Abstract
A rapid, simple and reproducible dot blot method is described for quantitating the amounts of specific messages in small numbers of intact cells. The method has been used to accurately determine the number of histone H4 mRNA molecules in growing (approximately 40,000) and in starved (approximately 1600) Tetrahymena thermophila, and to measure the amount of message contributed by an E. coli plasmid containing part of the S10 ribosomal operon. Use of the method is illustrated to optimize in situ hybridization protocols and to measure mRNA amounts in cell lysates. Preliminary studies also indicate that the method can be used to detect mRNA in intact yeast cells.
Full text
PDF







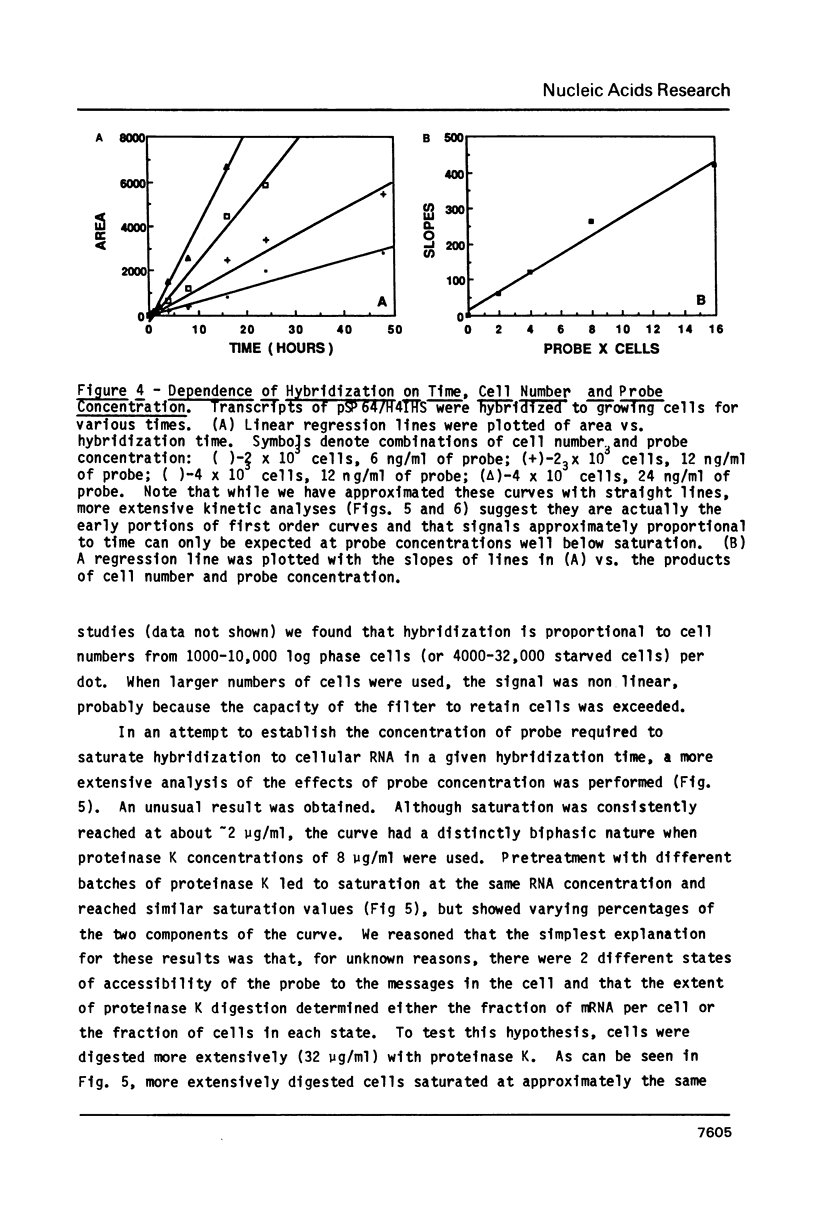
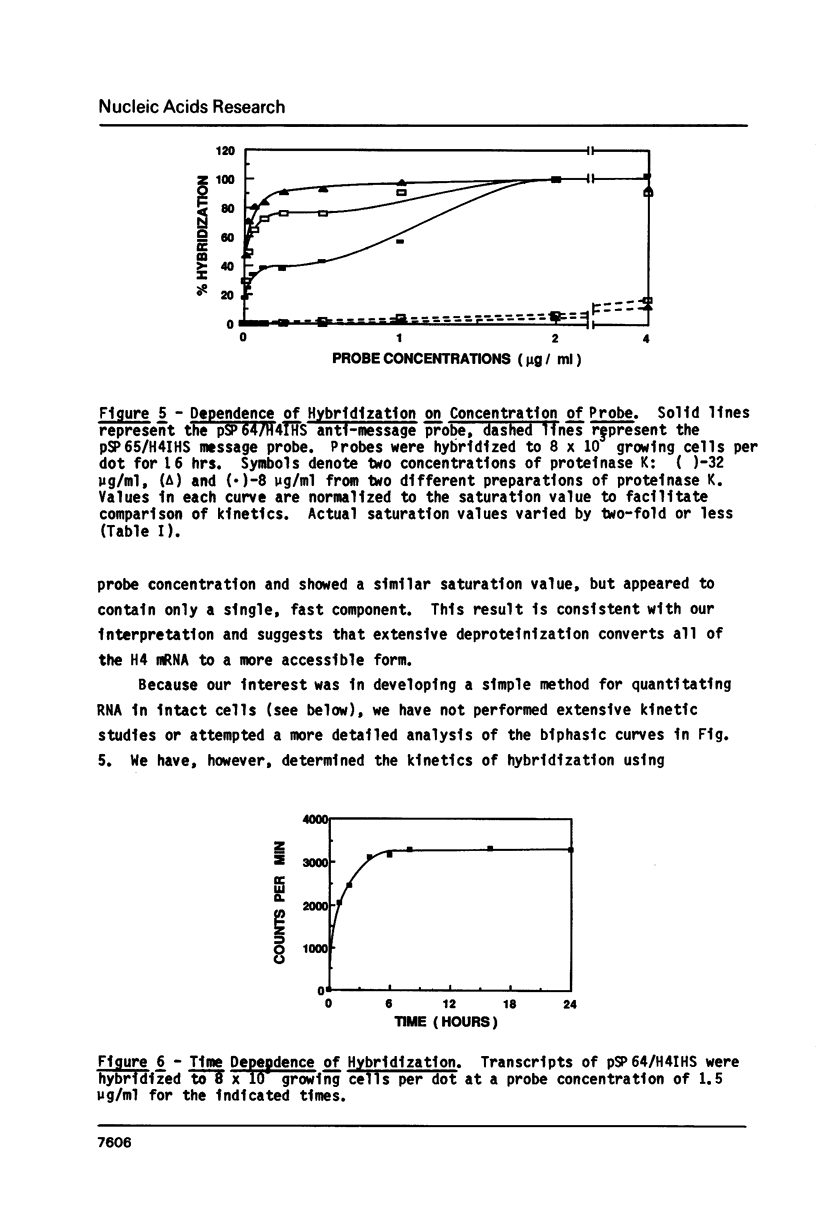







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angerer L. M., Angerer R. C. Detection of poly A+ RNA in sea urchin eggs and embryos by quantitative in situ hybridization. Nucleic Acids Res. 1981 Jun 25;9(12):2819–2840. doi: 10.1093/nar/9.12.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon G. A., Bowen J. K., Yao M. C., Gorovsky M. A. Tetrahymena H4 genes: structure, evolution and organization in macro- and micronuclei. Nucleic Acids Res. 1984 Feb 24;12(4):1961–1975. doi: 10.1093/nar/12.4.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahic M., Haase A. T. Detection of viral sequences of low reiteration frequency by in situ hybridization. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6125–6129. doi: 10.1073/pnas.75.12.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzone F. J., Angerer R. C., Gorovsky M. A. Regulation of protein synthesis in Tetrahymena. Quantitative estimates of the parameters determining the rates of protein synthesis in growing, starved, and starved-deciliated cells. J Biol Chem. 1983 Jun 10;258(11):6887–6898. [PubMed] [Google Scholar]
- Calzone F. J., Angerer R. C., Gorovsky M. A. Regulation of protein synthesis in Tetrahymena: isolation and characterization of polysomes by gel filtration and precipitation at pH 5.3. Nucleic Acids Res. 1982 Mar 25;10(6):2145–2161. doi: 10.1093/nar/10.6.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael G. G., McMaster G. K. The analysis of nucleic acids in gels using glyoxal and acridine orange. Methods Enzymol. 1980;65(1):380–391. doi: 10.1016/s0076-6879(80)65049-6. [DOI] [PubMed] [Google Scholar]
- Cox K. H., DeLeon D. V., Angerer L. M., Angerer R. C. Detection of mrnas in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984 Feb;101(2):485–502. doi: 10.1016/0012-1606(84)90162-3. [DOI] [PubMed] [Google Scholar]
- DeLeon D. V., Cox K. H., Angerer L. M., Angerer R. C. Most early-variant histone mRNA is contained in the pronucleus of sea urchin eggs. Dev Biol. 1983 Nov;100(1):197–206. doi: 10.1016/0012-1606(83)90211-7. [DOI] [PubMed] [Google Scholar]
- Edwards M. K., Wood W. B. Location of specific messenger RNAs in Caenorhabditis elegans by cytological hybridization. Dev Biol. 1983 Jun;97(2):375–390. doi: 10.1016/0012-1606(83)90094-5. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A. Genome organization and reorganization in Tetrahymena. Annu Rev Genet. 1980;14:203–239. doi: 10.1146/annurev.ge.14.120180.001223. [DOI] [PubMed] [Google Scholar]
- Hallberg R. L., Bruns P. J. Ribosome biosynthesis in Tetrahymena pyriformis. Regulation in response to nutritional changes. J Cell Biol. 1976 Nov;71(2):383–394. doi: 10.1083/jcb.71.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J. B., Singer R. H. Quantitative analysis of in situ hybridization methods for the detection of actin gene expression. Nucleic Acids Res. 1985 Mar 11;13(5):1777–1799. doi: 10.1093/nar/13.5.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L., Archer R., Zengel J. M. Transcription of the S10 ribosomal protein operon is regulated by an attenuator in the leader. Cell. 1983 May;33(1):241–248. doi: 10.1016/0092-8674(83)90353-7. [DOI] [PubMed] [Google Scholar]
- Lindahl L., Zengel J. M. Operon-specific regulation of ribosomal protein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6542–6546. doi: 10.1073/pnas.76.12.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister L. B., Scheller R. H., Kandel E. R., Axel R. In situ hybridization to study the origin and fate of identified neurons. Science. 1983 Nov 18;222(4625):800–808. doi: 10.1126/science.6356362. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985 Feb;40(2):405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- Singer R. H., Ward D. C. Actin gene expression visualized in chicken muscle tissue culture by using in situ hybridization with a biotinated nucleotide analog. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7331–7335. doi: 10.1073/pnas.79.23.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA transferred or dotted nitrocellulose paper. Methods Enzymol. 1983;100:255–266. doi: 10.1016/0076-6879(83)00060-9. [DOI] [PubMed] [Google Scholar]