Abstract
Balancing the capacity for protein maturation with changes in protein flux through the endoplasmic reticulum (ER) is crucial for maintaining ER homeostasis. In this issue, Merksamer et al. (2008) exploit a redox-sensitive fluorescent protein to monitor the environment inside the ER of living yeast, illuminating how this organelle responds to different perturbations.
Many cellular mRNAs code for secretory and membrane proteins that must mature in the endoplasmic reticulum (ER) before being transported to their final destinations. ER homeostasis in the face of this constant substrate flux is crucially dependent on three processes: chaperone-mediated protein folding, protein quality control, and misfolded protein degradation. A diminished capacity in any of these three pathways or an increase in the amount of substrate trafficking through the ER can lead to the persistent presence of misfolded proteins. Cells sense the accumulation of misfolded proteins as “ER stress” and initiate the unfolded protein response (UPR) to alleviate the problem. The capacity to maintain ER homeostasis influences the production and fate of misfolded proteins, the accumulation of which is associated with many diseases (Yoshida, 2007). Merksamer et al. (2008) now present an elegant reporter system that directly monitors changes in the ER environment of the budding yeast Saccharomyces cerevisiae at the single-cell level, thereby opening the door to a greater understanding of how different facets of the UPR contribute to ER homeostasis.
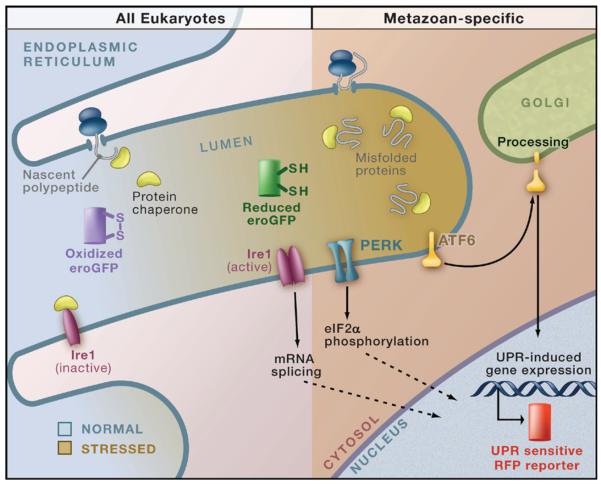
In metazoans, the UPR is a complex multipathway system (Figure 1) that simultaneously initiates several signaling cascades, reduces protein translation, mediates selective degradation of certain mRNAs, and attenuates translocation of certain proteins into the ER (Ron and Walter, 2007). The net effect of these responses is to temporarily reduce substrate burden in the ER until the cell can increase its capacity for protein maturation and degradation. The budding yeast uses a simplified version of the metazoan UPR response, which is initiated solely by the ER-localized transmembrane kinase Ire1 (Figure 1). Upon activation, the cytosolic domain of Ire1 removes an intron from HAC1 mRNA, allowing production of Hac1 protein, a transcription factor that upregulates numerous genes involved in ER biosynthesis and function (Travers et al., 2000). Although the downstream signaling pathways in response to ER stress are extensively studied, their direct relationships to protein homeostasis in the ER lumen are poorly understood. For example, it is largely unclear how quickly normal protein maturation is restored after the ER is subjected to stress, or how much heterogeneity in the ability to adapt exists within a population of cells.
Figure 1. Monitoring ER Homeostasis and UPR Activity.
The environment of the endoplasmic reticulum (ER) and activation of the unfolded protein response (UPR) can be monitored using a redox-sensitive green fluorescent protein (eroGFP) probe and a UPR-responsive red fluorescent protein (RFP) expression reporter, respectively (Merksamer et al., 2008). The eroGFP probe changes its excitation spectrum depending on the oxidation state of two engineered cysteines. Under normal conditions (left), the protein maturation capacity of the ER can meet the poly-peptide substrate load. During ER stress (right), the processing capacity of the ER is saturated, resulting in an accumulation of misfolded proteins (bound to chaperones) that initiates the UPR. Merksamer et al. (2008) find that accumulation of misfolded proteins can alter the redox state of eroGFP, making it a useful reporter of the ER environment. The UPR pathways include the universally conserved stress sensor Ire1 and the metazoan-specific stress sensors PERK (PKR-like ER resident kinase) and ATF6. Each of these functions by different mechanisms to initiate signaling pathways that activate transcription of numerous UPR-responsive genes (Ron and Walter, 2007).
A major obstacle to answering these fundamental questions about ER homeostasis has been the dearth of tools to directly measure misfolded proteins (i.e., the input for the UPR) and thus ER functionality. To address this issue, Merksamer and colleagues hypothesized that the many seemingly unrelated ER perturbations that activate the UPR may also perturb the redox state of the normally oxidizing ER lumen. The authors direct a previously described redox-sensitive green fluorescent protein (GFP) (Hanson et al., 2004) to the ER of yeast cells to enable monitoring of the lumen's redox state. This GFP variant (termed eroGFP) changes its excitation spectrum depending on the oxidation status of two engineered cysteines (Figure 1). Thus, eroGFP provides a proximal sensor for one aspect of ER homeostasis (redox). The authors also engineered the eroGFP-expressing yeast cells to harbor a UPR-responsive red fluorescent protein (RFP) reporter to provide a distal measure of UPR activity (Figure 1). Importantly, this dual reporter system is amenable to rapid quantitative analyses (by fluorescence-activated cell sorting) of large numbers of individual cells, which can be sampled automatically at regular intervals from a bioreactor culture (Chin et al., 2008). Thus, this reporter system can be used to measure not only temporal changes in the proximal and distal sensors of ER homeostasis in living cells but also the heterogeneities within a population.
Using this reporter, the authors set out to explore the relationship between different types of ER stress, the UPR response, and ER redox homeostasis in budding yeast. Merksamer et al. show that chemical reduction of the ER's oxidative capacity leads to a rapid (within minutes) change in the eroGFP reporter to the reduced state. After a short lag, they observe activation of the UPR-responsive RFP reporter due to the accumulation of misfolded proteins that cannot form disulfide bonds under reducing conditions. Importantly, treatment of the yeast cells with tunicamycin (a protein glycosylation inhibitor that results in misfolded protein accumulation) also detectably shifts eroGFP to its reduced state. In this case, the deflection from homeostasis is delayed (hours rather than minutes) and significantly muted, presumably because the effect on redox status is indirectly caused by misfolded protein accumulation. This finding validates the authors' initial hypothesis that the redox state of the ER lumen is a sensitive and direct indicator of altered ER homeostasis (caused even by non-redox perturbations), thereby providing a new tool for exploring the ER.
By measuring the redox state of the ER lumen, the authors make several intriguing observations about cells deficient in UPR signaling, oxidative ER protein folding, or ER protein degradation. Although these three classes of mutant cells all exhibit various altered UPR-induced gene expression phenotypes, Merksamer et al. show that their ER lumen redox states are completely normal. This suggests that the cells achieve an “adapted” state in which, at least under optimal growth conditions, ER homeostasis (as indicated by eroGFP oxidation state) is indistinguishable from that of wild-type cells despite altered UPR activity. However, differences in ER functionality among these adapted mutant cells become apparent when they are challenged with a chemical stress. In the UPR-deficient mutants, both oxidative and glycosylation stress lead to larger deviations of the eroGFP reporter than seen in normal cells, indicating a more severe departure from ER homeostasis. Indeed, most of the cells fail to recover and simply stop growing. In contrast, when glycosylation stress is applied to the mutant cells defective in oxidative ER protein folding and ER protein degradation, there is less of a change in eroGFP oxidation relative to normal cells. As these mutant cells activate UPR even when unstressed, Merksamer et al. suggest that basal UPR activation (which results in elevated levels of folding and maturation factors in the ER) partially buffers cells from certain chemical stressors. This is reminiscent of the classic notion of “preconditioning,” where a mild initial stress buffers against subsequent (even unrelated) stresses by pre-emptive upregulation of stress-responsive proteins. Indeed, a very similar effect is seen in mammalian cells that are subjected to chronic low-level chemical stress (Rutkowski et al., 2006). This may explain the tissue-type-dependent differences in sensitivity to ER stress that are observed in metazoans—cell types with vastly different substrate fluxes through the ER may differ markedly in their baseline “adapted” states.
Other insights into ER homeostasis come from using the reporter system to do population analyses at the single-cell level. One striking example of population heterogeneity in the ER stress response is seen when the yeast cells are deprived of inositol. In wild-type yeast, inositol starvation induces UPR signaling, even though there is no detectable change in eroGFP oxidation in any of the cells. However, a subpopulation of yeast cells defective in UPR signaling shows marked eroGFP activation, indicating loss of ER homeostasis. Remarkably, further analysis reveals that these persistently stressed cells are predominantly mother cells, whereas the newly budded daughter cells appear to be essentially normal. This intriguing observation suggests a previously unsuspected asymmetry in the segregation of the ER between mother and daughter cells under conditions of stress. How the stressed ER is selectively retained in the mother cell is an exciting area of future exploration.
The study by Merksamer et al. illustrates that eroGFP is a sensitive detector of ER homeostasis because numerous ER-based pathways seem to be interlinked with redox regulation (through as yet unknown mechanisms). This new molecular tool can now be combined with the vast collections of yeast strains (deletion, hypomorphic, overexpression, and other mutations) to reveal previously unanticipated pathways that intersect with ER homeostasis. The system can also be adapted to the more complex and nuanced metazoan system to understand the relative contributions of each of the various branches of the UPR. Most intriguingly, the capacity to simultaneously monitor multiple parameters at the single-cell level may be especially important in understanding how cells choose between adaptation and apoptosis during chronic protein misfolding stress. This may help to tease out the basis of the heterogeneous and seemingly arbitrary cell death observed in many diseases associated with chronic ER stress.
REFERENCES
- Chin CS, Chubukov V, Jolly ER, DeRisi J, Li H. PLoS Biol. 2008;6:e146. doi: 10.1371/journal.pbio.0060146. 10.1371/journal.pbio.0060146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. J. Biol. Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- Merksamer PI, Trusina A, Papa FR. Cell, this issue. 2008 doi: 10.1016/j.cell.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, Mori K, Sadighi Akha AA, Raden D, Kaufman RJ. PLoS Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Yoshida H. FEBS J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]