Abstract
Ovarian carcinomas are thought to arise from cells of the ovarian surface epithelium by mechanisms that are poorly understood. Molecules associated with neoplasia are potentially immunogenic, but few ovarian tumor antigens have been identified. Because ovarian carcinomas can elicit humoral responses in patients, we searched for novel tumor antigens by immunoscreening a cDNA expression library with ovarian cancer patient serum. Seven clones corresponding to the homeobox gene HOXB7 were isolated. ELISAs using purified recombinant HOXB7 protein revealed significant serologic reactivity to HOXB7 in 13 of 39 ovarian cancer patients and in only one of 29 healthy women (P < 0.0001). Ovarian carcinomas were found to express HOXB7 at markedly higher levels than normal ovarian surface epithelium, suggesting that immunogenicity of HOXB7 in patients could be associated with its elevated expression in ovarian carcinomas. Overexpression of HOXB7 in immortalized normal ovarian surface epithelial cells dramatically enhanced cellular proliferation. Furthermore, HOXB7 overexpression increased intracellular accumulation and secretion of basic fibroblast growth factor, a potent angiogenic and mitogenic factor. These results reveal HOXB7 as a tumor antigen whose up-regulated expression could play a significant role in promoting growth and development of ovarian carcinomas.
Carcinomas that arise from the ovarian surface epithelium (OSE) are the most lethal of gynecologic malignancies, and the majority of patients present with disseminated disease (1). The molecular mechanisms involved in ovarian carcinogenesis are poorly understood. Although it is known that ovarian carcinomas elicit immune responses in patients (2), few ovarian tumor antigens have been identified. These include the lysosomal protease cathepsin D (2), HER-2/neu, an antigen well studied in breast cancer (3), and NY-ESO-1 and MAGE-1, antigens originally identified in esophageal squamous cell carcinoma and melanoma, respectively (4, 5).
Serologic screening of cDNA expression libraries with patient sera has allowed relatively unbiased searches for molecules that elicit high-titer IgG antibody responses (4, 6–8). This methodology, termed SEREX, has identified several antigens, notably NY-ESO-1, whose restricted expression patterns and ability to elicit cell-mediated as well as humoral immune responses have made them ideal candidates for immunotherapy (9). Various molecules associated with carcinogenesis elicit humoral and cell-mediated immune responses, two examples being p53 and HER-2/neu (10). In this study, we applied SEREX methodology using ovarian cancer patient serum and isolated HOXB7, a homeobox gene. Ovarian carcinomas were found to express HOXB7 at markedly higher levels than normal OSE. Overexpression of HOXB7 in immortalized normal OSE cells up-regulated expression of basic fibroblast growth factor (bFGF), a potent mitogenic and angiogenic factor, and dramatically increased OSE cell proliferation. These results reveal HOXB7 as a tumor antigen whose up-regulated expression could play a significant role in growth of ovarian carcinomas.
Materials and Methods
Human Tissues and Sera.
Tumor tissues excess to diagnosis were snap-frozen in liquid nitrogen. OSE was scraped from normal ovaries. Sera were obtained from patients with primary ovarian carcinoma and from healthy female donors. Tissue and sera were collected with the informed consent of patients (protocol no. RPN98–03-02–01). Patients had disease that extended to the uterus and fallopian tubes (Stage II, n = 1), to the abdomen and lymph nodes (Stage III, n = 30), or involved distant metastasis (Stage IV, n = 8).
Cell Lines.
The IOSE-29 cell line was kindly provided by Nelly Auersperg (University of British Columbia, Vancouver) and cultured as previously described (11). OV-1063 (12) and OVCAR-3 (13) cell lines were obtained from American Type Culture Collection and cultured according to their specifications.
Western Blot Analysis.
Ten micrograms of protein lysate, prepared from tissue and cultured cells by using M-PER reagent (Pierce), were separated by SDS/PAGE and transferred to membranes that were incubated with diluted patient serum (1:500). Reactivity of serum antibodies was detected by using peroxidase-conjugated antibody to human IgG and LumiGLO chemiluminescent substrate (Kirkegaard & Perry Laboratories).
RNA Isolation and cDNA Expression Library Construction.
Total RNA was isolated from OV-1063 cells by using TRIZOL (Life Technologies, Rockville, MD). Poly(A)+ mRNA was purified by using oligo-dT cellulose (Qiagen, Chatsworth, CA). cDNA was prepared by using XhoI site-tagged oligo-dT primer, ligated to EcoRI adaptors and cloned into EcoRI/XhoI sites of the λZAP-Express vector (Stratagene). Ligated cDNA was packaged into phage particles by using Gigapack III Gold packaging extract (Stratagene), which were used to infect Escherichia coli strain XLI-Blue MRF′. A primary library of >800,000 recombinants was amplified and used for immunoscreening.
Immunoscreening of cDNA Expression Library.
Library screening with diluted patient serum (1:500) was performed as previously described (6, 8). Positive phage plaques were purified to monoclonality by repeated screening. False-positive plaques were eliminated by screening with alkaline phosphatase-conjugated anti-human IgG secondary antibody alone (Kirkegaard & Perry Laboratories). pBK-CMV phagemids were obtained by coinfection of E. coli with recombinant λ phage and M13 helper phage and cDNA inserts sequenced.
Expression and Purification of Recombinant HOXB7 Protein.
Full-length HOXB7 coding sequences were amplified by PCR from pBK-CMV phagemids and cloned into the pPROEXHTb vector (Life Technologies). E. coli transformed with plasmid pPROEXHTb-HOXB7 were grown to an OD600 of 0.6 and protein expression induced by isopropylthio-β-galactoside (1 mM). His-tagged protein was purified on nickel-nitrilotriacetic acid resin columns (Qiagen).
ELISA.
One hundred nanograms per well of purified recombinant HOXB7 was adsorbed to 96-well plates overnight. Control wells were coated with purified recombinant capsid protein L2 of bovine papillomavirus (14). After washing and blocking wells with 2% BSA/PBS, 100 μl of diluted human serum was added and incubated for 1 h at 4°C. Sera were tested at dilutions ranging from 1:100 to 1:50,000. Wells were washed and incubated for 1 h with peroxidase-conjugated antibody to human IgG, followed by reaction with TMB substrate (Dako) and measurement at an optical density of 450 nm.
Reverse-Transcribed–PCR (RT-PCR) Analysis of Antigen Expression.
Reverse transcription was performed by using 1 μg of DNase I-treated total RNA, 500 ng of oligo(dT) and Superscript II reverse transcriptase (Life Technologies). Amplification of cDNAs for HOXB7 and for β-actin were performed as described by others (15, 16) by using Platinum Taq DNA polymerase (Life Technologies). Briefly, amplification was performed with a 2 min start at 94°C, denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min, for 35 cycles for HOXB7 and 25 cycles for β-actin. Titrations were performed to ensure a linear range of amplification. Primers were the same as used by others (15, 16) and were as follows: for HOXB7 5′ AGAGTAACTTCCGGATCTA-3′ and 5′-TCTGCTTCAGCCCTGTCTT-3′, and for β-actin 5′-ATGATATCGCCGCGCTCG-3′ and 5′-CGCTCGGTGAGGATCTTCA-3′. Southern blot analysis of RT-PCR products was conducted by using 32P-labeled β-actin cDNA (CLONTECH) and HOXB7 cDNA. Hybridization signals were quantified by PhosphorImager analysis (Molecular Dynamics).
Transfection of IOSE-29 Cells with HOXB7.
Full-length HOXB7 cDNA was subcloned from pPROEXHTb-HOXB7 into mammalian expression vectors pBK-CMV (Stratagene) and pIRESpuro2 (CLONTECH). In addition, full-length HOXB7 cDNA, cloned in pcDNA3 (17) and provided by Alain Chariot (University of Liege, Liege, Belgium), was subcloned into pcDNA3.1 (Invitrogen). Subconfluent cultures of IOSE-29 cells were transfected with linearized DNA by using Lipofectamine PLUS reagent (Life Technologies). Cells transfected with pBK-CMV and pcDNA3.1 constructs were selected with G148 (400 μg/ml), and cells transfected with pIRESpuro2 constructs were selected with puromycin (1 μg/ml). Experiments were performed by using lines established from single colonies.
Proliferation Assays.
Stably transfected IOSE-29 cells were seeded at 2,000 cells/200 μl per well in 96-well plates. Thymidine incorporation was measured in cultures pulsed for 3 h with 1 μCi of [3H]methylthymidine (60 Ci/mmol) (ICN) after 1, 2, 3, and 4 days of culture. Cells were also seeded in wells coated with poly 2-hydroxyethylmethacrylate [poly(HEMA)] (Sigma) and pulsed for 18 h with [3H]methylthymidine.
Assays of bFGF Production.
Cells were seeded in 25-cm2 flasks containing 5 ml of medium. Culture supernatants were harvested when cells reached a density of 2 × 104 cells/cm2. Protein lysates were prepared by using M-PER reagent at 105 cells/10 μl. bFGF levels were assayed in culture supernatants and cell lysates by using the Quantikine human bFGF immunoassay (R&D Systems). Monoclonal anti-human bFGF (Sigma, clone FB-8) was used for Western blots.
Results
Isolation of HOXB7 cDNAs by Serologic Screening.
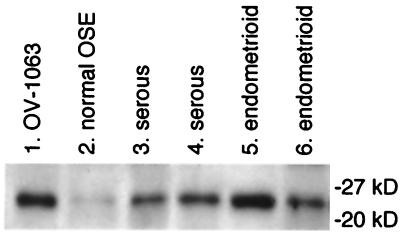
Serum antibodies of a patient with ovarian carcinoma were found by Western blot analysis to react with a common protein ≈24 kDa in mass that was present at low levels in normal OSE and at markedly higher levels in several ovarian carcinoma specimens and in the OV-1063 cell line (Fig. 1). We originally used this cell line because it was reported to be derived from an ovarian carcinoma (12). Subsequent studies have shown that the line contains a Y chromosome (http://www.atcc.org/phage/probline.html), indicating that its origin is extremely unlikely to be truly ovarian. Nevertheless, this cell line has been recently revealed by serial analysis of gene expression to have a global profile of gene expression similar to those found in ovarian carcinoma specimens (18). A λZAP cDNA expression library was constructed from OV-1063 cells. After screening ≈200,000 recombinant phage plaques with the patient serum, four positive clones were isolated. One of these clones encoded the mitochondrial iron transporter ABC7 (GenBank accession no. AF133659) (19). The other three clones encoded the homeodomain protein HOXB7, which has a predicted mass of 24 kDa (20). Another four HOXB7 clones were isolated in further screening. Coding sequences of the HOXB7 clones were identical to the original published sequence (20) (GenBank no. NM004502), except for two nucleotide substitutions G → C and G → A that respectively altered residue 53 from Gly to Ala and residue 173 from Ala to Thr. These substitutions were also present in other published HOXB7 clones (17, GenBank no. XM008559). HOXB7 is a member of the HOX family of homeobox genes that encode transcription factors that regulate normal cellular proliferation and differentiation during development (21, 22). HOX genes have been well-studied in their control of hematopoiesis, and their aberrant expression in leukemias and other cancers has implicated their involvement in tumorigenesis (22–24). It was therefore clearly of interest to investigate the role of HOXB7 in ovarian carcinogenesis.
Figure 1.
Detection of a protein common to OV-1063 cells and ovarian carcinomas by Western blot analysis by using patient serum. Protein lysates (10 μg per lane) prepared from OV-1063 cells (lane 1), normal OSE (lane 2), and ovarian carcinomas of the serous (lanes 3 and 4) and endometrioid (lanes 5 and 6) histotypes were probed with diluted serum (1:500) of a patient with Stage III serous ovarian carcinoma. Shown are bands corresponding to a common reactive species. Positions of protein size markers are indicated.
Prevalence of Antibody Responses to HOXB7 Among Ovarian Carcinoma Patients.
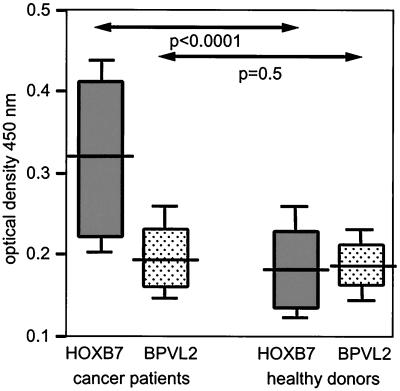
Many serologically identified antigens are reactive with antibodies of healthy donors as well as those of cancer patients (7, 8). Serologic responses to HOXB7 of ovarian cancer patients were compared with those of healthy female donors in ELISAs by using purified recombinant HOXB7 protein and, as a negative control protein, recombinant bovine papillomavirus capsid protein L2 (BPVL2) (14). As shown in Fig. 2, there was no significant difference between the low serologic responses to L2 observed among healthy women and among patients. In contrast, a significant difference was observed between serologic responses to HOXB7 of patients and of healthy women (P < 0.0001) (Fig. 2). Thirteen of 39 patients, but only one of 29 healthy women, were found to generate anti-HOXB7 antibodies where a positive reaction is defined as an optical density value that exceeds the mean optical density value of sera of healthy donors by three standard deviations. Although sera from patients with early-stage organ-confined disease were not available for analysis, there was no obvious correlation between serologic responses to HOXB7 and disease stage (II–IV). Preliminary studies showed reactivity to the ABC7 protein by the patient serum used to immunoscreen the library, but not by nine other patient sera (data not shown).
Figure 2.
Serologic responses to HOXB7 as assessed by ELISA. BPVL2 protein was used as a negative control protein. Sera were diluted 1:500. Shown are values of statistical significance for differences in optical density at 450 nm of sera of patients (n = 39) and of healthy women (n = 29) as determined by the Mann–Whitney u test. Statistical significance for differences in responses to HOXB7 and to BPVL2 were assessed by the Wilcoxon's signed-rank test for paired data and were found to be P < 0.0001 for patients and P = 0.4 for healthy women. Horizontal bars indicate median values
HOXB7 Expression Patterns in Ovarian Carcinomas and Normal OSE.
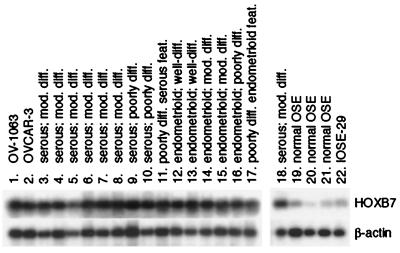
Low levels of HOXB7 expression were detected by semiquantitative RT-PCR analysis in normal OSE and in IOSE-29 cells, a nontumorigenic cell line established by immortalizing normal OSE cells with SV40 large T antigen (11) (Fig. 3). However, markedly higher levels of HOXB7 expression were detected in primary ovarian carcinomas. Such elevated levels were consistent between specimens of carcinomas which varied widely in their degree and type of histologic differentiation, and also in stage of disease. OV-1063 cells and the ovarian carcinoma cell line OVCAR-3 also expressed HOXB7 at levels similar to those in tumor tissue specimens (Fig. 3). These observations indicate that elevation of HOXB7 expression is a common feature of ovarian carcinomas and may render HOXB7 immunogenic.
Figure 3.
Semiquantitative RT-PCR analysis of HOXB7 expression. Shown are Southern blots of HOXB7 and β-actin RT-PCR products in OV-1063, OVCAR-3 and IOSE-29 cells (lanes 1, 2, and 22), specimens of normal OSE (lanes 19–21) and ovarian carcinomas (lanes 3–18). Histology of carcinomas ranged from poorly differentiated (diff.) with either serous or endometrioid features (feat.) to moderately (mod.) and well-differentiated serous and endometrioid. The specimen used for analysis shown in lane 18 is the same as that in lane 6.
Effect of HOXB7 Overexpression on OSE Cell Proliferation.
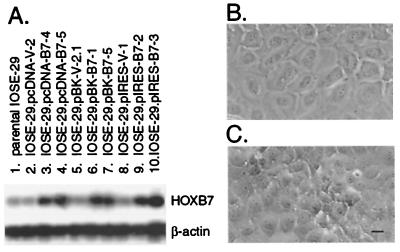
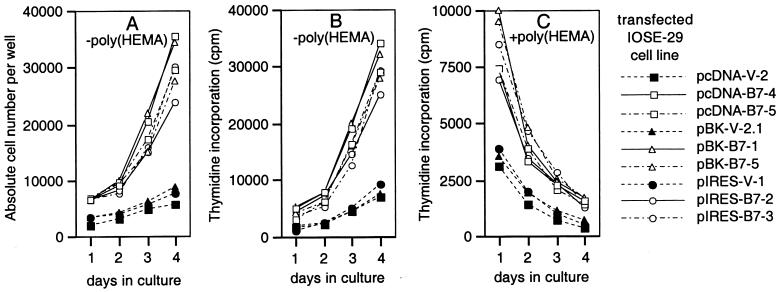
Because HOXB7 expression was markedly higher in carcinomas than in normal OSE and HOXB7 regulates proliferation of several other cell types (25–27), we investigated the possibility that HOXB7 overexpression increases proliferation of OSE cells. IOSE-29 cells were stably transfected with HOXB7 cloned in three different expression vectors. The resulting cell lines expressed HOXB7 at similar levels, 3- to 4-fold the level in parental IOSE-29 cells and in IOSE-29 cells stably transfected with vector DNA clone (Fig. 4A). Vector-transfected IOSE-29 cells grew in flat monolayers similar to the parental line (Fig. 4B). In contrast, cultures of HOXB7 transfectants exhibited islands of multi-layered overgrowth (Fig. 4C). The dramatically enhanced growth of HOXB7 transfectants was evidenced by increases in absolute cell number (Fig. 5A) and in thymidine incorporation (Fig. 5B), which were 3- to 4-fold the levels observed in vector-transfected cells. Growth was also examined under conditions where adherence to substratum was inhibited. Equivalent numbers of cells of vector- and HOXB7-transfected lines were seeded in wells coated with poly(HEMA) and proliferative activities monitored by measuring thymidine incorporation. Levels of incorporated thymidine in vector-transfected cells progressively declined, reaching almost background levels by Day 4 (Fig. 5C). A similar rate of decline in thymidine incorporation in HOXB7 transfectants was observed (≈50% decrease in levels per day), although levels of incorporated thymidine in HOXB7 transfectants were consistently higher than levels in vector-transfectants on any given day (Fig. 5C). An initial increase in numbers of HOXB7-transfected cells during the first 24 h after seeding in poly(HEMA)-coated wells could explain their higher levels of thymidine incorporation, but HOXB7 overexpression in these cells does not appear to permit sustained anchorage-independent growth.
Figure 4.
HOXB7 expression levels and morphology of transfected IOSE-29 cells. (A) RT-PCR analysis detected HOXB7 expression levels in HOXB7-transfected cells (lanes 3, 4, 6, 7, 9, 10) that were markedly higher than in cells transfected with vector DNA alone (lanes 2, 5, 8) and in the parental cell line (lane 1). Phase-contrast microscopy revealed that IOSE-29 cells transfected with vector DNA grew in flat monolayers (B), whereas cultures of HOXB7-transfected cells exhibited islands of multilayered overgrowth (C). (Bar = 10 μm.)
Figure 5.
Growth characteristics of transfected cells. Vector- and HOXB7-transfected IOSE-29 cells were seeded at 2,000 cells per well in uncoated 96-well plates (A and B) and wells coated with poly(HEMA) (C). Total numbers of cells in each uncoated well were counted daily (A). After 1, 2, 3, and 4 days in culture, thymidine incorporation was measured in cells pulsed with [3H]methylthymidine for 3 h in uncoated wells (B), and for 18 h in poly(HEMA)-coated wells (C). Shown are the mean values of three to four independent experiments. Differences in cell numbers and thymidine incorporation levels of HOXB7-transfected cells, as compared with corresponding vector-transfected cells, at each time point were found to be statistically significant (P < 0.001).
Effect of HOXB7 Overexpression on bFGF Production.
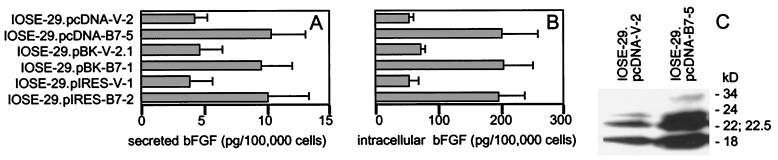
Growth factor autocrine loops represent a key mechanism regulating tumor cell growth. bFGF has been found by several studies to be expressed in ovarian carcinomas and is widely believed to stimulate their growth (28–30). We therefore investigated whether overexpression of HOXB7 in OSE cells could up-regulate bFGF production. ELISAs revealed levels of bFGF in culture supernatants of HOXB7-transfected IOSE-29 cells that were approximately 3-fold higher than levels in supernatants collected from equivalent numbers of vector-transfected cells (Fig. 6A). The intracellular bFGF content of HOXB7 transfectants was also approximately 3-fold higher than that of vector-transfected cells (Fig. 6B). Surprisingly, when the amount of secreted and intracellular bFGF were compared on a per cell basis, the vast majority (≈95%) of total bFGF produced by a cell was intracellular (compare Fig. 6 A with B). Human bFGF is produced naturally in several isoforms (18, 22, 22.5, 24, and 34 kD) that originate from alternative translation initiation sites within a single mRNA species (31, 32). Western blot analysis of cell lysates revealed increased levels of each of these bFGF isoforms in HOXB7-transfected cells (Fig. 6C), indicating that overexpression of HOXB7 up-regulates total bFGF production in OSE cells.
Figure 6.
Increased bFGF production by HOXB7-transfected cells. bFGF levels in (A) culture supernatants collected from equivalent numbers of vector- and HOXB7-transfected IOSE-29 cells and (B) lysates of these cells were assessed by ELISA. Shown are mean values of two to three independent experiments. (C) Western blot analysis of lysates (10 μg per lane) of vector-transfected (IOSE-29.pcDNA-V-2) and HOXB7-transfected (IOSE-29.pcDNA-B7–5) IOSE-29 cells. Molecular weights of bFGF isoforms are indicated.
Discussion
Tumor antigens have been increasingly identified by using the antibody repertoire of cancer patients, and recent attention has focused on their immunotherapeutic potential. However, the biological relevance to neoplasia of most SEREX-defined antigens is unclear. In this study, we identified HOXB7, a product of a homeobox gene, as a tumor antigen by serologic screening with ovarian cancer patient serum. Homeobox genes have been described as master control genes acting at the top of genetic hierarchies regulating cell growth, differentiation, and development (21, 22). Approximately 170 different vertebrate homeobox genes have been identified, of which 39 belong to the HOX family (21, 22). Although it is well established that HOX proteins function as transcription factors, the vast majority of their targets have yet to be elucidated. It has been reported that HOXA5 regulates p53 transcription (33), and that p21 is a target of HOXA10 (34). Expression of cell adhesion molecules is regulated by several HOX proteins (23). Auto- and crossregulatory interactions within the homeotic network have also been described (23). Our observations indicate that HOXB7 overexpression in OSE cells stimulates their proliferation and up-regulates production of bFGF. bFGF is expressed by a wide variety of tissues and regulates proliferation, differentiation, survival, and migration of cells of mesodermal, endodermal, and ectodermal origin (35). A number of studies have detected bFGF expression in ovarian carcinomas and elevated levels of bFGF in ascites and sera of ovarian cancer patients (28, 29, 36). bFGF is believed to promote growth of ovarian carcinomas (30) and various other tumors such as hepatomas (37), melanomas (25), and breast carcinomas (26). Proliferation of melanoma and breast carcinoma cells is reportedly controlled by HOXB7 and likewise involves its up-regulation of bFGF expression (25, 26). HOXB7 has been shown to directly transactivate the bFGF gene through at least one of five putative homeodomain-binding sites in its promoter (25). Although HOXB7 may control OSE cell proliferation through modulating targets in addition to bFGF, these results suggest that HOXB7 could stimulate growth of ovarian carcinomas by up-regulating bFGF production.
The vast majority (≈95%) of bFGF produced by OSE cells was intracellular, suggesting that bFGF could act via an intracrine mechanism. In unpublished work, we observed only partial inhibition of OSE cell proliferation by bFGF-neutralizing antibodies, consistent with studies of other bFGF-expressing cell types (38, 39). Intracellular accumulation of bFGF and the existence of functionally active intracellular bFGF receptors occurs in a wide variety of normal and tumor cell types (26, 37, 38, 40–42). Several isoforms of bFGF are generated through utilization of in-frame alternative translation initiation sites (31, 32). The smallest isoform (18 kDa) is primarily cytosolic and secreted, whereas the higher molecular weight isoforms (22, 22.5, 24, and 34 kDa) contain nuclear localization sequences and preferentially localize in nuclei and nucleoli (39–43). Despite considerable evidence that the high molecular weight isoforms of bFGF stimulate cell proliferation in an intracrine fashion, their signaling pathways are poorly understood. In this study, HOXB7 overexpression in OSE cells up-regulated levels of all isoforms of bFGF. This suggests that elevated HOXB7 expression, which commonly occurs in ovarian carcinomas, could promote their growth by triggering both intracrine and autocrine bFGF growth stimulatory pathways. Because bFGF is a potent stimulator of angiogenesis and cell migration (35), bFGF could additionally contribute to ovarian carcinoma growth and metastasis through paracrine effects.
The coordinated spatial and temporal expression of HOX genes is critical to their regulatory function, and aberrant HOX gene expression observed in various cancers has implicated their involvement in neoplasia (21–24). Although it is not surprising that such master control genes can contribute to neoplasia, it is as yet unclear whether transforming ability is an intrinsic and universal property of HOX genes. It has been reported that HOXB7 and several other HOX genes exhibit transforming ability in NIH 3T3 fibroblasts, a cell line prone to transformation (44). However, HOXB7 overexpression in hematopoietic stem cells promotes myeloid differentiation (27) and in multipotent mesenchymal cells promotes differentiation to smooth muscle cells (45). In this study, we failed to find strong evidence that HOXB7 overexpression in immortalized normal OSE cells promotes anchorage-independent growth, although increased proliferation and reduced contact inhibition was observed. This suggests that elevated HOXB7 expression levels detected in ovarian carcinomas may be associated with higher proliferative activity in tumors, rather than representing a step in the transformation process. Indeed, HOXB7 is constitutively expressed in melanomas and in proliferating but not quiescent normal melanocytes (25). However, the possibility that HOXB7 functions in cell transformation by acting cooperatively with other homeoproteins cannot be excluded. Coactivation of HoxA9 and Meis1, another homeobox gene, in mouse bone marrow cells has been reported to rapidly induce acute myeloid leukemia, an effect not observed with overexpression of these homeobox genes alone (46). Interestingly, selective overexpression of the high molecular weight isoforms of bFGF in NIH 3T3 cells promotes growth in low serum and increases saturation cell density, whereas overexpression of the 18-kDa isoform does not (43, 47). However, selective overexpression of the larger bFGF isoforms in cardiac myocytes induced binucleation (40) and in smooth muscle cells increased proliferation (39). As for HOXB7, there is as yet no firm evidence that any of the bFGF isoforms have intrinsic transforming ability in normal cells.
The majority of SEREX-defined antigens are intracellular, and their release by necrotic tumor cells may render such proteins immunogenic (7, 8, 48). In this study, we found HOXB7 to be expressed in normal OSE cells, and others have detected HOXB7 expression in normal tissues such as kidney and colon (24). HOXB7 immunogenicity in ovarian cancer patients could be ascribed, at least in part, to its elevated expression in their tumors. Overexpressed genes are believed to elicit immune responses by overriding thresholds critical for maintenance of tolerance. Although tumor-restricted expression is an obviously desirable property of an immunotherapeutic target, over-expressed self antigens have served as targets for active and passive immunotherapy, e.g., HER-2/neu and Melan A, a differentiation antigen present in melanoma and normal melanocytes (9). Gene amplification is a major mechanism of overexpression that can contribute to immunogenicity. Amplification and resulting overexpression of the HER-2/neu oncogene occurs in 30% of breast cancers and 20% of ovarian cancers (3). The translation initiation factor eIF-4 γ, identified by SEREX in squamous cell lung carcinoma, is encoded by an amplified gene (49). It has been recently reported that TGIF2, a novel homeobox gene, is amplified and overexpressed in ovarian cancer cell lines (50). However, it is unlikely that HOXB7 overexpression in ovarian carcinomas can be attributed to gene amplification, as comparable signal intensities of the HOXB7 gene have been detected by PCR analysis in equivalent amounts of genomic DNA isolated from ovarian carcinomas and from peripheral blood mononuclear cells of three patients (H.N., unpublished observations). Little is known of the precise mechanisms that could up-regulate HOX gene expression, although several HOX genes appear to be regulated by DNA methylation and by hormones (51, 52). Furthermore, mechanisms other than or in addition to overexpression could contribute to the immunogenicity of HOXB7, e.g., tumor-associated posttranslational modification and alterations in antigen processing and/or presentation by tumor cells. Surprisingly, peptide portions of Antennapedia, a Drosophila homeobox gene related to HOXB7, are efficiently internalized by cells in culture in a receptor-independent manner (53). Although the physiological significance of such specific uptake is unclear, it is tempting to speculate that HOXB7 could be efficiently taken up by antigen-presenting cells.
The generation of circulating autologous antibodies against tumor antigens can be regarded as the systemic amplification by the host immune system of a signal that indicates the presence of the tumor. Numerous studies have evaluated the prognostic relevance and diagnostic potential of autologous antitumor antibodies as serum biomarkers, particularly for detection of small, early-stage lesions (reviewed in ref. 54). Twenty to 40% of patients with a p53 mutation have been found to generate detectable levels of serum anti-p53 antibodies (55). The prevalence of antibody responses to SEREX-defined antigens has been likewise found to be quite low. For example, frequencies range from 14 to 27% in colon cancer (7) and from 5 to 25% in renal cancer (8). In this study, we found significant serologic reactivity to HOXB7 in 13 of 39 ovarian cancer patients and in only one of 29 healthy women. The use of serum tumor biomarkers for early detection of ovarian carcinoma has been limited by their insufficient specificity and sensitivity. Our preliminary observations need to be validated in larger case/control studies with particular attention being drawn to correlating titers of anti-HOXB7 antibodies with stage of disease. However, our data raise the possibility that serologic detection of autologous anti-HOXB7 antibodies could have diagnostic potential.
Acknowledgments
We thank Drs. Drew Pardoll, Hyam Levitsky, Venu Raman, T.-C. Wu, and Brigette Ronnett (Johns Hopkins University School of Medicine) for valuable discussions. Special thanks to Sean Patrick for her enthusiasm and inspiration. This work was supported by the Cancer Research Foundation of America (H.N.), the Richard TeLinde Foundation (R.K.), a pilot grant from the Johns Hopkins Oncology Center (R.R.), and generous donations to http://ovariancancer.jhmi.edu (Johns Hopkins University).
Abbreviations
- bFGF
basic fibroblast growth factor
- OSE
ovarian surface epithelium
- poly(HEMA)
poly 2-hydroxyethylmethacrylate
- RT-PCR
reverse-transcribed–PCR
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rosenthal A, Jacobs I. Semin Oncol. 1998;25:315–325. [PubMed] [Google Scholar]
- 2.Taylor D D, Gercel-Taylor C. Oncol Rep. 1998;5:1519–1524. doi: 10.3892/or.5.6.1519. [DOI] [PubMed] [Google Scholar]
- 3.Slamon D J, Godolphin W, Jones L A, Holt J A, Wong S G, Keith D E, Levin W J, Stuart S G, Udove J, Ullrich A, Press M F. Science. 1989;24:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y-T, Scanlan M J, Sahin U, Tureci O, Gure A O, Tsang S, Williamson B, Stockert E, Pfreundschuh M, Old L J. Proc Natl Acad Sci USA. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russo V, Dalerba P, Ricci A, Bonazzi C, Leone B E, Mangioni C, Allavera P, Bordignon C, Traversari C. Int J Cancer. 1996;67:457–460. doi: 10.1002/(SICI)1097-0215(19960729)67:3<457::AID-IJC24>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Sahin U, Tureci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Proc Natl Acad Sci USA. 1992;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scanlan M J, Chen Y-T, Williamson B, Gure A O, Stockert E, Gordan J D, Tureci O, Sahin U, Pfreundschuh M, Old L J. Int J Cancer. 1998;76:652–658. doi: 10.1002/(sici)1097-0215(19980529)76:5<652::aid-ijc7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 8.Scanlan M J, Gordan J D, Williamson B, Stockert E, Bander N H, Vongeneel V, Gure A O, Jager D, Jager E, Knuth A, Chen Y-T, Old L J. Int J Cancer. 1999;83:456–464. doi: 10.1002/(sici)1097-0215(19991112)83:4<456::aid-ijc4>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 9.Jager E, Jager D, Knuth A. Cancer Metastasis Rev. 1999;18:143–150. doi: 10.1023/a:1006220707618. [DOI] [PubMed] [Google Scholar]
- 10.Disis M L, Cheever M A. Curr Opin Immunol. 1996;8:637–642. doi: 10.1016/s0952-7915(96)80079-3. [DOI] [PubMed] [Google Scholar]
- 11.Maines-Bandiera S L, Kruk P A, Auersperg N. Am J Obstet Gynecol. 1992;167:729–735. doi: 10.1016/s0002-9378(11)91579-8. [DOI] [PubMed] [Google Scholar]
- 12.Horowitz A T, Treves A J, Voss R, Okon E, Fuks Z, Davidson L, Biran S. Oncology. 1985;42:332–337. doi: 10.1159/000226056. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton T C, Young R C, McKoy W M, Grotzinger K R, Green J A, Chu E W, Whang-Peng J, Rogan A M, Green W R, Ozols R F. Cancer Res. 1983;43:5379–5389. [PubMed] [Google Scholar]
- 14.Roden R B, Weissinger E M, Henderson D W, Booy F, Kirnbauer R, Mushinski J F, Lowy D R, Schiller J T. J Virol. 1994;68:7570–7574. doi: 10.1128/jvi.68.11.7570-7574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alami Y, Castronovo V, Belotti D, Flagiello D, Clausse N. Biochem Biophys Res Commun. 1999;257:738–745. doi: 10.1006/bbrc.1999.0516. [DOI] [PubMed] [Google Scholar]
- 16.Nakajima-Iijima S, Hamada H, Reddy P, Kakunaga T. Proc Natl Acad Sci USA. 1985;82:6133–6137. doi: 10.1073/pnas.82.18.6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chariot A, Princen F, Gielen J, Merville M-P, Franzoso G, Brown K, Siebenlist U, Bours V. J Biol Chem. 1999;274:5318–5325. doi: 10.1074/jbc.274.9.5318. [DOI] [PubMed] [Google Scholar]
- 18.Hough C D, Sherman-Baust C A, Pizer E S, Montz F J, Im D D, Rosenshein N B, Cho K R, Riggins G J, Morin P J. Cancer Res. 2000;60:6281–6287. [PubMed] [Google Scholar]
- 19.Allikmets R, Raskind W H, Hutchinson A, Schueck N D, Dean M, Koeller D M. Hum Mol Genet. 1999;8:743–749. doi: 10.1093/hmg/8.5.743. [DOI] [PubMed] [Google Scholar]
- 20.Simeone A, Mavilio F, Acampora D, Giampaolo A, Faiella A, Zappavigna V, D'Esposito M, Pannese M, Russo G, Boncinelli E, Peschle C. Proc Natl Acad Sci USA. 1987;84:4914–4918. doi: 10.1073/pnas.84.14.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mark M, Rijli F M, Chambon P. Pediatr Res. 1997;42:421–429. doi: 10.1203/00006450-199710000-00001. [DOI] [PubMed] [Google Scholar]
- 22.Cillo C, Faiella A, Cantile M, Boncinelli E. Exp Cell Res. 1999;248:1–9. doi: 10.1006/excr.1999.4451. [DOI] [PubMed] [Google Scholar]
- 23.Van Oostveen J W, Bijl J J, Raaphorst F H, Walboomers J J M, Meijer C J LM. Leukemia. 1999;13:1675–1690. doi: 10.1038/sj.leu.2401562. [DOI] [PubMed] [Google Scholar]
- 24.Cillo C. Invasion Metastasis. 1995;14:38–49. [PubMed] [Google Scholar]
- 25.Care A, Silvani A, Meccia E, Mattia G, Stoppacciaro A, Parmiani G, Peschle C, Colombo M. Mol Cell Biol. 1996;16:4842–4851. doi: 10.1128/mcb.16.9.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Care A, Silvani A, Meccia E, Mattia G, Peschle C, Colombo M. Oncogene. 1998;16:3285–3289. doi: 10.1038/sj.onc.1201875. [DOI] [PubMed] [Google Scholar]
- 27.Care A, Valtieri M, Mattia G, Mecchia E, Masella B, Luchetti L, Felicetti F, Colombo M P, Peschle C. Oncogene. 1999;18:1993–2001. doi: 10.1038/sj.onc.1202498. [DOI] [PubMed] [Google Scholar]
- 28.Fujimoto J, Ichigo S, Hori M, Hirose R, Sakaguchi H, Tamaya T. Eur J Gynecol Oncol. 1997;18:349–352. [PubMed] [Google Scholar]
- 29.Obermair A, Speiser P, Reisenberger K, Ullrich R, Czerwenka K, Kaider A, Zeillinger R, Miksche M. Cancer Lett. 1998;130:69–76. doi: 10.1016/s0304-3835(98)00119-0. [DOI] [PubMed] [Google Scholar]
- 30.DiBlasio A M, Carniti C, Vigano P, Vignali M. J Steroid Biochem Mol Biol. 1995;53:375–379. doi: 10.1016/0960-0760(95)00082-b. [DOI] [PubMed] [Google Scholar]
- 31.Florkiewicz R Z, Sommer A. Proc Natl Acad Sci USA. 1989;86:3978–3981. doi: 10.1073/pnas.86.11.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnaud E, Touriol C, Boutonnet C, Gensac M-C, Vagners S, Prats H, Prats A-C. Mol Cell Biol. 1999;19:505–514. doi: 10.1128/mcb.19.1.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raman V, Martensen S A, Reisman D, Evron E, Odenwald W F, Jaffee E, Marks J, Sukumar S. Nature (London) 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 34.Bromleigh V G, Freedman L P. Genes Dev. 2000;14:2581–2586. doi: 10.1101/gad.817100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bikfalvi A, Klein S, Pintucci G, Rifkin D B. Endocr Rev. 1997;18:26–45. doi: 10.1210/edrv.18.1.0292. [DOI] [PubMed] [Google Scholar]
- 36.Barton D P, Cai A, Wendt K, Young M, Gamero A, De Cesare S. Clin Cancer Res. 1997;3:1579–1586. [PubMed] [Google Scholar]
- 37.Maret A, Galy B, Arnaud E, Bayard F, Prats H. Cancer Res. 1995;55:5075–5079. [PubMed] [Google Scholar]
- 38.Berger W, Setinek U, Mohr T, Kindas-Mugge I, Vetterlein M, Dekan G, Eckersberg F, Caldas C, Micksche M. Int J Cancer. 1999;83:415–423. doi: 10.1002/(sici)1097-0215(19991029)83:3<415::aid-ijc19>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 39.Davis M G, Zhou M, Ali S, Coffin J D, Doetschman T, Dorn G W. J Mol Cell Cardiol. 1997;29:1061–1072. doi: 10.1006/jmcc.1997.0383. [DOI] [PubMed] [Google Scholar]
- 40.Pasumarthi K B S, Kardami E, Cattini P A. Circ Res. 1996;78:126–136. doi: 10.1161/01.res.78.1.126. [DOI] [PubMed] [Google Scholar]
- 41.Biro S, Yu Z-X, Fu Y-M, Smale G, Sasse J, Sanchez J, Ferrans V J, Casscells W. Circ Res. 1994;74:485–494. doi: 10.1161/01.res.74.3.485. [DOI] [PubMed] [Google Scholar]
- 42.Stachowiak M K, Moffet J, Joy A, Puchacz E, Florkiewicz R, Stachowiak E K. J Cell Biol. 1994;127:203–223. doi: 10.1083/jcb.127.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arese M, Chen Y, Florkiewicz R Z, Gualandris A, Shen B, Rifkin D B. Mol Cell Biol. 1999;10:1429–1444. doi: 10.1091/mbc.10.5.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maulbecker C, Gruss P. Cell Growth Differ. 1993;4:431–441. [PubMed] [Google Scholar]
- 45.Bostrom K, Tintut Y, Kao S C, Stanford W P, Demer L L. J Cell Biochem. 2000;78:210–221. doi: 10.1002/(sici)1097-4644(20000801)78:2<210::aid-jcb4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 46.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg A M, Sauvageau G. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bikfalvi A, Klein S, Pintucci G, Quatro N, Mignatti P, Rifkin D B. J Cell Biol. 1995;129:233–243. doi: 10.1083/jcb.129.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sahin U, Tureci O, Pfreundschuh M. Cur Opin Immunol. 1997;9:709–716. doi: 10.1016/s0952-7915(97)80053-2. [DOI] [PubMed] [Google Scholar]
- 49.Brass N, Racz A, Bauer C, Heckel D, Sybrecht G, Meese E. Blood. 1999;93:2158–2166. [PubMed] [Google Scholar]
- 50.Imoto I, Pimkhaokham A, Watanabe T, Saito-Ohara F, Soeda E, Inazawa J. Biochem Biophys Res Commun. 2000;276:264–270. doi: 10.1006/bbrc.2000.3449. [DOI] [PubMed] [Google Scholar]
- 51.Flagiello D, Poupan M-F, Cillo C, Dutrillaux B, Malfroy B. FEBS Lett. 1996;380:103–107. doi: 10.1016/0014-5793(96)00017-8. [DOI] [PubMed] [Google Scholar]
- 52.Friedmann Y, Daniel C A, Strickland P, Daniel C W. Cancer Res. 1994;54:5981–5985. [PubMed] [Google Scholar]
- 53.Derossi D, Calvet S, Trembleau A, Brunissen A, Chassaing G, Prochiantz A. J Biol Chem. 1996;271:18188–18193. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 54.Canevari S, Pupa S M, Menard S. Ann Oncol. 1996;7:227–232. doi: 10.1093/oxfordjournals.annonc.a010564. [DOI] [PubMed] [Google Scholar]
- 55.Soussi T. Cancer Res. 2000;60:1777–1788. [PubMed] [Google Scholar]