Abstract
Analysis of mouse brain gene expression, using strains that differ in alcohol consumption, provided a number of novel candidate genes that potentially regulate alcohol consumption. We selected six genes [beta-2-microglobulin (B2m), cathepsin S (Ctss), cathepsin F (Ctsf), interleukin 1 receptor antagonist (Il1rn), CD14 molecule (Cd14) and interleukin 6 (Il6)] for behavioral validation using null mutant mice. These genes are known to be important for immune responses but were not specifically linked to alcohol consumption by previous research. Null mutant mice were tested for ethanol intake in three tests: 24 hr two-bottle choice, limited access two-bottle choice and limited access to one bottle of ethanol. Ethanol consumption and preference were reduced in all the null mutant mice in the 24 hr two-bottle choice test, the test that was the basis for selection of these genes. No major differences were observed in consumption of saccharin in the null mutant mice. Deletion of B2m, Ctss, Il1rn, Cd14 and Il6 also reduced ethanol consumption in the limited access two bottle choice test for ethanol intake; with the Il1rn and Ctss null mutants showing reduced intake in all three tests (with some variation between males and females). These results provide the most compelling evidence to date that global gene expression analysis can identify novel genetic determinants of complex behavioral traits. Specifically, they suggest a novel role for neuroimmune signaling in regulation of alcohol consumption.
Keywords: alcohol, neuroimmune genes, proinflammatory, drinking, null mutant, mouse
INTRODUCTION
Although mechanisms of excessive alcohol consumption have been explored using brain gene expression profiling, a causal link between changes in expression of specific genes or gene networks and ethanol consumption has not been established (Spanagel, 2009). Even in the larger realm of behavioral neuroscience, there are only a few studies that have validated microarray targets with subsequent behavioral analysis (Kong et al., 2010; Flint and Mott, 2008). Other examples include the role of glyoxalase-1 and glutathione reductase 1 in anxiety (Hovatta et al., 2005), prodynorphin and FK506bp in morphine withdrawal (McClung et al., 2005) and Cdk5 in adaptation to cocaine (Bibb et al., 2001).
Recent microarray studies identified hundreds of genes and functional pathways that may be involved in regulation of alcohol drinking in mice (Mulligan et al., 2006; Tabakoff et al., 2008), rats (Kimpel et al., 2007) or humans (Liu et al., 2006, 2007; Flatscher-Bader et al., 2008), providing numerous candidates for functional (behavioral or biological) validation. The studies in mice and rats identified innate differences between alcohol-preferring and -nonpreferring animals based on voluntary ethanol intake in a two-bottle choice paradigm, while the human studies examined differences in gene expression between human alcoholics and controls. Because multiple genes were identified, prioritization of targets for functional validation is challenging. Our approach was to select candidates based on converging validity of genes and pathways commonly regulated across different related data sets, with the goal of discovering novel genes not previously implicated in the behavior. This informatics analysis led to the selection of six genes related to neuroimmune pathways.
There is some evidence linking brain neuroimmune or proinflammatory signaling to alcohol action. Long lasting increases in levels of several cytokines, including MCP-1 (Ccl2) in mouse brain, were found after chronic pretreatment with high doses of ethanol followed by injection of lipopolysaccharide (LPS) (Qin et al., 2008). Interestingly, a similar increase in MCP-1 (Ccl2) was found in the brain of human alcoholics (He and Crews, 2008) and deletion of Ccl2 or its receptor in mice decreased alcohol consumption (Blednov et al., 2005). Recently, Kong et al. (2010) found up-regulation of genes in the Toll and Imd signaling pathways in Drosophila exposed to ethanol, extending the link between alcohol and immune mediators to an invertebrate species.
Because different tests of alcohol consumption may have distinct genetic determinants (Blednov and Harris, 2008), we selected three tests for behavioral validation of the selected genes: 24 hour two-bottle choice (2 BC), limited access to one bottle (1B-Drinking in the Dark, DID) and limited access to two bottles (2B-DID). Preference for non-alcohol tastants was also evaluated. We found that all the null mutations reduced alcohol drinking, and the amount of the decrease was dependent on the the null mutant and, the specific drinking test and sex. These results suggest a key role for neuroimmune signaling in the regulation of alcohol consumption.
MATERIALS AND METHODS
Microarray data sets
Results from these studies (Mulligan et al., 2006; Kimpel et al., 2007; Liu et al., 2006) were obtained from either the publishers’ web sites or the authors. Flatscher-Bader et al. (2008) published only a small subset of the genes available on their microarray platform, therefore, the complete dataset was downloaded from Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/; GSE9058) and gene expression values in alcoholics and controls were compared using a two-tail t-test.
Animals
Null B2m (B6.129P2-B2mtm1Unc/J; Stock # 002087), Cd14 (B6.129S-cd14tm1Frm/J; Stock # 003726), Il1rn (B6.129S-IL1rntm!Dih/J; Stock # 004754) and Il6 (B6.129S2-IL6tm1kopf/J; Stock # 002650) mice were purchased from Jackson Laboratory (Bar Harbor, Maine). Breeding pairs of Ctss and Ctsf knockout mice were kindly provided by Dr. Chapman (University of California San Francisco). Il1rn and Ctsf null mice were maintained on original mixed C57Bl/6J × 129/SvJ genetic background by heterozygous breeding. Because B2m, Cd14, Il6 and Ctss knockout mice were backcrossed on C57Bl/6J genetic background more than 10 times, these four strains were maintained by homozygous breeding and C57Bl/6J mice from a colony maintained at the University of Texas at Austin (original breeders were purchased from Jackson Laboratories, Bar Harbor, ME) were used as the control or wild-type strain. For Il1rn and Ctsf null mice wild type littermates were used as a control. All of the mutant strains showed normal fertility and there was no evidence of any health problems. To minimize the potential variability from rearing conditions, all breeding colonies were contained in the same room of vivarium. Mice were group-housed four or five to a cage based on gender and litter. Food and water were available ad libitum. The vivarium was maintained on a 12:12 hr light:dark cycle with lights on at 7.00. The temperature and humidity of the room were kept constant. Behavioral testing began when the mice were at least two months old. All experiments were conducted in isolated behavioral testing rooms in the animal facility to avoid external distractions. All experiments were approved by the Institutional Animal Care and Use Committee.
Ethanol drinking – 24 hr two-bottle choice
The two-bottle choice protocol was carried out as previously described (Blednov et al., 2001). Tubes were weighed every other day. Food was available ad libitum, and mice were weighed every 4 days. Tube positions were changed daily to control for position preferences. Ethanol consumption (g/kg body weight/24 hr) was calculated for each mouse and values summarized for every concentrations of ethanol. The range of ethanol concentrations was varied from % to 18%.
Preference for non-alcohol tastants - 24 hr two-bottle choice
Mice were tested for saccharin and quinine consumption. One tube always contained water and the other contained the tastant solution. Mice were serially offered saccharin (0.033% and 0.066%) or quinine hemisulfate (0.03 mM and 0.06 mM) and intake was calculated. Each concentration was offered for 4 days, with bottle position changed every day. For each tastant, the low concentration was always presented first, followed by the higher concentration.
Ethanol drinking - limited access drinking in the dark phase (one-bottle DID)
This method for consumption of ethanol (20% solution) under conditions of limited access achieves pharmacologically significant levels of ethanol (Rhodes et al., 2005). Briefly, starting 3 hr after lights off, water bottles were replaced with bottles containing a 20% ethanol solution. The ethanol bottles remained in place for either 2 (first 3 days) or 4 hr (day 4) and then were replaced with water bottles. Except for this short period of time of ethanol drinking, mice had unlimited access to water. This procedure was repeated for 4 consecutive days. The ethanol bottles were weighed before placement and after removal from the experimental cage.
Ethanol drinking – limited access in the dark phase (two-bottle choice DID)
This was similar to the one-bottle DID test described above by Rhodes et al. (2005) except that two bottles (instead of one) containing either 20% ethanol or water were used (Blednov and Harris, 2008). The ethanol and water bottles remained in place for 3 hr. After their removal, mice had unlimited access to one bottle of water. Bottle positions during 3 hr access were changed daily to avoid potential side preferences. The ethanol and water bottles were weighed before placing and after removal from experimental cages.
Statistical Analysis
Data are reported as the mean ± S.E.M. The statistics software program GraphPad Prizm (Jandel Scientific, Costa Madre, CA) was used throughout. To evaluate differences between groups, analysis of variance (two-way ANOVA for repeated measurement) with post-hoc Bonfferoni corrections and Student’s t-tests were carried out. To evaluate changes in preference for ethanol in dependence on time, analysis of variance (one-way ANOVA for repeated measurement) was carried out.
RESULTS
Gene Selection
We began with a meta-analysis of gene expression changes associated with genetic predisposition to high alcohol consumption in mice (based on the 24 hr two-bottle choice test) (Mulligan et al., 2006). Initial examination of this data set showed statistically significantly regulation of functional pathways related to immune/inflammatory responses, including IL-1, IL-2, IL-6, NF-kB, Toll-like receptor and TNF receptor 1 signaling pathways. We then compared functional groups and individual genes identified in mice with expression studies in rats with genetic predisposition to high alcohol consumption (Kimpel et al., 2007) and in human alcoholics (Liu et al., 2006, 2007; Flatscher-Bader et al., 2008), Examination of data from these four gene expression studies showed that a number of genes differentially expressed in each study could be classified into a functional category broadly defined as “immune/inflammatory/defense/stress response” and we chose this pathway for our validation study. We applied five criteria for selection of genes from this functional category: 1) Statistical differences in gene expression (individual gene or over-represented pathway) related to predisposition to alcohol consumption in mice (Mulligan et al., 2006); 2) Statistical differences in gene expression in a rat or human study (Flatscher-Bader et al., 2008 Kimpel et al., 2007; Liu et al., 2006); 3) Availability of knockout mice; 4) Functional commonality and 5) Absence of publications linking the genes to alcohol consumption. This resulted in four genes that fulfilled all criteria and two genes that fulfilled three criteria and were found only in the Mulligan et al. (2006) data (Table 1). Thus, we evaluated null mutant mice for six genes: B2m (beta-2 microglobulin), Cd14 (cluster of differentiation 14 or CD14 antigen), Il1rn (interleukin 1 receptor antagonist, also known as IL-1ra and IRAP), Il6 (interleukin 6), Ctss (cathepsin S) and Ctsf (cathepsin F).
Table 1.
“Immune” candidate genes for functional validation.
| Gene Symbol: Gene Name | Chromosome (cM) | Evidence | Reference |
|---|---|---|---|
|
B2m: Beta-2-microglobulin |
2 (69.0) |
B2m is upregulated in brain of alcohol-preferring mice B2m is downregulated in brain of alcohol-preferring rats B2m is upregulated in VTA of human alcoholics |
Mulligan et al., 2006 Kimpel et al., 2007 Flatscher-Bader et al., 2008 |
| Il1rn: Interleukin 1 receptor antagonist | 2 (10.0) |
Il1rn is downregulated in brain of alcohol-preferring mice and is a part of over-represented “Signal Transduction Through IL1R” pathway Interleukin 1 receptor (IL1R1) gene is upregulated in VTA of human alcoholics |
Mulligan et al., 2006 Flatscher-Bader et al., 2008 |
|
Il6: Interleukin 6 |
5 (17.0) | “IL6 Signaling Pathway” is over-represented in alcohol-preferring mice Interleukin 6 receptor (Il6r) gene is upregulated in brain of alcohol-preferring rats |
Mulligan et al., 2006 Kimpel et al., 2007 |
|
Cd14: CD14 antigen |
18 (31.0) | Cd14 is downregulated in brain of alcohol-preferring mice and is a part of over-represented “Toll-Like Receptor Pathway” | Mulligan et al., 2006 |
|
Ctss: cathepsin S |
3 (42.7) |
Ctss is upregulated in brain of alcohol-preferring mice Ctss is downregulated in Frontal Cortex of human alcoholics |
Mulligan et al., 2006 Liu et al., 2006 |
|
Ctsf: cathepsin F |
19 (Syntenic) | Ctsf is upregulated in brain of alcohol-preferring mice | Mulligan et al., 2006 |
cM – centiMorgans.
One question that arises is whether we should expect a relationship between gene expression changes in human alcoholics and high drinking mice. An important consideration is that the human cases used in the gene expression studies were chosen for levels of alcohol consumption, not for alcohol dependence (Liu et al. 2006). In addition, we do not know which rodent behaviors or tests are consilent with human alcoholism (Crabbe, 2010) but there is evidence that chromosomal regions important for human alcohol dependence are syntenic with regions that influence alcohol consumption in mice (Ehlers et al., 2010).
Ethanol consumption in 24-hr two-bottle choice paradigm
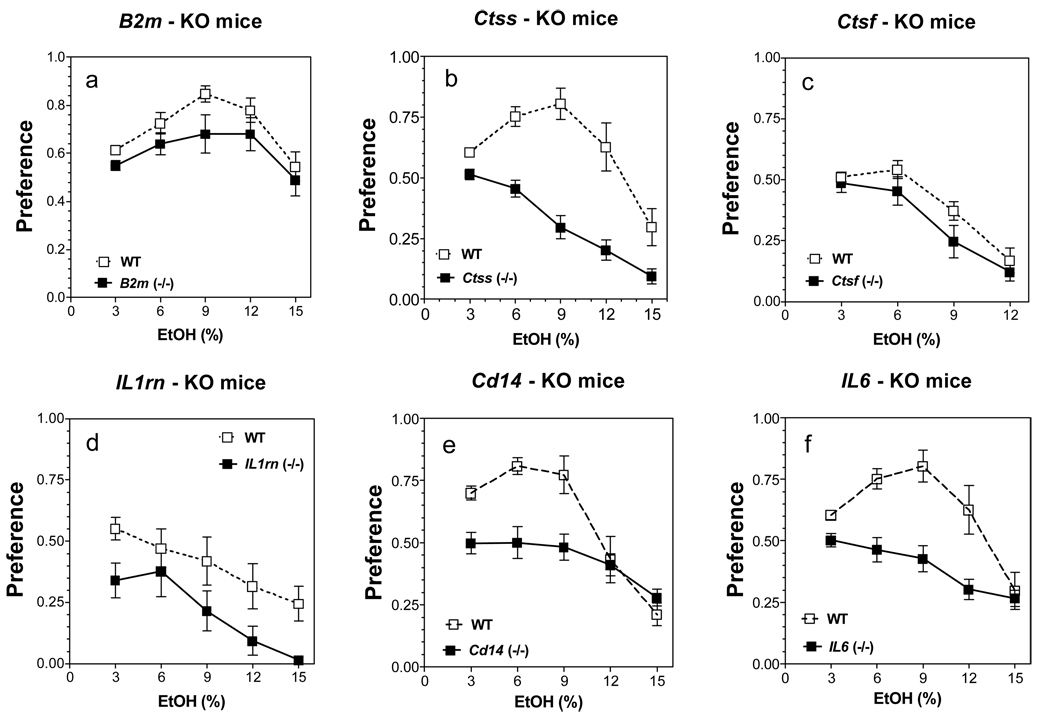
In a two-bottle free-choice paradigm in which mice could drink either water or a series of increasing ethanol concentrations, the preference for ethanol was significantly reduced in all six mutant strains of male mice as shown in Fig. 1 [main effect of genotype: F(1,80) = 7.4, p<0.01 for B2m mice; F(1,90) = 81, p<0.001 for Ctss mice; F(1,64) = 5.2, p<0.05 for Ctsf mice; F(1,58) = 15, p<0.001 for Il1rn mice; F(1,90) = 19, p<0.001 for Cd14 mice; F(1,90) = 41, p<0.001 for Il6 mice]. The amount of ethanol consumed was also significantly reduced in B2m, Ctss, Ctsf, Il1rn and Il6 knockout male mice. Total fluid intake was reduced in B2m and Ctss null male mice and increased in Cd14 knockout male mice (for complete results see Supplemental Fig. 1 and Fig. 2; for detailed statistics see Supplemental Table 1).
Figure 1. Lack of B2m, Ctss, Ctsf, Il1rn, Il6 or Cd14 reduces ethanol preference in two-bottle choice test in male mice.
A. B2m knockout mice, (p<0.01, effect of genotype). B. Ctss knockout mice, (p<0.001, effect of genotype). C. Ctsf knockout mice. (p<0.05, effect of genotype). D. Il1rn knockout mice, (p<0.001, effect of genotype). E. Cd14 knockout mice, (p<0.001, effect of genotype). F. Il6 knockout mice, (p<0.001, effect of genotype). n = 9 – 10 for all groups.
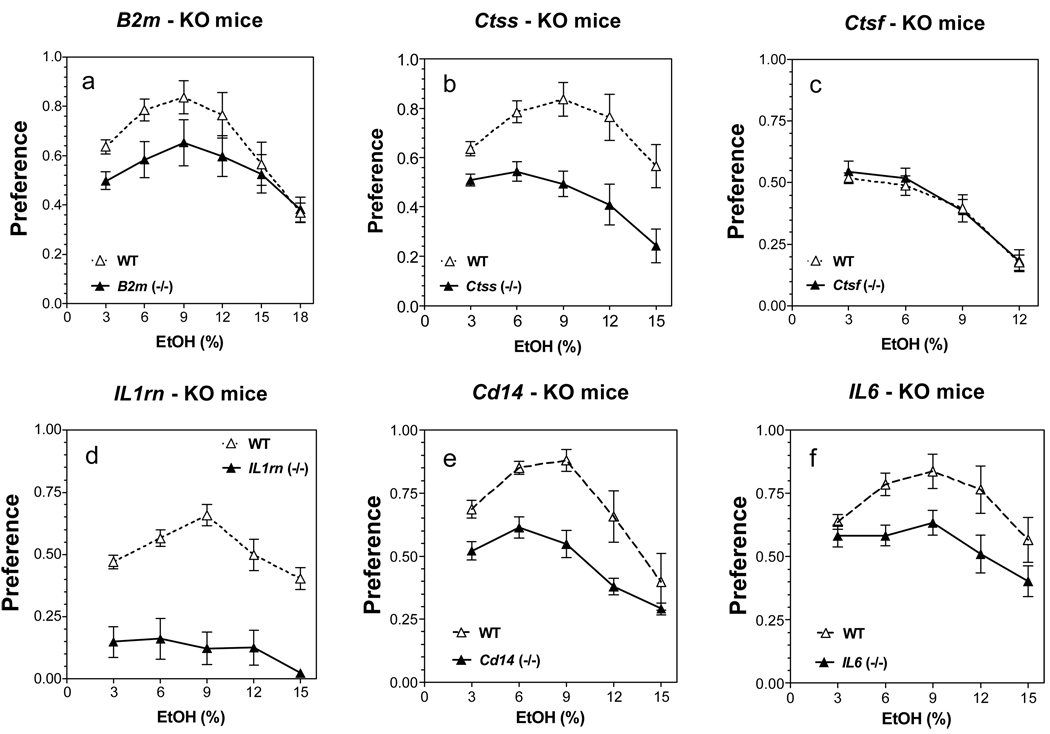
Similar results were obtained in female mice. Preference for ethanol was significantly reduced in five of six mutant strains of female mice as shown in Fig. 2 [main effect of genotype: F(1,96) = 9.2, p<0.01 for B2m mice; F(1,85) = 47, p<0.001 for Ctss mice; F(1,63) = 130, p<0.001 for Il1rn mice; F(1,80) = 40, p<0.001 for Cd14 mice; F(1,90) = 20, p<0.001 for Il6 mice]. The amount of ethanol consumed was also significantly reduced in these same five strains of knockout female mice. However, no differences in preference for ethanol or amount of ethanol consumed were found between wild type and Ctsf female mice. Total fluid intake was reduced in B2m and Ctss null female mice (as it was for the male mice in these two strains) and increased in Cd14 and Il6 knockout female mice (for complete results see Supplemental Fig. 3 and Fig. 4; for detailed statistics see Supplemental Table 2).
Figure 2. Lack of B2m, Ctss, Il1rn, Il6 or Cd14 reduces ethanol preference in two-bottle choice test in female mice.
A. B2m knockout mice, (p<0.01, effect of genotype). B. Ctss knockout mice, (p<0.001, effect of genotype). C. Ctsf knockout mice, (p>0.05, effect of genotype). D. Il1rn knockout mice, (p<0.001, effect of genotype). E. Cd14 knockout mice, (p<0.001, effect of genotype). F. Il6 knockout mice, (p<0.001, effect of genotype). n = 9 – 10 for all groups.
Preference for non-alcohol tastants
Altered alcohol consumption in mutant mice can be related to changes in taste (Blednov et al., 2008). We studied consumption of saccharin and quinine in the mutant mice in a 24 hr two bottle choice test to determine if taste could account for the reduced alcohol consumption. Of the six knockout strains, only B2m null male mice showed a slight change (reduction) in preference for saccharin. Total fluid intake was reduced in B2m and Ctss null male mice and increased in Cd14 and Il6 knockout male mice. Only Ctss null male mice showed slightly reduced avoidance (increased preference) for quinine. Ctss null male mice also demonstrated reduced total fluid intake whereas in Cd14 knockout males, total fluid intake was increased (for complete results see Supplemental Table 3; for detailed statistics see Supplemental Table 5).
For females, Ctss and Il1rn null mice showed slightly reduced preference for saccharin whereas Ctsf knockout mice demonstrated slightly increased preference for the sweet taste. Total fluid intake was reduced in Ctss and Il1rn null female mice and increased in Cd14 knockout females. Similar to male mice, only Ctss null female mice showed reduced avoidance (increased preference) for quinine. Total fluid intake was reduced in B2m, Ctss and Il1rn null females and increased in Cd14 and Il6 knockout female mice (for complete results see Supplemental Table 4; for detailed statistics see Supplemental Table 6).
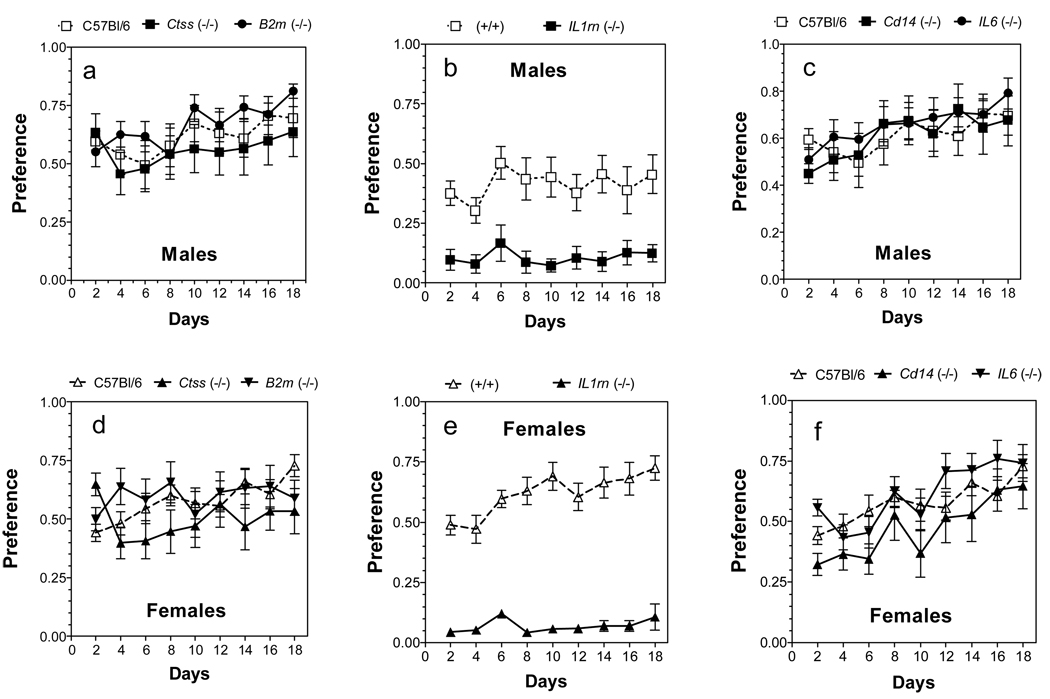
Ethanol consumption in limited access in the dark phase (two-bottle choice DID)
In a two-bottle free-choice paradigm with limited access to ethanol, the preference for ethanol was significantly reduced only in Il1rn null male mice [main effect of genotype: F(1,135) = 85, p<0.001] (Fig. 3b). Amount of ethanol consumed was significantly reduced in Ctss and Il1rn mutant male mice. Total fluid intake was reduced in B2m, Ctss and Il6 null male mice and increased in Cd14 and Il1rn knockout male mice (for complete results see Supplemental Fig. 5; for detailed statistics see Supplemental Table 7).
Figure 3. Effect of deletion of B2m, Ctss, Il1rn, Cd14 or Il6 on preference for ethanol in two-bottle DID procedure (2B-DID).
Males: A. B2m and Ctss knockout mice. B. Il1rn knockout mice. (p<0.001, effect of genotype). C. Cd14 and Il6 knockout mice. Females: D. B2m and Ctss knockout mice. (p<0.05, effect of genotype for the Ctss knockout mice) E. Il1rn knockout mice. (p<0.001, effect of genotype) F. Cd14 and Il6 knockout mice. (p<0.01, effect of genotype for the Cd14 knockout mice) n = 10 – 14 for all groups. Ethanol was used in concentration 20%.
In female mice, the amount of ethanol consumed was significantly reduced in all six null strains. Ctss, Il1rn and CD14 null strains showed reduced preference for ethanol in the 2B-DID test [main effect of genotype: F(1,162) = 5.7, p<0.05 for Ctss mice; F(1,126) = 490, p<0.001 for Il1rn mice; F(1,162) = 9.1, p<0.01 for Cd14 mice] (Fig. 3 d,e,f). Total fluid intake was significantly reduced in B2m, Ctss and Il6 null female mice and increased in Il1rn and Cd14 knockout female mice (for complete results see Supplemental Fig. 6; for detailed statistics see Supplemental Table 8). Some strains appeared to increase their alcohol consumption over days, but this was not a consistent finding for either the wild-type or mutant mice (statistics in Supplemental Table 9). Because Ctsf null mice of both sexes showed very low preference for ethanol in the 24 hr two-bottle choice test, these mice were not tested in either 2B-DID or 1B-DID.
Ethanol consumption in limited access in the dark phase (one-bottle DID)
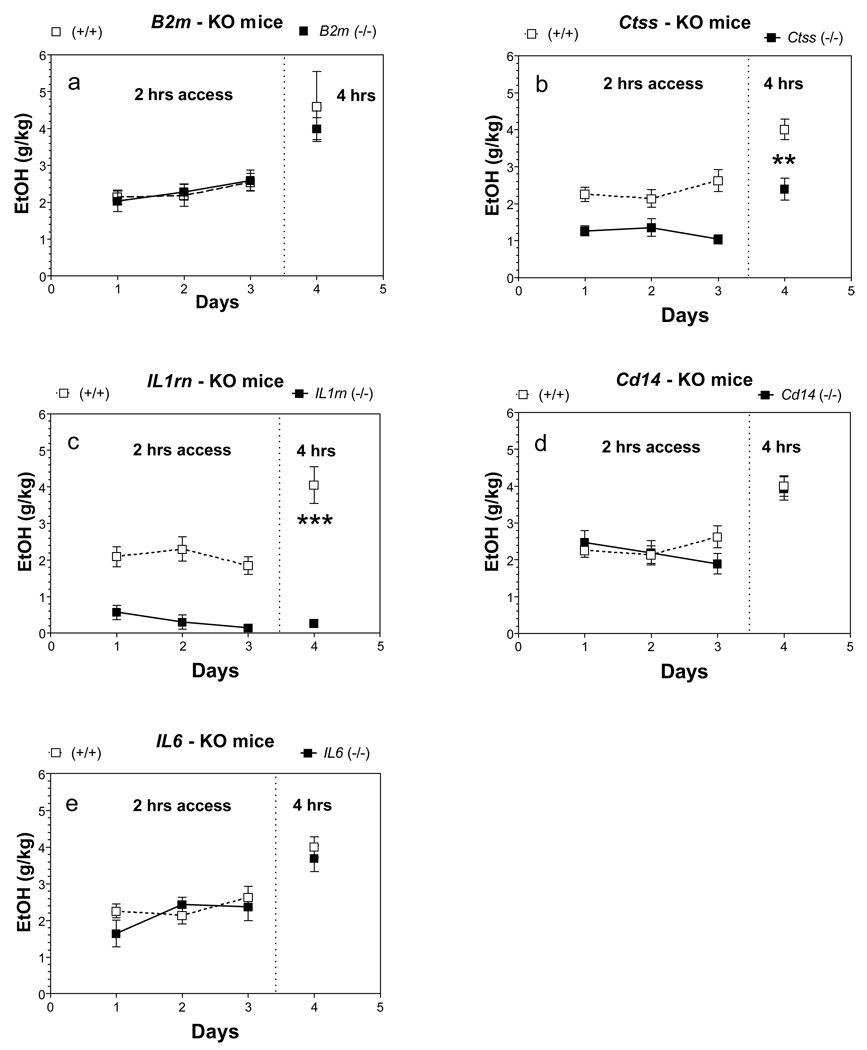
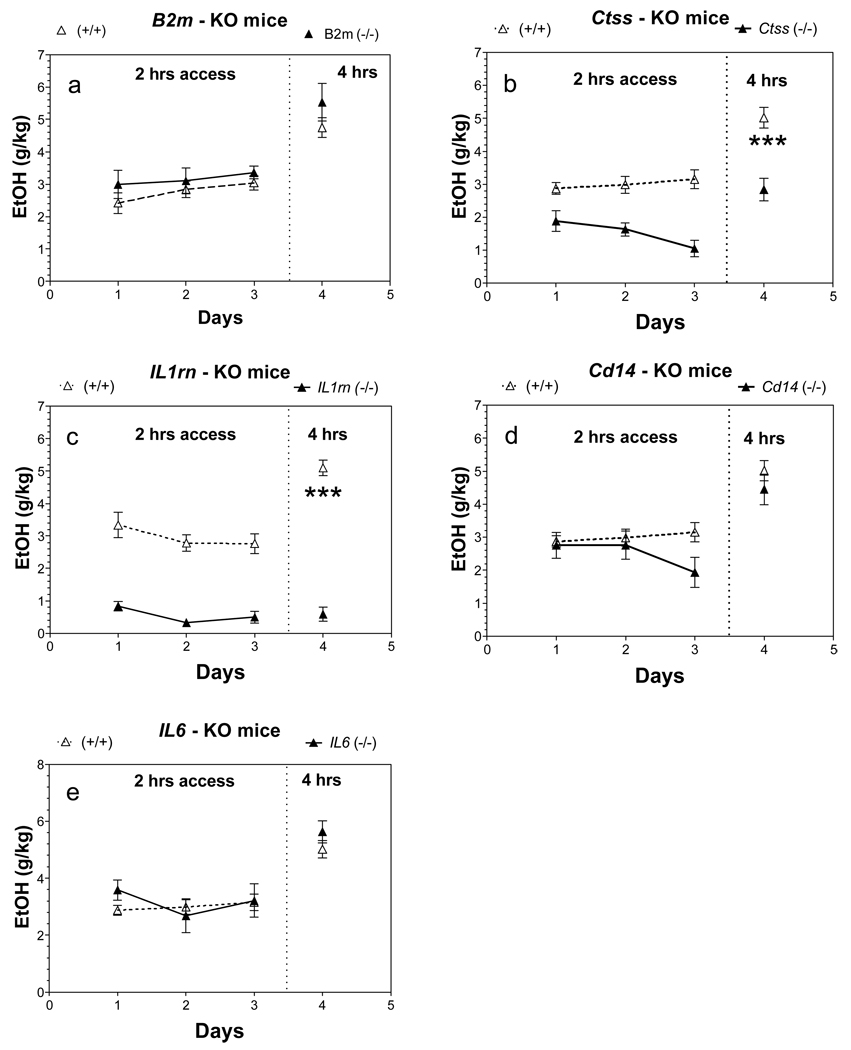
Only Ctss and Il1rn null male mice showed reduced ethanol intake during three consecutive days with 2 hr access to ethanol [main effect of genotype: F(1,39) = 43, p<0.001 for Ctss mice; F(1,36) = 71, p<0.001 for Il1rn mice] (Fig. 4 b,c). During 4 hr access to ethanol, male mice of both mutant strains also demonstrated a large reduction in amount of ethanol consumed.
Figure 4. Effect of deletion of B2m, Ctss, Il1rn, Cd14 or Il6 on preference for ethanol in one bottle DID procedure (1B-DID) in male mice.
A. B2m knockout mice. B. Ctss knockout mice, (p<0.001, effect of genotype for the period with 2 hrs access). C. Il1rn knockout mice, (p<0.001, effect of genotype for the period with 2hrs access). D. Cd14 knockout mice. E. Il6 knockout mice. ** - p<0.01, *** - p<0.001 – statistically significant differences between mutant and correspondent wild type mice for the period with 4 hrs access (Student’s t-test). n = 7 – 9 for all groups. Ethanol was used in concentration 20%.
As seen in males, only Ctss and Il1rn null females showed reduced ethanol intake during three consecutive days with 2 hr access to ethanol [main effect of genotype: F(1,39) = 52, p<0.001 for Ctss mice; F(1,36) = 111, p<0.001 for Il1rn mice] (Fig. 5 b,c). During 4 hr access to ethanol, female mice in both mutant strains also demonstrated reduced consumption of ethanol. Because only one-bottle is offered at a time, ethanol preference can't be measured in this test.
Figure 5. Effect of deletion of B2m, Ctss, Il1rn, Cd14 or Il6 on preference for ethanol in one bottle DID procedure (1B-DID) in female mice.
A. B2m knockout mice. B. Ctss knockout mice, (p<0.001, effect of genotype for the period with 2hrs access). C. Il1rn knockout mice, (p<0.001, effect of genotype for the period with 2hrs access). D. Cd14 knockout mice. E. Il6 knockout mice. *** - p<0.001 – statistically significant differences between mutant and correspondent wild type mice for the period with 4 hrs access (Student’s t-test). n = 6 – 8 for all groups. Ethanol was used in concentration 20%.
DISCUSSION
Remarkably, all six genes originally selected from large and diverse gene expression datasets were associated with changes in alcohol consumption in mice. Apart from the gene expression studies, none of these genes had previously been implicated in alcohol actions and most are ‘immune/inflammatory’ genes with limited visibility in neurobiology. These findings support the validity of our strategy for selection of gene targets from microarray data using convergent statistical, informatics and functional approaches. A unique aspect of the present study is the behavioral validation for use of multiple microarray datasets to predict targets important for alcohol action and indeed the actual validation of this approach for any behavioral trait. It is important to note that the most consistent and robust changes occurred in the 24 hr two-bottle choice test, the test that was the basis for selection of these genes. The 24 hr two-bottle choice test showed reduced alcohol preference and consumption in most male and female null mutant mice. However, in limited access 1B-DID tests, only deletion of the Il1rn and Ctss genes decreased ethanol consumption, indicating differences in the genetic control of these behaviors (Supplemental Table 10).
The high success rate (6 out of 6) of our validation raises the question of whether a global KO of any gene with a significant brain function will result in behavioral disturbance that would to, at least, some degree affect alcohol consumption. This is not consistent with the literature as a review of 93 null mutants tested for alcohol responses noted that only about 1/3 showed a decrease in alcohol consumption (Crabbe et al., 2006), A more specific test of this hypothesis is whether negative microarray findings would be followed by negative behavioral results. Our data and results of others indicate that this is indeed the case. For example, genetic deletion of some key brain receptors, such as the beta2 subunit of GABAA receptors or metabotropic glutamate receptors (mGluR4, mGluR5) did not change alcohol consumption (Blednov et al., 2003; Blednov et al., 2004; Blednov and Harris, 2008;) and none of these genes passed the statistical threshold of significant regulation in Mulligan et al. (2006). On the other hand, genetic differences in GABAA alpha2 subunit expression were found (Mulligan et al., 2006) and these differences corresponded to the altered drinking in Gabra2 null mutants (Boehm et al., 2004).
All six genes selected for validation have important roles in peripheral immune and inflammatory signaling and global deletion of these genes may affect multiple tissues. For example, mice deficient in β2-microglobulin fail to express major histocompatibility complex (MHC) class I products (Rothenberg and Voland, 1996), and Cd14-deficient mice are resistant to LPS (Schütt, 1999). Ctss is a key enzyme controlling MHC class II-mediated antigen presentation by epithelial cells in vivo (Beers et al., 2005), and inflammatory responses are compromised in Il6-deficient mice (Kopf et al., 1994). In addition, we found that deletion of related proinflammatory mediators (the chemokines Ccl1 and Ccl2 or the chemokine receptor Ccr2) reduced alcohol preference and intake and also reduced the motivational effect of ethanol in a conditioned taste aversion paradigm (Blednov et al., 2005). Thus, we now have nine genes with roles in immune and inflammatory processes that regulate alcohol consumption. However, as discussed below, these genes should not be considered as exclusively ‘immune’ or ‘inflammatory’ as their role in normal brain signaling is an area of intense investigation. This raises the possibility that the altered alcohol consumption produced by deletion of these ‘immune’ genes is due to changes that occur in brain. Studies of some of these immune mediators in central nervous system function are just beginning, but roles for each of these genes in brain are emerging.
In humans, alcohol abuse is associated with disruption of immune defenses against infections (Nelson and Kolls, 2002) and increases in serum proinflammatory cytokines (McClain and Cohen, 1989; McClain et al., 1999). It is not known if voluntary alcohol consumption in mice changes these immune and inflammatory processes, and it is possible that alcohol's action on these pathways is altered in the null mutants.
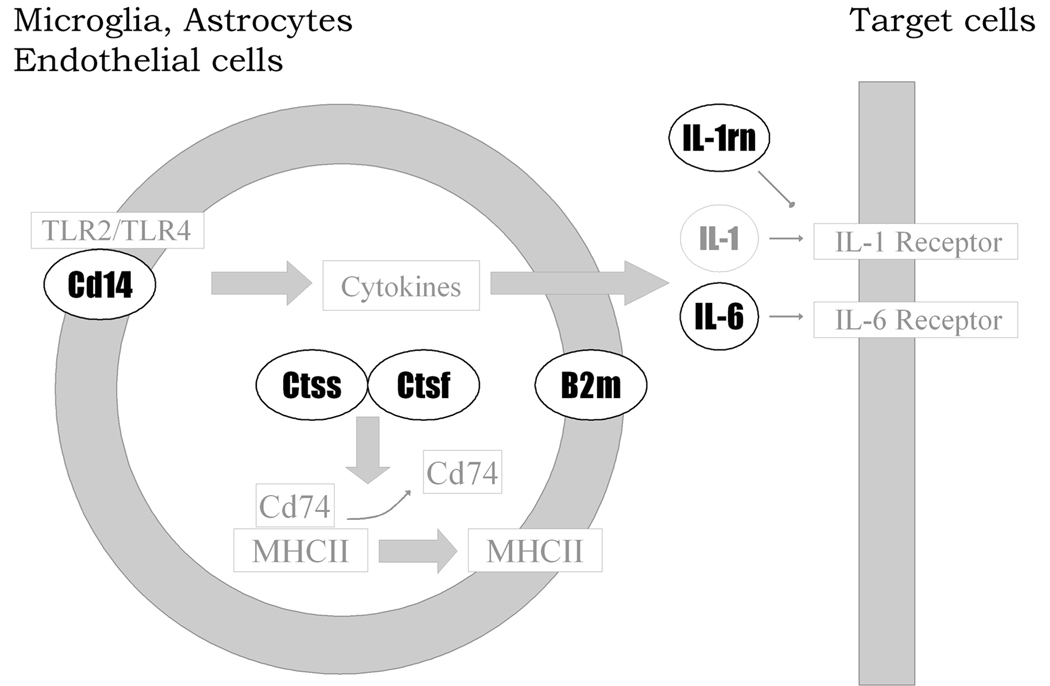
The immune proteins have important roles in neuronal plasticity and some are found in neurons as well as microglia or astrocytes (Boulanger, 2009; Rivest, 2003), and we will briefly review the role of the six proteins tested in this study in brain function (shown schematically in Fig. 6). CD14 is critical for the function of two Toll-Like receptors, TLR 2 and 4, which are expressed by microglia as well as astrocytes and neurons (Bsibsi et al., 2002; van Noort and Bsibsi, 2009). Cathepsins, including Ctsf and Ctss, are found in microglia where they are important for the function of MHC class II molecules and are also secreted from microglia and may be involved in cytokine processing (Nakanishi, 2003; Wang et al., 1998). B2m, which is part of the MHC class I complex, is found in many neurons, including dopaminergic neurons, and is critical for neuronal plasticity, including long-term potentiation (Corriveau et al., 1998; Huh et al., 2000; Linda et al., 1999). IL-1 is one of the most prevalent and important cytokines and its actions are regulated by an endogenous antagonist, IL-1rn. Both are released from microglia and glia and act on receptors on neurons and glia (Molina-Holgado et al., 2003; Tsakiri et al., 2008), and elevated levels of IL-1rn interfere with memory consolidation (Spulber et al., 2009). IL-1 also stimulates the release of IL-6 from neurons and glia (Tsakiri et al., 2008). IL-6 acts on neuronal receptors to suppress inhibitory transmission by reducing GABA-A and glycine receptor function (Kawasaki et al., 2008). Deletion of Il1rn produced some of the most pronounced changes in drinking in both tests and it is of interest to note that activation of cannabinoid (CB1 and CB2) receptors mediate release of IL-1rn and that IL-1rn disrupts BDNF-ERK1/2 signaling (Molina-Hidalgo et al, 2003: Spulber et al., 2009). Thus, deletion of Il1rn should reduce some effects of endocannabioids and enhance BDNF signaling. Importantly, treatments that reduce CB1 receptor signaling or enhance BDNF activity reduce alcohol consumption (Wang et al., 2003; Jeanblanc et al., 2009), suggesting mechanisms by which deletion of Il1rn may reduce drinking. Thus, the six genes and gene products manipulated in our study are critically involved in signaling among microglia, glia and neurons (Fig. 6) and most of them have been implicated in synaptic plasticity.
Figure 6.
Schematic diagram linking genes that were targeted for functional validation using null mutant mice. The diagram is based on existing pathways from KEGG (http://www.genome.jp/kegg/) and BioCarta (http://www.biocarta.com/) as well as published literature (see Discussion for references). Cd14 is a key component of the Toll-like receptor pathway resulting in production of proinflammatory cytokines, such as IL-1 and IL-6. IL1RN is an antagonist for IL-1 receptor. Cathepsins S and F are cysteine proteases that degrade the invariant polypeptide of major histocompatibility complex, class II (Cd74) and promote the presentation of antigens by antigen-presenting cells. This function is served by microglia and astrocytes in brain. B2m also plays a role in antigen presentation.
It is somewhat surprising that deletion of each of these genes caused either a decrease or no change (depending on the test) in drinking and none of the deletions increased drinking. This is particularly true for deletion of Il1rn which should cause an increase in IL-1 signaling, whereas the other deletions might be expected to decrease cytokine activity. However, given the complexity of this signaling system, this is likely an incorrect over-simplification. It is important not to assume that levels of mRNA and protein are always positively correlated and that the direction of mRNA changes reflects the direction of functional changes. Although in many cases changes in levels of mRNA predict changes in protein, no correlation or even a negative correlation has been observed because transcription, translation and mRNA and protein degradation are intrinsically dynamic processes regulated by independent mechanisms and time constraints (Lewandowski and Small; 2005; Jayapal et al., 2008). For example, activation of microglia or macrophages increases the secretion of cathepsin S, but decreases the level of the transcript (Liuzzo et al., 1999). The important point is the the current experiments validated previous microarray findings by confirming a phenomenological link between molecular and behavioral changes. An understanding of the mechanisms responsible for the decreased alcohol consumption will require studies of neuronal or cellular function.
Our results demonstrate the validity of convergent genomic analyses to nominate genes that might be important for genetic regulation of complex behaviors. The subtlety of such regulation is emphasized by the observation that although three different tests of alcohol consumption show effects of deletion of some of the genes, only the 24 hr two-bottle choice test, which was the primary basis of gene selection, showed effects of all six genes. In addition, our findings indicate a novel and unexpected role for proinflammatory signaling in regulation of alcohol consumption. Although the brain proinflammatory system has most often been studied in relation to neuropathology, emerging evidence shows that it represents a signaling network that regulates normal behavior (Boulanger, 2009; Rihel et al., 2010). Our results raise the possibility that rewarding (or aversive) effects of alcohol may be determined by cytokine signaling. This may be related to the observation that activation of cytokine signaling (e.g., by lipopolysaccharide) produces a ‘sickness’ behavior that includes decreases in reward function (measured by intracranial self-stimulation) (Borowski et al., 1998; Henry et al., 2008) similar to that observed in alcoholism. A possible neurochemical mechanism for neuroimmune regulation of drinking behavior is activation of glutamatergic transmission as Crews et al. (2006) proposed that cytokines can induce a hyperglutamatergic state and Spanagel et al. (2005) proposed that a hyperglutamate state drives drinking behavior. Reversal of the dysregulation of glutamatergic and reward function produced by alcohol and other drugs by neuroimmune signals may provide new opportunities for pharmacotherapy of alcoholism and other addictions.
Several caveats apply to studies of null mutant mice, including the fact that most gene-targeted mice, including those used in this study, are developed on a mixed genetic background, usually C57Bl/6J and a 129 substrain and these parental strains differ in responses to ethanol and many other drugs (Phillips et al., 1999). Four of the lines of knockout mice used in this study were backcrossed on the C57Bl/6J strain for many generations, reducing the potential confounding effects of the mixed genetic background, but genetic background may have a role in the phenotypes of Ctsf (−/−) and Il1rn (−/−) which were not backcrossed. Another concern is that homologous recombination places additional "hitchhiking" (e.g., 129 strain) DNA near the disrupted gene (Crusio, 1996; Gerlai, 1996). The complete replacement of 129 alleles is limited by the probability of recombination; even after 12 backcrosses a segment of 129-derived chromosome (about 16 centiMorgans) remains surrounding the mutated gene (Festing, 1992) and these neighboring genes can potentially influence the phenotype of the null mice. To distinguish the role of null genes and "hitchhiking" genes, it is necessary to determine the overlap between the known QTLs for alcohol consumption and chromosomal regions of derived from the parental strains. The behavioral QTLs for alcohol preference were compiled by Saba et al. (2006) and are on Chr2 (9–49 cM and 107 cM), Chr3 (48–83 cM), Chr4 (59–75 cM), Chr5 (44 cM) and Chr9 (9–61 cM). For genes tested in our study, Cd14 and Ctsf are not on any of these chromosomes (Table 1). Il6 is located on Chr5 but is far away from the suggested QTL. B2m is located on Chr2 and Ctss is located on Chr3, but out of suggested QTL regions for these chromosomes. One gene – IL1rn - is located within a suggested QTL region on Chr2. Thus, the strong reduction of drinking seen in IL1rn mutants may be result of a combination of gene deletion and "hitchhiking" genes derived from the 129 strain.
In summary, these findings provide links between a group of genes regulating immunity and voluntary alcohol drinking. This initial validation allows prioritization of targets for the future mechanistic studies of proimmune signaling in alcohol reward.
Supplementary Material
Acknowledgments
This study was supported by NIH grants for the Integrated Neuroscience Initiative on Alcoholism (INIA) (AA13518, AA13520, AA012404 and AA06399). The authors would like to thank Jody Mayfield for critical editorial assistance and Virginia Bleck and Danielle Walker for excellent technical work.
Footnotes
Authors Contribution. YAB, and RAH were responsible for the study concept and design. YAB, IP and CG contributed to the acquisition of data. YAB, IP and RAH assisted with data analysis and interpretation of findings. YAB drafted the manuscript. IP, SB, GFK and RAH provided critical revision of the manuscript for important intellectual content. YAB, RAH, GFK and SB obtained funding for the project. RAH supervised the study. All authors critically reviewed content and approved final version submitted for publication.
REFERENCES
- Beers C, Burich A, Kleijmeer MJ, Griffith JM, Wong P, Rudensky AY. Cathepsin S controls MHC class II-mediated antigen presentation by epithelial cells in vivo. J Immunol. 2005;174:1205–1212. doi: 10.4049/jimmunol.174.3.1205. [DOI] [PubMed] [Google Scholar]
- Bibb JA, Chen J, Taylor JR, Svenningsson P, Nishi A, Snyder GL, Yan Z, Sagawa ZK, Ouimet CC, Nairn AC, Nestler EJ, Greengard P. Effects of chronic exposure to cocaine are regulated by the neuronal protein Cdk5. Nature. 2001;410:376–380. doi: 10.1038/35066591. [DOI] [PubMed] [Google Scholar]
- Blednov YA, Bergeson SE, Walker D, Ferreira VM, Kuziel WA, Harris RA. Perturbation of chemokine networks by gene deletion alters the reinforcing actions of ethanol. Behav Brain Res. 2005;165:110–125. doi: 10.1016/j.bbr.2005.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Harris RA. Metabotropic glutamate receptor 5 (mGluR5) regulation of ethanol sedation, dependence and consumption: relationship to acamprosate actions. Int J Neuropsychopharmacol. 2008;11:775–793. doi: 10.1017/S1461145708008584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Stoffel M, Chang SR, Harris RA. Potassium channels as targets for ethanol: studies of G-protein-coupled inwardly rectifying potassium channel 2 (GIRK2) null mutant mice. J Pharmacol Exp Ther. 2001;298:521–530. [PubMed] [Google Scholar]
- Blednov YA, Walker D, Alva H, Creech K, Findlay G, Harris RA. GABAA receptor alpha 1 and beta 2 subunit null mutant mice: behavioral responses to ethanol. J Pharmacol Exp Ther. 2003;305:854–863. doi: 10.1124/jpet.103.049478. [DOI] [PubMed] [Google Scholar]
- Blednov YA, Walker D, Martinez M, Levine M, Damak S, Margolskee RF. Perception of sweet taste is important for voluntary alcohol consumption in mice. Genes Brain Behav. 2008;7:1–13. doi: 10.1111/j.1601-183X.2007.00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blednov YA, Walker D, Osterndorf-Kahanek E, Harris RA. Mice lacking metabotropic glutamate receptor 4 do not show the motor stimulatory effect of ethanol. Alcohol. 2004;34:251–259. doi: 10.1016/j.alcohol.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Boehm SL, 2nd, Ponomarev I, Jennings AW, Whiting PJ, Rosahl TW, Garrett EM, Blednov YA, Harris RA. gamma-Aminobutyric acid A receptor subunit mutant mice: new perspectives on alcohol actions. Biochem Pharmacol. 2004;68:1581–1602. doi: 10.1016/j.bcp.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Borowski T, Kokkinidis L, Merali Z, Anisman H. Lipopolysaccharide, central in vivo biogenic amine variations, and anhedonia. Neuroreport. 1998;9:3797–3802. doi: 10.1097/00001756-199812010-00006. [DOI] [PubMed] [Google Scholar]
- Boulanger LM. Immune proteins in brain development and synaptic plasticity. Neuron. 2009;64:93–109. doi: 10.1016/j.neuron.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Bsibsi M, Ravid R, Gveric D, van Noort JM. Broad expression of Toll-like receptors in the human central nervous system. J Neuropathol Exp Neurol. 2002;61:1013–1021. doi: 10.1093/jnen/61.11.1013. [DOI] [PubMed] [Google Scholar]
- Crabbe JC. Consilience of rodent and human phenotypes relevant for alcohol dependence. Addict Biol. 2010;15:103–108. doi: 10.1111/j.1369-1600.2009.00188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF. Alcohol-related genes: contributions from studies with genetically engineered mice. Addict Biol. 2006;11:195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- Crews FT, Bechara R, Brown LA, Guidot DM, Mandrekar P, Oak S, Qin L, Szabo G, Wheeler M, Zou J. Cytokines and alcohol. Alcohol Clin Exp Res. 2006;30:720–730. doi: 10.1111/j.1530-0277.2006.00084.x. [DOI] [PubMed] [Google Scholar]
- Crusio WE. Gene-targeting studies: new methods, old problems. Trends Neurosci. 1996;19:186–187. doi: 10.1016/s0166-2236(96)20023-2. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Walter NA, Dick DM, Buck KJ, Crabbe JC. A comparison of selected quantitative trait loci associated with alcohol use phenotypes in humans and mouse models. Addict Biol. 2010;15:185–199. doi: 10.1111/j.1369-1600.2009.00195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festing MFW. From character to gene. Some strategies for identifying single genes controlling behavioral characters. In: Goldowiz D, Wahlsten D, Wimer RE, editors. Techniques for the Genetic Analysis of Brain and Behavior: Focus on the Mouse. Amsterdam: Elsevier; 1992. pp. 17–38. [Google Scholar]
- Flatscher-Bader T, Zuvela N, Landis N, Wilce PA. Smoking and alcoholism target genes associated with plasticity and glutamate transmission in the human ventral tegmental area. Hum Mol Genet. 2008;17:38–51. doi: 10.1093/hmg/ddm283. [DOI] [PubMed] [Google Scholar]
- Flint J, Mott R. Applying mouse complex-trait resources to behavioural genetics. Nature. 2008;456:724–727. doi: 10.1038/nature07630. [DOI] [PubMed] [Google Scholar]
- Gerlai R. Gene-targeting studies of mammalian behavior: is it the mutation or the background genotype? Trends Neurosci. 1996;19:177–181. doi: 10.1016/s0166-2236(96)20020-7. [DOI] [PubMed] [Google Scholar]
- He J, Crews FT. Increased MCP-1 and microglia in various regions of the human alcoholic brain. Exp Neurol. 2008;210:349–358. doi: 10.1016/j.expneurol.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry CJ, Huang Y, Wynne A, Hanke M, Himler J, Bailey MT, Sheridan JF, Godbout JP. Minocycline attenuates lipopolysaccharide (LPS)-induced neuroinflammation, sickness behavior, and anhedonia. J Neuroinflammation. 2008;5:15. doi: 10.1186/1742-2094-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovatta I, Tennant RS, Helton R, Marr RA, Singer O, Redwine JM, Ellison JA, Schadt EE, Verma IM, Jayapal KP, Philp RJ, Kok YJ, Yap MG, Sherman DH, Griffin TJ, Hu WS. Uncovering genes with divergent mRNA-protein dynamics in Streptomyces coelicolor. PLoS One. 2008;3:e2097. doi: 10.1371/journal.pone.0002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanblanc J, He DY, Carnicella S, Kharazia V, Janak PH, Ron D. Endogenous BDNF in the dorsolateral striatum gates alcohol drinking. J Neurosci. 2009;29:13494–13502. doi: 10.1523/JNEUROSCI.2243-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Barlow C. Glyoxalase 1 and glutathione reductase 1 regulate anxiety in mice. Nature. 2005;438:662–666. doi: 10.1038/nature04250. [DOI] [PubMed] [Google Scholar]
- Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, Shatz CJ. Functional requirement for class I MHC in CNS development and plasticity. Science. 2000;290:2155–2159. doi: 10.1126/science.290.5499.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki Y, Zhang L, Cheng JK, Ji RR. Cytokine mechanisms of central sensitization: distinct and overlapping role of interleukin-1beta, interleukin-6, and tumor necrosis factor-alpha in regulating synaptic and neuronal activity in the superficial spinal cord. J Neurosci. 2008;28:5189–5194. doi: 10.1523/JNEUROSCI.3338-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpel MW, Strother WN, McClintick JN, Carr LG, Liang T, Edenberg HJ, McBride WJ. Functional gene expression differences between inbred alcohol-preferring and -non-preferring rats in five brain regions. Alcohol. 2007;41:95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong EC, Allouche L, Chapot PA, Vranizan K, Moore MS, Heberlein U, Wolf FW. Ethanol-regulated genes that contribute to ethanol sensitivity and rapid tolerance in Drosophila. Alcohol Clin Exp Res. 2010;34:302–316. doi: 10.1111/j.1530-0277.2009.01093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M, Baumann H, Freer G, Freudenberg M, Lamers M, Kishimoto T, Zinkernagel R, Bluethmann H, Köhler G. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature. 1994;368:339–342. doi: 10.1038/368339a0. [DOI] [PubMed] [Google Scholar]
- Lewandowski NM, Small SA. Brain microarray: finding needles in molecular haystacks. J Neurosci. 2005;25:10341–10346. doi: 10.1523/JNEUROSCI.4006-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linda H, Hammarberg H, Piehl F, Khademi M, Olsson T. Expression of MHC class I heavy chain and beta2-microglobulin in rat brainstem motoneurons and nigral dopaminergic neurons. J Neuroimmunol. 1999;101:76–86. doi: 10.1016/s0165-5728(99)00135-6. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Dodd PR, Mayfield RD. Altered gene expression profiles in the frontal cortex of cirrhotic alcoholics. Alcohol Clin Exp Res. 2007;31:1460–1466. doi: 10.1111/j.1530-0277.2007.00444.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, Mayfield RD. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006;31:1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Liuzzo JP, Petanceska SS, Moscatelli D, Devi LA. Inflammatory mediators regulate cathepsin S in macrophages and microglia: A role in attenuating heparan sulfate interactions. Mol Med. 1999;5:320–333. [PMC free article] [PubMed] [Google Scholar]
- McClain CJ, Barve S, Deaciuc I, Kugelmas M, Hill D. Cytokines in alcoholic liver disease. Semin Liver Dis. 1999;19:205–219. doi: 10.1055/s-2007-1007110. [DOI] [PubMed] [Google Scholar]
- McClain CJ, Cohen DA. Increased tumor necrosis factor production by monocytes in alcoholic hepatitis. Hepatology. 1989;9:349–351. doi: 10.1002/hep.1840090302. [DOI] [PubMed] [Google Scholar]
- McClung CA, Nestler EJ, Zachariou V. Regulation of gene expression by chronic morphine and morphine withdrawal in the locus ceruleus and ventral tegmental area. J. Neurosci. 2005;25:6005–6015. doi: 10.1523/JNEUROSCI.0062-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Holgado F, Pinteaux E, Moore JD, Molina-Holgado E, Guaza C, Gibson RM, Rothwell NJ. Endogenous interleukin-1 receptor antagonist mediates anti-inflammatory and neuroprotective actions of cannabinoids in neurons and glia. J Neurosci. 2003;23:6470–6474. doi: 10.1523/JNEUROSCI.23-16-06470.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci USA. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H. Microglial functions and proteases. Mol Neurobiol. 2003;27:163–176. doi: 10.1385/MN:27:2:163. [DOI] [PubMed] [Google Scholar]
- Corriveau RA, Huh GS, Shatz CJ. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron. 1998;21:505–520. doi: 10.1016/s0896-6273(00)80562-0. [DOI] [PubMed] [Google Scholar]
- Nelson S, Kolls JK. Alcohol, host defence and society. Nat Rev Immunol. 2002;2:205–209. doi: 10.1038/nri744. [DOI] [PubMed] [Google Scholar]
- van Noort JM, Bsibsi M. Toll-like receptors in the CNS: implications for neurodegeneration and repair. Prog Brain Res. 2009;175:139–148. doi: 10.1016/S0079-6123(09)17509-X. [DOI] [PubMed] [Google Scholar]
- Qin L, He J, Hanes RN, Pluzarev O, Hong JS, Crews FT. Increased systemic and brain cytokine production and neuroinflammation by endotoxin following ethanol treatment. J Neuroinflammation. 2008;5:1–17. doi: 10.1186/1742-2094-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes JS, Best K, Belknap JK, Finn DA, Crabbe JC. Evaluation of a simple model of ethanol drinking to intoxication in C57BL/6J mice. Physiol Behav. 2005;84:53–63. doi: 10.1016/j.physbeh.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Rihel J, Prober DA, Arvanites A, Lam K, Zimmerman S, Jang S, Haggarty SJ, Kokel D, Rubin LL, Peterson RT, Schier AF. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science. 2010;327:348–351. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Hen R, Crabbe JC. Complications associated with genetic background effects in research using knockout mice. Psychopharmacology. 1999;147:5–7. doi: 10.1007/s002130051128. [DOI] [PubMed] [Google Scholar]
- Rivest S. Molecular insights on the cerebral innate immune system. Brain Behav Immun. 2003;17:13–19. doi: 10.1016/s0889-1591(02)00055-7. [DOI] [PubMed] [Google Scholar]
- Rothenberg BE, Voland JR. β2 knockout mice develop parenchymal iron overload: a putative role for class I genes of the major histocompatibility complex in iron metabolism. Proc Natl Acad Sci USA. 1996;93:1529–1534. doi: 10.1073/pnas.93.4.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba L, Bhave SV, Grahame N, Bice P, Lapadat R, Belknap J, Hoffman PL, Tabakoff B. Candidate genes and their regulatory elements: alcohol preference and tolerance. Mamm Genome. 2006;17:669–688. doi: 10.1007/s00335-005-0190-0. [DOI] [PubMed] [Google Scholar]
- Schütt C. CD14. Int J Biochem Cell Biol. 1999;31:545–549. doi: 10.1016/s1357-2725(98)00153-8. [DOI] [PubMed] [Google Scholar]
- Spanagel R. Alcoholism: a systems approach from molecular physiology to addictive behavior. Physiol Rev. 2009;89:649–705. doi: 10.1152/physrev.00013.2008. [DOI] [PubMed] [Google Scholar]
- Spanagel R, Pendyala G, Abarca C, Zghoul T, Sanchis-Segura C, Magnone MC, Lascorz J, Depner M, Holzberg D, Soyka M, Schreiber S, Matsuda F, Lathrop M, Schumann G, Albrecht U. The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nat Med. 2005;11:35–42. doi: 10.1038/nm1163. [DOI] [PubMed] [Google Scholar]
- Spulber S, Mateos L, Oprica M, Cedazo-Minguez A, Bartfai T, Winblad B, Schultzberg M. Impaired long term memory consolidation in transgenic mice overexpressing the human soluble form of IL-1ra in the brain. J Neuroimmunol. 2009;208:46–53. doi: 10.1016/j.jneuroim.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Kechris K, Hu W, Bhave SV, Finn DA, Grahame NJ, Hoffman PL. The genomic determinants of alcohol preference in mice. Mamm Genome. 2008;19:352–365. doi: 10.1007/s00335-008-9115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakiri N, Kimber I, Rothwell NJ, Pinteaux E. Differential effects of interleukin-1 alpha and beta on interleukin-6 and chemokine synthesis in neurones. Mol Cell Neurosci. 2008;38:259–265. doi: 10.1016/j.mcn.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Wang L, Liu J, Harvey-White J, Zimmer A, Kunos G. Endocannabinoid signaling via cannabinoid receptor 1 is involved in ethanol preference and its age-dependent decline in mice. Proc Natl Acad Sci U S A. 2003;100:1393–1398. doi: 10.1073/pnas.0336351100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Shi GP, Yao PM, Li Z, Chapman HA, Brömme D. Human cathepsin F. Molecular cloning, functional expression, tissue localization, and enzymatic characterization. J Biol Chem. 1998;273:32000–32008. doi: 10.1074/jbc.273.48.32000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.