Abstract
CpG island methylation (CIM) is an epigenetic mechanism for transcriptional silencing that occurs at various stages of colon tumorigenesis. CIM has been found in serrated adenomas and hyperplastic polyps. There is also evidence for hypermethylation in aberrant crypt foci (ACF) that are found in resected colons from cancer patients. Our study addresses promoter methylation of a tumor suppressor gene, RASSF1A, within the colonic epithelium of subjects undergoing screening colonoscopies in the absence of synchronous tumors. Patients included in this study were at elevated risk for colorectal cancer (CRC) based on family history, but without a previously occurring or synchronous colon carcinoma. ACF were identified using close-focus magnifying chromendoscopy and collected by biopsy in situ. We isolated ACF and adjacent normal colonic epithelium by laser capture microdissection (LCM) and studied methylation of the RASSF1A promoter region in ACF and in adjacent normal mucosa. Expression of RASSF1A was verified using quantitative real-time polymerase chain reaction (QRT–PCR). We found that 8.6% (3 out of 35) of ACF had K-ras mutations and 24% (6 out of 25) had RASSF1A hypermethylation. Our results demonstrate that RASSF1A hypermethylation and K-ras mutations are not mutually exclusive and are present in patients at elevated risk of CRC. Importantly, CIM of RASSF1A is an early epigenetic aberration, occurring in the absence of synchronous colon tumors and is not accompanied by field effects into the surrounding epithelium.
Introduction
Aberrant crypt foci (ACF) are microscopic surface abnormalities that represent an early precursor lesion to colorectal cancer (CRC) (1,2). The association of ACF with CRC is supported by shared histological features with colonic polyps and adenomas (3,4). ACF exhibit many of the molecular and genetic abnormalities that form the basis for the adenoma-carcinoma sequence in CRC (4,5). Genetic alterations in ACF include mutations in the APC tumor suppressor gene and the K-ras proto-oncogene, and microsatellite instability (MSI) (5–8). With the advent of new endoscopic instrumentation, it is possible to visualize ACF in situ at high resolution using colonoscopy (4,9). The histopathology of ACF is variable, but is generally classified into two major categories: hyperplastic and dysplastic (Figure 1) (10). Hyperplastic ACF display characteristics similar to hyperplastic polyps and to adenomas and are more common in sporadic cases than in FAP patients (3,4). They are characterized by frequent K-ras mutations and lack APC mutations (7,8). On the other hand, dysplastic ACF display abnormal proliferation within upper regions of the crypts and, in sporadic cases, generally have mutations in K-ras and lack APC mutations, although dysplastic ACF from FAP patients have frequent APC mutations (7,8). While dysplastic ACF are generally accepted as precursors to CRC, the significance of hyperplastic ACF in tumor progression is less well established (3,4). Therefore, it is important to fully characterize the genetic alterations that are present in hyperplastic ACF to better understand the distinct molecular pathways leading to CRC.
Fig. 1.
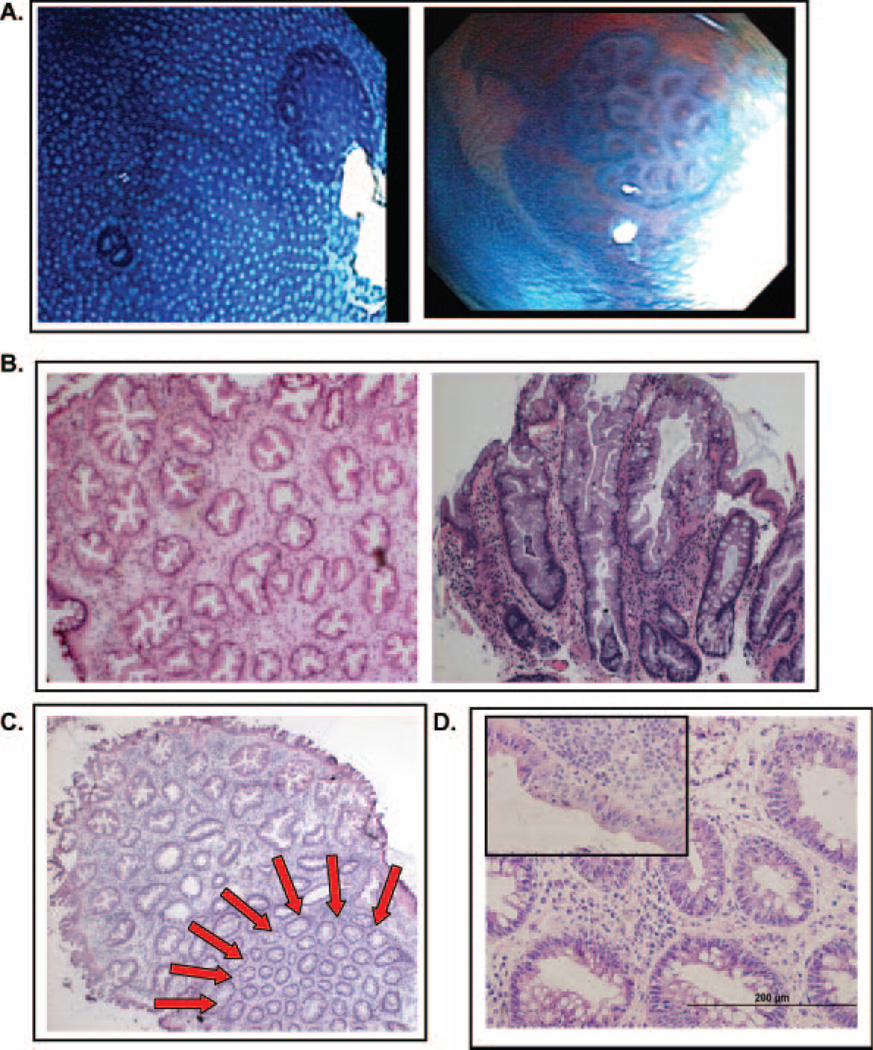
Macroscopic and histological analysis of ACF. (A) Gross views of ACF visualized through the Olympus prototype close-focus endoscope at a magnification of ×60. The colon epithelium was stained with 0.5% methylene blue. (B) H&E stained frozen sections of hyperplastic ACF, cross-sectional (left, ×200) and longitudinal (right, ×400) views. (C) H&E-stained frozen section of hyperplastic ACF with adjacent normal colonic mucosa indicated by the red arrows (×200). (D) H&E-stained frozen section of dysplastic ACF, cross-sectional view (×400). The inset in the upper left shows a region of surface dysplasia from the same lesion.
Epigenetic silencing caused by DNA CpG island methylation (CIM) is a mechanism for transcriptional silencing (11). CIM has been identified during several key stages in colon tumorigenesis, including ACF, sporadic serrated adenomas, hyperplastic polyps and tumors (12–16). In sporadic cases of CRC, studies have proposed that DNA hypermethylation mediates a ‘field defect’ whereby large regions of colonic epithelium have sustained genetic aberrations (17,18). In CRC, a panel of genes that have been shown to be silenced by promoter hypermethylation include p16, hMLH1, MGMT, and MINT 1, 2, 12 and 31 (14–16). Carcinomas and adenomas that are methylated at multiple loci (two or more) are referred to as having the CpG island methylator phenotype high status (CIMP high) (14–16).
RASSF1A is a putative tumor suppressor gene located at chromosome 3p21.3, a region that frequently exhibits loss of heterozygosity in human tumors. RASSF1A is a major isoform of the RASSF1 gene that is generated by alternative splicing. The C-terminus of RASSF1 encodes a Ras association domain. RASSF1A transcripts are often missing in cancer cell lines and in tumors (19,20). In addition, RASSF1A knockout mice show increased tumor multiplicity and size (21). In sporadic CRC, reported RASSF1A inactivation through promoter hypermethylation is variable (~30% of cases) (22–24). Since RASSF1A and K-ras share a common signaling pathway, it has been proposed that hypermethylation of RASSF1A and mutations in K-ras are mutually exclusive (23–25). However, more recent data suggest that RASSF1A inactivation and K-ras mutations are concurrent in MSI sporadic CRC, suggesting a synergistic effect (22).
In the following study, ACF were identified in and biopsied in situ from patients at elevated risk for CRC using a prototype high-resolution, close-focus magnifying endoscope. Aberrant crypts were isolated from the surrounding normal mucosa by laser capture microdissection (LCM). We investigated the methylation status of RASSF1A within ACF and compared this epigenetic change within abutting normal colonic mucosa. Correlations were made to K-ras mutations, ACF histology and clinicopathological features. The functional consequence of hypermethylated RASSF1A was confirmed by expression analysis. Our data support the evidence that epigenetic silencing can occur within ACF in the absence of a ‘field effect’ within surrounding normal mucosa, and in the absence of synchronous tumors.
Materials and methods
Subject selection
All patients included in this study were considered to be at elevated risk for CRC based on a positive family history (at least one first- or second-degree relative with the disease) and/or a positive personal history for benign polyps/adenomas. All patients meeting the Amsterdam criteria for HNPCC or FAP were excluded from this study. No subjects used in this study had a history of prior colon cancer nor had evidence of a synchronous colon tumor(s). Patients underwent total colonoscopy at the John Dempsey Hospital (JDH) at the University of Connecticut Health Center (UCHC) in accordance with institutional policies. This study was performed after approval by an Institutional Review Board and all subjects provided written informed consent.
ACF collection and characterization
ACF were isolated from grossly normal appearing colonic mucosa by biopsy in situ during the high-resolution close-focus chromendoscopy portion of total colonoscopy procedures performed in the JDH at UCHC. The cecum and, in addition, 20 cm of the distal colon (encompassing part rectum, part sigmoid colon) were stained with up to 40 ml of 0.5% methylene blue to identify ACF. ACF were visualized and photographed using an Olympus Prototype Close Focus Colonoscope (XCF-Q160ALE), which enables visualization from 2 to 100 mm at a magnifying power of ×60 (Figure 1A) (4). Under close-focus magnification, a finding was accepted as an ACF if two or more crypts had increased lumen diameter, thick crypt walls or abnormally shaped lumens. Biopsies of individual ACF were embedded immediately in tissue freezing medium (OCT) and stored at −80°C. Frozen serial sections of ACF were prepared at 5–7 microns on glass slides. Representative sections of ACF were stained with hematoxylin and eosin (H&E) for routine histological analysis by light microscopy of coded specimens by a pathologist (TR) (3,26).
DNA and RNA extraction
Frozen sections of ACF were stained and dehydrated using the Histogene staining kit (Arcturus, Mountain View, CA), according to the manufacturer’s instructions. LCM was performed on prepared sections using the Veritas LCM system (Arcturus). Whenever possible, adjacent normal mucosal (ANM) cells directly abutting the aberrant crypts were collected separately by laser capture. On average, 1500–3000 cells were collected from each sample. DNA was extracted using the Picopure DNA extraction kit (Arcturus) and quantified using Picogreen (Molecular Probes, Eugene, OR). RNA was extracted using the Picopure RNA isolation kit (Arcturus).
Cell lines and culture conditions
DNA and RNA were isolated from three cancer cell lines: A549, HEK293, and HCT116. A549 cells were grown in Ham’s F12K medium (Invitrogen, Carlsbad, CA), supplemented with 10% fetal bovine serum (FBS) and 5% penicillin/streptomycin. HEK293 and HCT116 cells were grown in Dulbecco’s modified Eagle’s medium (Invitrogen), supplemented with 10% FBS and 5% penicillin/streptomycin.
K-ras mutational analysis
Genomic DNA was extracted from microdissected tissue, precipitated with isopropanol and amplified using the GenomiPhi amplification kit (Amersham Biosciences, Piscataway, NJ). A 167 bp product spanning codons 12 and 13 of human K-ras was amplified in 25 µl volumes using 1 µl of amplified genomic DNA, 10× polymerase chain reaction (PCR) buffer, 50 mM MgCl2, 10 mM dNTPs, 0.5 U Platinum Taq and 2.5 pmoles forward and reverse primers (Invitrogen). Primers were designed using Primer3 software [5′- GCC TGC TGA AAA TGA CTG AA-3′ (sense) and 5′ -AGA AGT GTC CTG CAC CAG TAA-3′ (antisense)], and a 35-cycle PCR was performed using 58°C as the annealing temperature. PCR products were separated on a 2% agarose gel, excised and subjected to sequencing using the ABI Prism BigDye Terminator (Applied Biosystems, Foster City, CA), 2.0 µl 5× BigDye reaction buffer (Applied Biosystems) and 3.2 pmoles forward primer. Cycling conditions were as follows: 96°C for 2 min, 28 cycles of 96°C for 10 s, 50°C for 5 s and 60°C for 4 min. The reaction products were sequenced by capillary electrophoresis using an ABI Prism 3100 DNA Analyzer. Mutations were confirmed by sequencing using the reverse primer. HCT116 cells were used as a positive control for a mutation in codon 13, and HEK293 cells were used as a negative control.
RASSF1A methylation-specific PCR (MSP)
The methylation status of RASSF1A was determined by sodium bisulfite treatment of DNA (27) followed by MSP (24). Briefly, 1 µg of salmon sperm DNA was added as a carrier to 10–30 ng of microdissected genomic DNA, or 100 ng of genomic DNA from cell lines, and the total sample volume was brought to 50 µl with nuclease-free H2O. DNA was denatured with 7.5 µl of 2M NaOH at 37°C for 10 min. DNA was incubated with 3 µl of 100 mM hydroquinone and 540 µl of 3M sodium bisulfite (pH 5.0) at 50–55°C for 16 h in darkness. After treatment, DNA was purified using the Wizard DNA Cleanup Kit (Promega, Madison, WI) and de-sulfonated with 4.4 µl of 3M NaOH at 37°C for 15 min. DNA was precipitated with 1/10 volume of 3M sodium acetate (pH 5.2) and 3 volumes of 100% ethanol, washed with 70% ethanol and resuspended in 25 µl nuclease-free water.
MSP primers were the same as those reported by van Engeland et al. (24). Nested PCR was performed to increase reaction sensitivity. A 144 bp product was amplified in 25 µl volumes using 6 µl bisulfite-treated genomic DNA, 10× PCR buffer, 50 mM MgCl2, 10 mM dNTPs, 0.5 U Platinum Taq and 2.5 pmoles forward and reverse primers. The primers used for the first round of PCR were 5′-GTT TAG TTT GGA TTT TGG GGG AG-3′ (sense) and 5′-CCC RCA ACT CAA TAA ACT CAA ACT-3′ (antisense). PCR reactions were performed using the following cycling conditions: 95°C for 5 min, 23 cycles of 95°C for 45 s, 59°C for 45 s and 72°C for 1 min, and extension at 72°C for 10 min. The resulting 144 bp fragment was used as a template for the MSP reaction. 1 : 100 dilutions of the first PCR reaction were made, and the methylated and unmethylated products were amplified using the same reaction mixture as above. A 76 bp methylated product was amplified using the primers 5′ -GGG TTC GTT TTG TGG TTT CGT TC-3′ (sense) and 5′ -TAA CCC GAT TAA ACC CGT ACT TCG-3′ (antisense). An 81 bp unmethylated product was amplified using the primers 5′-GGG GTT TGT TTT GTG GTT TTG TTT-3′ (sense) and 5′-AAC ATA ACC CAA TTA AAC CCA TAC TTC A-3′ (antisense). The cycling conditions were 95°C for 5 min, 30 cycles of 95°C for 45 s, 63°C (67°C for the methylated reaction) for 45 s and 72°C for 1 min, and 72°C for 10 min. All PCR reactions were performed with two controls for methylation (A549 and HEK293 cells), one control for no methylation (HCT116 cells) and a H2O control. Twenty-five microliters of each PCR reaction was loaded onto a 2% agarose gel, stained with ethidium bromide and visualized under UV illumination.
RASSF1A mRNA expression
LCM was performed on frozen sections of ACF, and RNA was extracted as described above. Eleven microliters of microdissected RNA was reverse-transcribed into cDNA using Sensiscript RT (Qiagen). RNA from cell lines was reverse-transcribed into cDNA using Superscript II RT (Invitrogen). mRNA expression was determined by quantitative real-time PCR (QRT–PCR) using an ABI 7500 real-time PCR instrument (Applied Biosciences Foster city, CA). QRT–PCR reactions were performed in duplicate in 25 µl volumes using 1.25 µl 20× RASSF1 TaqMan Assay on Demand (ID = Hs00945257_m1) (ABI), 3.5 µl cDNA template and 12.5 µl 2× TaqMan Universal Master Mix. The cycling conditions were 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 s and 60°C for 1 min. QRT–PCR reactions were also performed in duplicate for each sample with an endogenous control gene, huHPRT1. All QRT–PCR reactions were performed with two negative controls for RASSF1A mRNA expression (A549 and HEK293 cells), one positive control for RASSF1A mRNA expression (HCT116 cells) and a H2O control.
Statistical analysis
For comparison of K-ras mutations, RASSF1A hypermethylation and clinicopathological features, analysis was carried out for two-sided P-values using the χ2-test and Web Chi Squared Calculator (http://www.georgetown.edu/faculty/ballc/webtools/web_chi.html).
Results
The subjects included in this study did not have a past or current colon cancer. They were identified as being at elevated risk on the basis of a positive family history of CRC or a personal history of benign polyps or adenomas. As shown in Table I, 41.7% (5 out of 12) of subjects had a personal history of benign polyps or adenomas, 33.3% (4 out of 12) had a positive family history of CRC and 25% (3 out of 12) had a personal history of benign lesions as well as a family history of CRC. The patients were from 46 to 79 years of age (average = 63). All of the patients included in our study presented with multiple ACF at the time of colonoscopy (3–12 ACF each), with an average of 6 ACF per subject.
Table I.
Clinicopathological features of patients, K-ras mutations and RASSF1A methylation in ACF
| Patient | Sex | Age | Total ACF | Sample number | ACF histology |
K-ras mutation (codons 12 and13) |
RASSF1A methylation |
|
|---|---|---|---|---|---|---|---|---|
| ACF | ANMa | |||||||
| 1b | M | Unknown | 5 | 042004-1130-3 | Hyperplastic | WT | Uc | U |
| 2b | M | Unknown | 5 | 030904-1130-5 | HNDd | WT | X | X |
| 3b | F | 74 | 4 | 112003-1045-4 | Hyperplastic | WT | U | NDe |
| 112003-1045-6 | HND | WT | X | X | ||||
| 112003-1045-7 | Hyperplastic | WT | X | X | ||||
| 4b | M | 49 | 6 | 021904-1000-2 | Hyperplastic | WT | U | X |
| 021904-1000-4 | Hyperplastic | WT | U | X | ||||
| 021904-1000-5 | Hyperplastic | WT | U | X | ||||
| 5a,f | F | 50 | 3 | 121103-1100-1 | Dysplastic | WT | U | U |
| 121103-1100-2 | Hyperplastic | WT | U | X | ||||
| 121103-1100-3 | Hyperplastic | Mutation (12) | Mg | U | ||||
| 6a,f | M | 65 | 5 | 042604-1130-1 | Hyp. & dysp. | WT | ND | ND |
| 042604-1130-2 | HND | WT | ND | X | ||||
| 042604-1130-3 | Hyperplastic | WT | M | U | ||||
| 042604-1130-4 | Hyperplastic | WT | U | X | ||||
| 042604-1130-5 | Hyperplastic | WT | X | X | ||||
| 7a | M | 73 | 9 | 032904-230-2 | Hyperplastic | WT | M | U |
| 032904-230-3 | Hyperplastic | WT | M | X | ||||
| 032904-230-5 | Hyperplastic | WT | M | U | ||||
| 032904-230-7 | Hyperplastic | WT | U | X | ||||
| 032904-230-8 | Hyperplastic | WT | X | X | ||||
| 8b | F | 51 | 7 | 042904-1200-2 | Hyperplastic | Mutation (12) | M | X |
| 042904-1200-5 | HND | WT | X | X | ||||
| 042904-1200-6 | Hyperplastic | WT | U | X | ||||
| 042904-1200-7 | HND | WT | X | X | ||||
| 9b | M | Unknown | 12 | 101804-1100-2 | Hyperplastic | WT | U | X |
| 101804-1100-4 | Hyperplastic | WT | U | X | ||||
| 101804-100-6 | Hyperplastic | WT | U | X | ||||
| 101804-100-7 | Hyperplastic | WT | U | X | ||||
| 101804-1100-9 | Hyperplastic | WT | U | X | ||||
| 101804-1100-10 | Hyperplastic | WT | U | X | ||||
| 101804-1100-11 | Hyperplastic | WT | U | X | ||||
| 101804-1100-12 | Hyperplastic | WT | X | X | ||||
| 10b | F | 79 | 7 | 101104-1130-1 | Hyperplastic | Xh | U | X |
| 11b,f | F | 76 | 5 | 052704-1130-4 | Hyperplastic | X | U | X |
| 052704-1130-5 | Hyperplastic | Mutation (13) | U | X | ||||
| 12f | M | 46 | 3 | 071204-1130-3 | Hyperplastic | WT | ND | X |
All ACF assayed for mutations/methylation were located in the sigmoid colon. Total ACF refers to the number of ACF identified during the chromendoscopy procedure.
ANM, adjacent normal mucosa.
Personal history (past benign polyps/adenomas).
U, unmethylated.
HND, histology not determined.
ND, none detected.
Family history (at least one first- or second-degree relative with CRC).
M, methylated.
X, assay not performed.
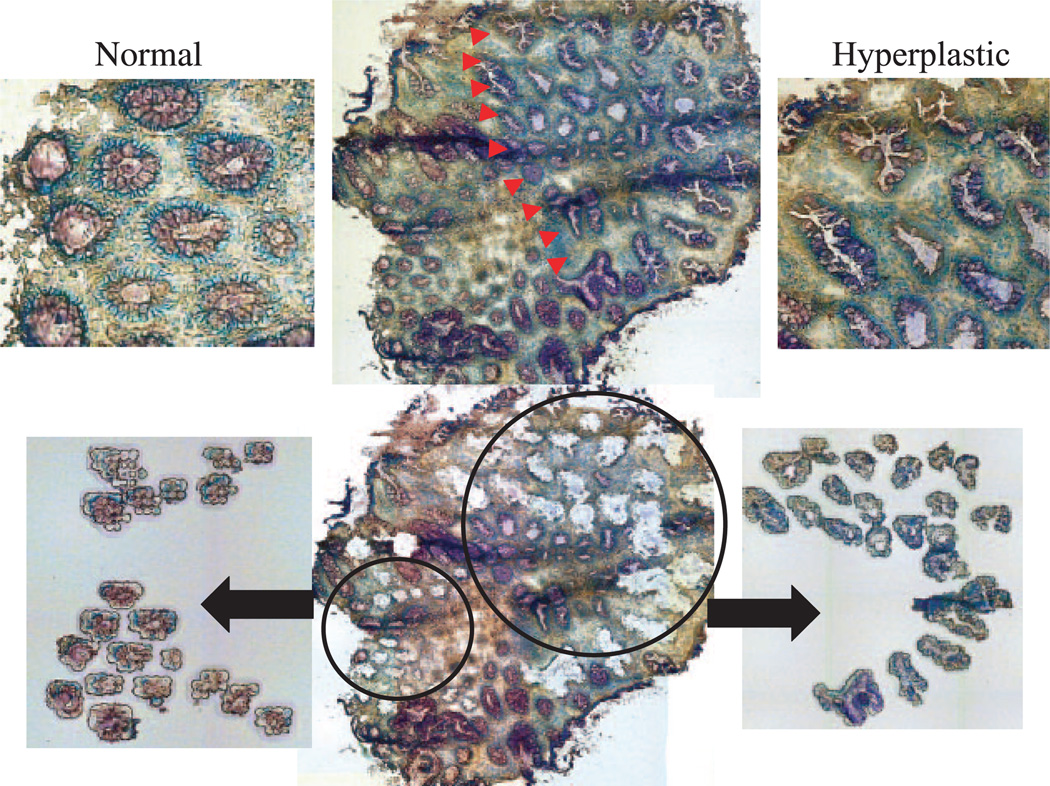
Of the total number of ACF identified during colonoscopy from all subjects, 56.3% (40 out of 71) displayed serrated crypts, normal nuclei and mucin depletion and were classified histologically as hyperplastic. Representative images of hyperplastic and dysplastic ACF collected during our study are shown in Figure 1. As shown in Figure 2, ACF were isolated from the surrounding colonic epithelium by LCM for (epi)genetic analysis. Whenever possible, ANM was captured separately by LCM (Figure 2). We assessed K-ras mutations in codons 12 and 13 by direct PCR sequencing. As shown in Table I, 8.6% (3 out of 35) of ACF were found to have a K-ras mutation in either codon 12 or codon 13 and were hyperplastic. A G-to-A transition mutation at the second nucleotide of codon 12 was present in two ACF and a G-to-C transversion at the second base of codon 13 was present in one ACF. Of the two dysplastic ACF examined, neither had K-ras mutations (Table I).
Fig. 2.
Representative LCM of a hyperplastic ACF and adjacent normal colonic epithelium. LCM was used to isolate aberrant crypts in the hyperplastic ACF (right side, indicated by arrows) from the ANM (left side). The lower images show two populations of microdissected crypts isolated separately from the surrounding stroma. All images are at ×200 magnification.
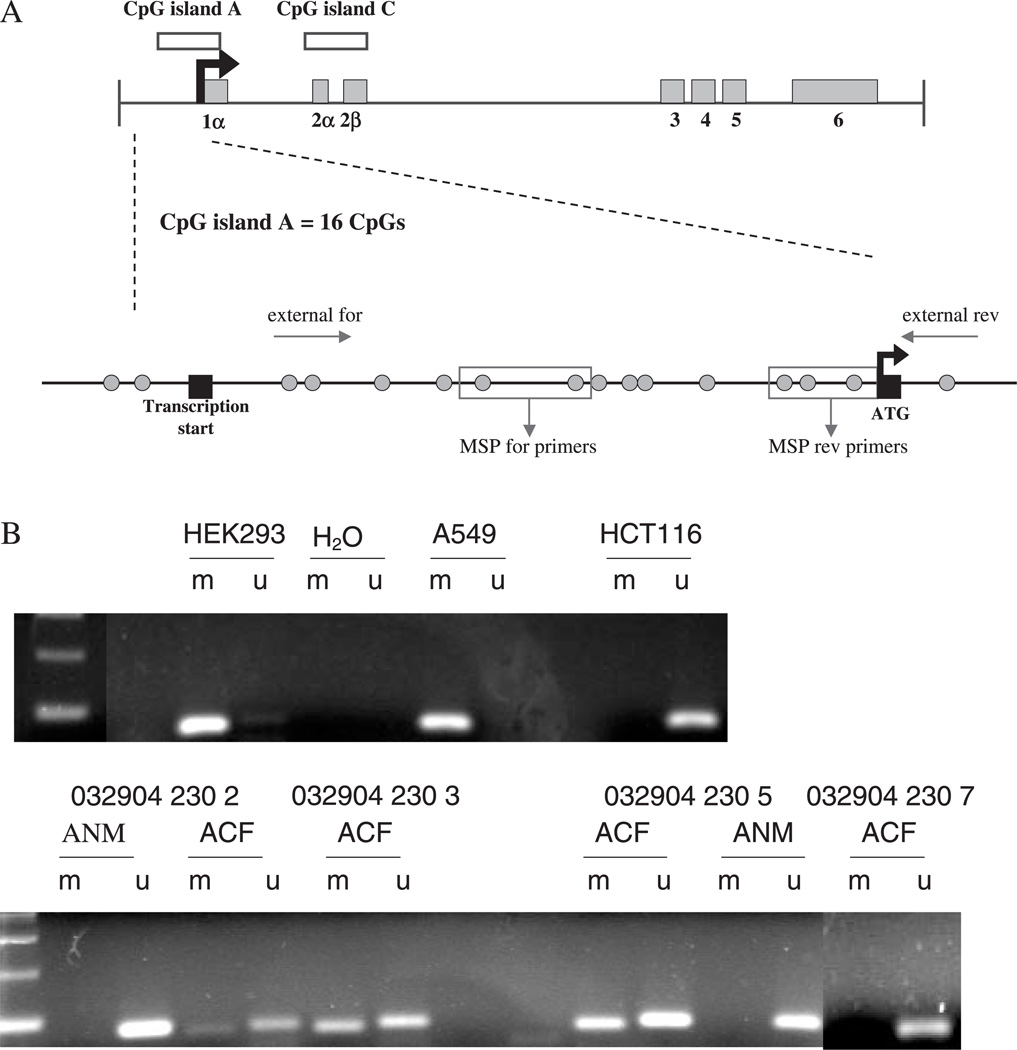
We measured RASSF1A promoter methylation in hyperplastic ACF and adjacent normal epithelium in 25 samples from 10 subjects by MSP. The locations of CpG sites and regions amplified by MSP primers within the RASSF1A promoter region are detailed in Figure 3A. Figure 3B shows representative examples of the RASSF1A MSP. Our study found that 24% of ACF (6 out of 25) from 10 patients exhibited RASSF1A methylation (Table I). In contrast, there was no evidence for RASSF1A hypermethylation within LCM-isolated adjacent normal colonic mucosa (0 out of 6). Although the number of normal samples that were analyzed was too small to yield statistical significance, these results indicate the absence of a methylation ‘field effect’ (Table I, Figure 3B).
Fig. 3.
The RASSF1A locus and MSP analysis. (A) Map of the RASSF1 locus and CpG island A that is used for analysis of RASSF1A promoter methylation by MSP. The three major isoforms (RASSF1A, RASSF1C and RASSF1F) are made by alternative splicing and promoter usage. RASSF1A is formed from exons 1α, 2αβ, 3, 4, 5, 6. The promoter region, which is located within CpG island A, is indicated by a black arrow. CpG island A contains 16 CpG dinucleotides (green circles). The flanking PCR reaction, indicated by ‘external for’ and ‘external rev’ primers, amplifies a 144 bp product that is used as a template for the RASSF1A MSP, indicated by the red boxes labeled ‘MSP for primers’ and ‘MSP rev primers’. (B) Representative MSP analysis of the promoter of RASSF1A in cell lines and in microdissected tissue. DNA was analyzed by a chemical modification with sodium bisulfite, and a subsequent flanking PCR reaction amplified all bisulfite-treated DNA followed by RASSF1A MSP, as described under Materials and methods. The 76 bp methylated product and 81 bp unmethylated product were run on a 2% agarose gel and visualized under UV. A549 and HEK293 cells were used as a control for methylated RASSF1A. HCT116 cells were used as a control for unmethylated RASSF1A. ANM = adjacent normal mucosa. All MSP reactions were also run with a H2O control in the flanking PCR and in the RASSF1A MSP. The presence of an unmethylated product in the ACF that are methylated is probably the result of normal contaminating tissue.
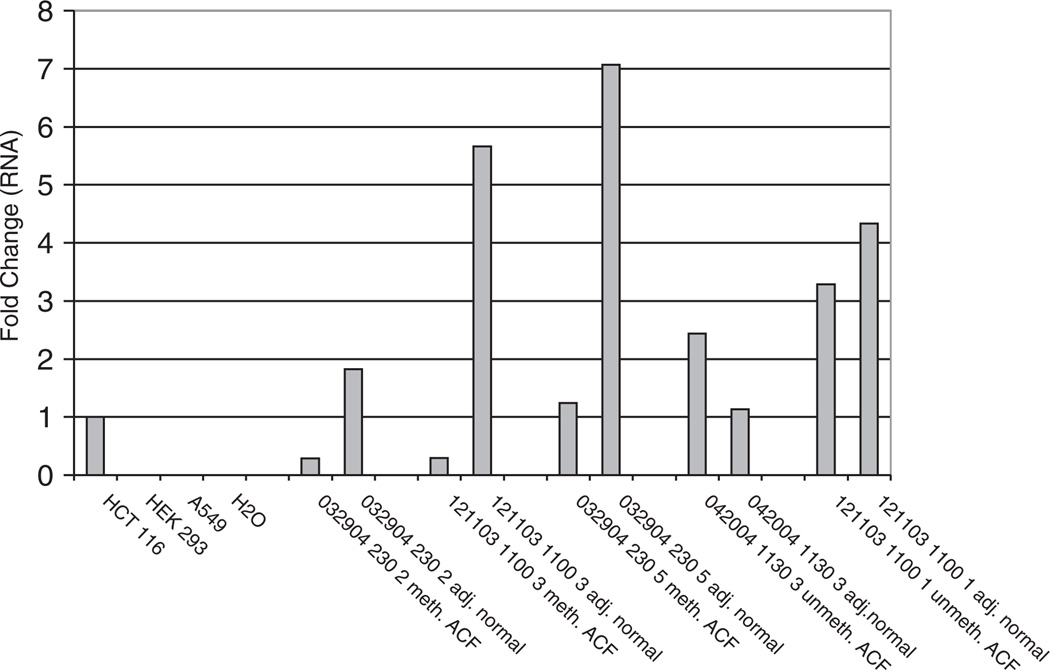
To confirm that RASSF1A hypermethylation is associated with decreased gene transcription, we measured the expression by QRT–PCR of RASSF1A mRNA in three ACF exhibiting methylation, as well as two ACF that were unmethylated. Corresponding ANM was also evaluated. As shown in Figure 4, RASSF1A expression ranged from 5.7 to 19.5-fold higher in adjacent non-methylated normal mucosa compared with the methylated ACF. In contrast, in the unmethylated ACF, RASSF1A expression was not significantly different compared with the adjacent unmethylated normal mucosa, ranging from 0.75- to 2.16-fold higher. Expression was absent in HEK293 and A549 cells that are both fully methylated at the RASSF1A promoter. However, HCT116 cells, which are characterized by an unmethylated RASSF1A promoter, showed RASSF1A expression (Figure 4).
Fig. 4.
QRT–PCR analysis of RASSF1A expression in cell lines and microdissected tissue. mRNA levels were quantified as described under Materials and methods. Concentrations were normalized to huHPRT1 to control for differences in cDNA input amounts. Fold differences were determined by using HCT116 as the calibrator sample (expression arbitrarily normalized to 1). (Fold difference = normalized quantity sample/normalized quantity HCT116). HCT116 cells were used as a positive control for RASSF1A mRNA expression, and HEK293 and A549 cells were used as a negative control. A H2O negative control was also included. 032904-230-2, 121103-1100-3 and 032904-230-5 were hyperplastic ACF that had methylated RASSF1A. 042004-1130-3 and 121103-1100-1 were hyperplastic ACF that had unmethylated RASSF1A. The ANM in all samples was unmethylated. All samples were assayed in duplicate.
Discussion
We have analyzed a subset of hyperplastic ACF identified by close-focus magnifying colonoscopy for RASSF1A methylation and K-ras mutations in patients at elevated risk for CRC. The subset of patients used in our study is unique since they do not have a personal history of CRC nor an existing cancer, but are considered at elevated risk because of a positive family history and/or a personal history of benign polyps or adenomas. In our study, ACF were identified during screening colonoscopy by in situ surface dye staining and video/biopsy chromendoscopy, affording a much higher level of resolution than traditional colonoscopy, and thus providing a more accurate method for ACF detection (9). For example, in a recent study, it was found that traditional flexible sigmoidoscopy was able to identify only 18% of significant flat lesions that were readily detectable by magnifying colonoscopy (9). In our study, ACF were visualized at ×60 by staining the colonic epithelium with methylene blue and identifying the areas exhibiting thicker epithelial lining and darker staining (Figure 1A). Since the ACF were identified in situ, rather than ex vivo within resected colons, it becomes feasible to monitor the progression of subsets of remaining small lesions over time.
The biological significance of hyperplastic ACF and their malignant potential is unclear. In contrast to high-grade adenomas and carcinomas, there is reportedly only a low frequency of APC mutations in hyperplastic and non-dysplastic ACF (8). Nonetheless, there is evidence that these lesions represent a pre-cancerous change since they show clonal genetic aberrations, including activating mutations in K-ras (8), DNA MSI (6) and chromosomal rearrangements (12). Recently, a ‘serrated’ pathway, encompassing hyperplastic ACF, hyperplastic polyps and serrated adenomas, has been proposed as an alternative pathway of sporadic colorectal neoplasia (28). The ‘serrated pathway’ is thought to be initiated by the silencing of DNA repair genes and the resulting resistance to DNA damage and inhibition of apoptosis (28). The serrated pathway has also been associated with epigenetic silencing of multiple genes (13,15). In fact, CIM has been demonstrated in hyperplastic ACF (12), and it is possible that this alteration could serve as a marker for progression potential.
The Ras pathway is an important mediator of cellular response to growth signals. K-ras activating mutations are found in up to 50% of patients with sporadic CRC and to a varying extent in ACF (5,29). ACF from patients with sporadic carcinomas frequently have CIM and K-ras mutations, while ACF from FAP patients often show dysplastic features, but lack CIM or K-ras mutations (8,12). The ACF examined in our study, however, are from patients who do not present with sporadic CRC and provide further insight into mutational frequency. The low frequency of K-ras mutations in ACF observed in our study (8.6%) contrasts with earlier studies showing high mutation rates (8), which is perhaps a reflection of the subset of patients selected for study.
K-ras signaling is associated with cell proliferation, and K-ras also directly controls a pro-apoptotic signaling pathway mediated by RASSF1A (19). RASSF1A may exert its effect in part by inhibiting cell cycle progression through suppression of cyclin D (30). These functions, together with the tumor phenotype observed following genetic deletion (25), support the role of RASSF1A as a putative tumor suppressor gene (19). Epigenetic silencing of RASSF1A by promoter hypermethylation has been described in a variety of human carcinomas, including CRC (20). However, the reported frequency of RASSF1A hypermethylation in adenomas and tumors is highly variable (2–80%) (23,24). To date, the methylation status of RASSF1A has not been analyzed in ACF. Our study found that 24% of ACF (6 out of 25) from 10 patients exhibited RASSF1A methylation. Importantly, there was no evidence for RASSF1A hypermethylation within laser-captured adjacent normal colonic mucosa, perhaps indicating the absence of a ‘field effect’ (Table I, Figure 3). The high frequency of methylation within this subset of ACF supports the concept that epigenetic silencing through promoter methylation is one of the earliest alterations within the ‘serrated’ pathway (28). These earlier observations, together with the low frequency of K-ras mutations found in the present study, imply that CIM of RASSF1A is an early event that may precede mutational activation of K-ras.
To confirm the functional significance of RASSF1A methylation, we measured RASSF1A expression in microdissected ACF and in abutting normal crypts (Figures 2 and 4). As anticipated, there was a clear reduction in RASSF1A expression within methylation-positive ACF, confirming decreased transcription of the gene. Although there was not a complete loss of RASSF1A mRNA expression within the methylated ACF, this may be explained in part by the presence of contaminating normal cells, or incomplete gene silencing. In contrast, RASSF1A expression in the unmethylated ACF was higher in one case than in the adjacent normal epithelium, and in the other case was only slightly lower than the normal mucosa. This also confirms the absence of a field effect within the surrounding normal colon epithelium, as there were high levels of RASSF1A expression in these cells in all cases.
Several studies have suggested an inverse association between the frequency of K-ras mutations and RASSF1A methylation based on the shared signaling pathway (23–25). However, a recent study found concordance between RASSF1A hypermethylation and K-ras mutations in MSI CRC (36%), suggesting a synergistic effect (22). Our study is consistent with this latter finding. RASSF1A methylation was present in 67% (2 out of 3) of ACF with K-ras mutations, but only 20% (4 out of 20) of ACF without K-ras mutations, P > 0.05. Although the correlation did not achieve statistical significance as a result of only a limited sample size, the observed trend does imply that the two events are not necessarily mutually exclusive. Additional analysis of MSI status, however, would be required to confirm whether this synergism is restricted to MMR-deficient lesions, or is dependent upon other factors, such as lesion histology.
In conclusion, our data provide the first evidence that an important tumor suppressor gene, RASSF1A, has undergone promoter hypermethylation within the ACF of patients undergoing surveillance colonoscopy. Importantly, this epigenetic alteration was identified within the colonic epithelium of normal subjects at elevated risk of CRC, but with no evidence of synchronous colon cancer. A significant consequence of this epigenetic event is transcriptional silencing of RASSF1A, occurring exclusively within the aberrant crypt cells, but not in abutting normal epithelium. These data argue against the possibility of a methylation ‘field effect’, and further suggest that epigenetic silencing may in fact represent one of the earliest alterations in the CRC pathway.
Acknowledgements
We thank the Olympus Corporation for generously providing the endoscopy instrumentation and the Yellin Golf Foundation for their generous support and NIH grant CA81428.
Abbreviations
- ACF
aberrant crypt foci
- ANM
adjacent normal mucosa
- CIM
CpG island methylation
- CRC
colorectal cancer
- FBS
fetal bovine serum
- H&E
hematoxylin and eosin
- LCM
laser capture microdissection
- MMR
mismatch repair
- MSI
microsatellite instability
- MSP
methylation-specific PCR
- PCR
polymerase chain reaction
- QRT–PCR
quantitative real-time PCR
Footnotes
Conflict of Interest Statement: None declared.
References
- 1.Bird RP. Observation and quantification of aberrant crypts in the murine colon treated with a colon carcinogen: preliminary findings. Cancer Lett. 1987;37:147–151. doi: 10.1016/0304-3835(87)90157-1. [DOI] [PubMed] [Google Scholar]
- 2.Pretlow TP, Barrow BJ, Ashton WS, O’Riordan MA, Pretlow TG, Jurcisek JA, Stellato TA. Aberrant crypts: putative preneoplastic foci in human colonic mucosa. Cancer Res. 1991;51:1564–1567. [PubMed] [Google Scholar]
- 3.Nucci MR, Robinson CR, Longo P, Campbell P, Hamilton SR. Phenotypic and genotypic characteristics of aberrant crypt foci in human colorectal mucosa. Hum. Pathol. 1997;28:1396–1407. doi: 10.1016/s0046-8177(97)90230-6. [DOI] [PubMed] [Google Scholar]
- 4.Takayama T, Katsuki S, Takahashi Y, Ohi M, Nojiri S, Sakamaki S, Kato J, Kogawa K, Miyake H, Niitsu Y. Aberrant crypt foci of the colon as precursors of adenoma and cancer. N. Engl. J. Med. 1998;339:1277–1284. doi: 10.1056/NEJM199810293391803. [DOI] [PubMed] [Google Scholar]
- 5.Jen J, Powell SM, Papadopoulos N, Smith KJ, Hamilton SR, Vogelstein B, Kinzler KW. Molecular determinants of dysplasia in colorectal lesions. Cancer Res. 1994;54:5523–5526. [PubMed] [Google Scholar]
- 6.Heinen CD, Shivapurkar N, Tang Z, Groden J, Alabaster O. Microsatellite instability in aberrant crypt foci from human colons. Cancer Res. 1996;56:5339–5341. [PubMed] [Google Scholar]
- 7.Otori K, Konishi M, Sugiyama K, Hasebe T, Shimoda T, Kikuchi-Yanoshita R, Mukai K, Fukushima S, Miyaki M, Esumi H. Infrequent somatic mutation of the adenomatous polyposis coli gene in aberrant crypt foci of human colon tissue. Cancer. 1998;83:896–900. doi: 10.1002/(sici)1097-0142(19980901)83:5<896::aid-cncr14>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 8.Takayama T, Ohi M, Hayashi T, et al. Analysis of K-ras, APC, and beta-catenin in aberrant crypt foci in sporadic adenoma, cancer, and familial adenomatous polyposis. Gastroenterology. 2001;121:599–611. doi: 10.1053/gast.2001.27203. [DOI] [PubMed] [Google Scholar]
- 9.Hurlstone DP, Karajeh M, Sanders DS, Drew SK, Cross SS. Rectal aberrant crypt foci identified using high-magnification-chromoscopic colonoscopy: biomarkers for flat and depressed neoplasia. Am. J. Gastroenterol. 2005;100:1283–1289. doi: 10.1111/j.1572-0241.2005.40891.x. [DOI] [PubMed] [Google Scholar]
- 10.Di Gregorio C, Losi L, Fante R, Modica S, Ghidoni M, Pedroni M, Tamassia MG, Gafa L, Ponz de Leon M, Roncucci L. Histology of aberrant crypt foci in the human colon. Histopathology. 1997;30:328–334. doi: 10.1046/j.1365-2559.1997.d01-626.x. [DOI] [PubMed] [Google Scholar]
- 11.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 12.Chan AO, Broaddus RR, Houlihan PS, Issa JP, Hamilton SR, Rashid A. CpG island methylation in aberrant crypt foci of the colorectum. Am. J. Pathol. 2002;160:1823–1830. doi: 10.1016/S0002-9440(10)61128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SJ, Rashid A, Lee JH, Kim SG, Hamilton SR, Wu TT. Frequent CpG island methylation in serrated adenomas of the colorectum. Am. J. Pathol. 2003;162:815–822. doi: 10.1016/S0002-9440(10)63878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rashid A, Shen L, Morris JS, Issa JP, Hamilton SR. CpG island methylation in colorectal adenomas. Am. J. Pathol. 2001;159:1129–1135. doi: 10.1016/S0002-9440(10)61789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wynter CV, Walsh MD, Higuchi T, Leggett BA, Young J, Jass JR. Methylation patterns define two types of hyperplastic polyp associated with colorectal cancer. Gut. 2004;53:573–580. doi: 10.1136/gut.2003.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc. Natl Acad. Sci. USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braakhuis BJ, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of Slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727–1730. [PubMed] [Google Scholar]
- 18.Shen L, Kondo Y, Rosner GL, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. J. Natl Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- 19.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat. Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 20.Pfeifer GP, Dammann R. Methylation of the tumor suppressor gene RASSF1A in human tumors. Biochemistry (Mosc) 2005;70:576–583. doi: 10.1007/s10541-005-0151-y. [DOI] [PubMed] [Google Scholar]
- 21.Tommasi S, Dammann R, Zhang Z, Wang Y, Liu L, Tsark WM, Wilczynski SP, Li J, You M, Pfeifer GP. Tumor susceptibility of RASSF1A knockout mice. Cancer Res. 2005;65:92–98. [PubMed] [Google Scholar]
- 22.Oliveira C, Velho S, Domingo E, Preto A, Hofstra RM, Hamelin R, Yamamoto H, Seruca R, Schwartz S. Concomitant RASSF1A hypermethylation and KRAS/BRAF mutations occur preferentially in MSI sporadic colorectal cancer. Oncogene. 2005;24:7630–7634. doi: 10.1038/sj.onc.1208906. [DOI] [PubMed] [Google Scholar]
- 23.Sakamoto N, Terai T, Ajioka Y, et al. Frequent hypermethylation of RASSF1A in early flat-type colorectal tumors. Oncogene. 2004;23:8900–8907. doi: 10.1038/sj.onc.1207993. [DOI] [PubMed] [Google Scholar]
- 24.van Engeland M, Roemen GM, Brink M, Pachen MM, Weijenberg MP, de Bruine AP, Arends JW, van den Brandt PA, de Goeij AF, Herman JGM. K-ras mutations and RASSF1A promoter methylation in colorectal cancer. Oncogene. 2002;21:3792–3795. doi: 10.1038/sj.onc.1205466. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Zhang Z, Dai Z, Popkie AP, Plass C, Morrison C, Wang Y, You M. RASSF1A promoter methylation and Kras2 mutations in non small cell lung cancer. Neoplasia. 2003;5:362–366. doi: 10.1016/S1476-5586(03)80029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nambiar PR, Nakanishi M, Gupta R, et al. Genetic signatures of high- and low-risk aberrant crypt foci in a mouse model of sporadic colon cancer. Cancer Res. 2004;64:6394–6401. doi: 10.1158/0008-5472.CAN-04-0933. [DOI] [PubMed] [Google Scholar]
- 27.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jass JR, Whitehall VL, Young J, Leggett BA. Emerging concepts in colorectal neoplasia. Gastroenterology. 2002;123:862–876. doi: 10.1053/gast.2002.35392. [DOI] [PubMed] [Google Scholar]
- 29.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 30.Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol. Cell Biol. 2002;22:4309–4318. doi: 10.1128/MCB.22.12.4309-4318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]