, Email: joerg.t.epplen@rub.de.
Larissa Arning, Email: larissa.arning@rub.de.
References
- The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is unstable on Huntington's disease chromosomes. Cell. 1993;6:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Snell RG, MacMillan JC, Cheadle JP, Fenton I, Lazarou LP, Davies P, MacDonald ME, Gusella JF, Harper PS, Shaw DJ. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington's disease. Nat Genet. 1993;6:393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- Andrew SE, Goldberg YP, Kremer B, Telenius H, Theilmann J, Adam S, Starr E, Squitieri F, Lin B, Kalchman MA. et al. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat Genet. 1993;6:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- Duyao M, Ambrose C, Myers R, Noveletto A, Persichetti F, Frontali M, Folstein S, Ross C, Franz M, Abbott M. et al. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat Genet. 1993;6:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- Gusella JF, Macdonald ME. Huntington's disease: the case for genetic modifiers. Genome Med. 2009;6:80. doi: 10.1186/gm80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresen JM, Gaya'n J, Cherny SS, Brocklebank D, Alkorta-Aranburu G, Addis EA. US-Venezuela Collaborative Research Group. Cardon LR, Housman DE, Wexler NS. Replication of twelve association studies for Huntington's disease residual age of onset in large Venezuelan kindreds. J Med Genet. 2007;6:44–50. doi: 10.1136/jmg.2006.045153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arning L, Kraus PH, Valentin S, Saft C, Andrich J, Epplen JT. NR2A and NR2B receptor gene variations modify age at onset in Huntington disease. Neurogenetics. 2005;6:25–28. doi: 10.1007/s10048-004-0198-8. [DOI] [PubMed] [Google Scholar]
- Arning L, Monté D, Hansen W, Wieczorek S, Jagiello P, Akkad DA, Andrich J, Kraus PH, Saft C, Epplen JT. ASK1 and MAP2K6 as modifiers of age at onset in Huntington's disease. J Mol Med. 2008;6:485–490. doi: 10.1007/s00109-007-0299-6. [DOI] [PubMed] [Google Scholar]
- Metzger S, Rong J, Nguyen HP, Cape A, Tomiuk J, Soehn AS, Propping P, Freudenberg-Hua Y, Freudenberg J, Tong L. et al. Huntingtin-associated protein-1 is a modifier of the ageat - onset of Huntington's disease. Hum Mol Genet. 2008;6:1137–1146. doi: 10.1093/hmg/ddn003. [DOI] [PubMed] [Google Scholar]
- Taherzadeh-Fard E, Saft C, Wieczorek S, Epplen JT, Arning L. Age at onset in Huntington's disease: replication study on the associations of ADORA2A, HAP1 and OGG1. Neurogenetics. 2010;6:435–439. doi: 10.1007/s10048-010-0248-3. [DOI] [PubMed] [Google Scholar]
- Weydt P, Soyal SM, Gellera C, Didonato S, Weidinger C, Oberkofler H, Landwehrmeyer GB, Patsch W. The gene coding for PGC-1alpha modifies age at onset in Huntington's disease. Mol Neurodegener. 2009;6:3. doi: 10.1186/1750-1326-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taherzadeh-Fard E, Saft C, Andrich J, Wieczorek S, Arning L. PGC-1alpha as modifier of onset age in Huntington disease. Mol Neurodegener. 2009;6:10. doi: 10.1186/1750-1326-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arning L, Haghikia A, Taherzadeh-Fard E, Saft C, Andrich J, Pula B, Höxtermann S, Wieczorek S, Akkad DA, Perrech M, Gold R, Epplen JT, Chan A. Mitochondrial haplogroup H correlates with ATP levels and age at onset in Huntington disease. J Mol Med. 2010;6:431–436. doi: 10.1007/s00109-010-0589-2. [DOI] [PubMed] [Google Scholar]
- Che HV, Metzger S, Portal E, Deyle C, Riess O, Nguyen HP. Localization of sequence variations in PGC-1α influence their modifying effect in Huntington disease. Mol Neurodegener. 2011;6:1. doi: 10.1186/1750-1326-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Róna-Vörös K, Weydt P. The role of PGC-1α in the pathogenesis of neurodegenerative disorders. Curr Drug Targets. 2010;6:1262–1269. doi: 10.2174/1389450111007011262. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jäger S, Vianna CR, Reznick RM. et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;6:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C. et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;6:101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;6:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Weydt P, Pineda VV, Torrence AE, Libby RT, Satterfield TF, Lazarowski ER, Gilbert ML, Morton GJ, Bammler TK, Strand AD. et al. Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab. 2006;6:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Chaturvedi RK, Adhihetty P, Shukla S, Hennessy T, Calingasan N, Yang L, Starkov A, Kiaei M, Cannella M, Sassone J, Ciammola A, Squitieri F, Beal MF. Impaired PGC-1alpha function in muscle in Huntington's disease. Hum Mol Genet. 2009;6:3048–3065. doi: 10.1093/hmg/ddp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi RK, Calingasan NY, Yang L, Hennessey T, Johri A, Beal MF. Impairment of PGC-1alpha expression, neuropathology and hepatic steatosis in a transgenic mouse model of Huntington's disease following chronic energy deprivation. Hum Mol Genet. 2010;6:3190–3205. doi: 10.1093/hmg/ddq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho DJ, Calingasan NY, Wille E, Dumont M, Beal MF. Resveratrol protects against peripheral deficits in a mouse model of Huntington's disease. Exp Neurol. 2010;6:74–84. doi: 10.1016/j.expneurol.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta. 2002;6:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta. 2010. in press . [DOI] [PMC free article] [PubMed]
- Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;6:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- Blesa JR, Prieto-Ruiz JA, Hernández JM, Hernández-Yago J. NRF-2 transcription factor is required for human TOMM20 gene expression. Gene. 2007;6:198–208. doi: 10.1016/j.gene.2006.12.024. [DOI] [PubMed] [Google Scholar]
- Chiu RW, Chan LY, Lam NY, Tsui NB, Ng EK, Rainer TH, Lo YM. Quantitative analysis of circulating mitochondrial DNA in plasma. Clin Chem. 2003;6:719–726. doi: 10.1373/49.5.719. [DOI] [PubMed] [Google Scholar]
- Coyle EF. Physical activity as a metabolic stressor. Am J Clin Nutr. 2000;6:512S–520S. doi: 10.1093/ajcn/72.2.512S. [DOI] [PubMed] [Google Scholar]
- He Z, Hu Y, Feng L, Li Y, Liu G, Xi Y, Wen L, Lucia A. NRF-1 genotypes and endurance exercise capacity in young Chinese men. Br J Sports Med. 2008;6:361–366. doi: 10.1136/bjsm.2007.042945. [DOI] [PubMed] [Google Scholar]
- Lucia A, Gómez-Gallego F, Barroso I, Rabadán M, Bandrés F, San Juan AF, Chicharro JL, Ekelund U, Brage S, Earnest CP. et al. PPARGC1A genotype (Gly482Ser) predicts exceptional endurance capacity in European men. J Appl Physiol. 2005;6:344–348. doi: 10.1152/japplphysiol.00037.2005. [DOI] [PubMed] [Google Scholar]
- Niemi AK, Majamaa K. Mitochondrial DNA and ACTN3 genotypes in Finnish elite endurance and sprint athletes. Eur J Hum Genet. 2005;6:965–969. doi: 10.1038/sj.ejhg.5201438. [DOI] [PubMed] [Google Scholar]
- Castro MG, Terrados N, Reguero JR, Alvarez V, Coto E. Mitochondrial haplogroup T is negatively associated with the status of elite endurance athlete. Mitochondrion. 2007;6:354–357. doi: 10.1016/j.mito.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Stefan N, Thamer C, Staiger H, Machicao F, Machann J, Schick F, Venter C, Niess A, Laakso M, Fritsche A. et al. Genetic variations in PPARD and PPARGC1A determine mitochondrial function and change in aerobic physical fitness and insulin sensitivity during lifestyle intervention. J Clin Endocrinol Metab. 2007;6:1827–1833. doi: 10.1210/jc.2006-1785. [DOI] [PubMed] [Google Scholar]
- Ahmetov II, Popov DV, Missina SS, Vinogradova OL, Rogozkin VA. Association of mitochondrial transcription factor (TFAM) gene polymorphism with physical performance in athletes. Human Physiology. 2010;6:229–233. doi: 10.1134/S0362119710020155. [DOI] [PubMed] [Google Scholar]
- Handschin C. The biology of PGC-1alpha and its therapeutic potential. Trends Pharmacol Sci. 2009;6:322–329. doi: 10.1016/j.tips.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Wu Z, Boss O. Targeting PGC-1 alpha to control energy homeostasis. Expert Opin Ther Targets. 2007;6:1329–1338. doi: 10.1517/14728222.11.10.1329. [DOI] [PubMed] [Google Scholar]
- Jagiello P, Gencik M, Arning L, Wieczorek S, Kunstmann E, Csernok E, Gross WL, Epplen JT. New genomic region for Wegener's granulomatosis as revealed by an extended association screen with 202 apoptosis-related genes. Hum Genet. 2004;6:468–477. doi: 10.1007/s00439-004-1092-z. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M. et al. The structure of haplotype blocks in the human genome. Science. 2002;6:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- He Z, Hu Y, Feng L, Lu Y, Liu G, Xi Y, Wen L, McNaughton LR. NRF2 genotype improves endurance capacity in response to training. Int J Sports Med. 2007;6:717–721. doi: 10.1055/s-2007-964913. [DOI] [PubMed] [Google Scholar]
- Kuningas M, Putters M, Westendorp RG, Slagboom PE, van Heemst D. SIRT1 gene, age-related diseases, and mortality: the Leiden 85-plus study. J Gerontol A Biol Sci Med Sci. 2007;6:960–965. doi: 10.1093/gerona/62.9.960. [DOI] [PubMed] [Google Scholar]