Abstract
Kinetics-based dose targeting is often conducted in hematopoietic cell transplant (HCT) patients conditioned with intravenous (IV) or oral busulfan to lower rates of rejection, nonrelapse mortality, and relapse. Using the candidate gene approach, we evaluated whether busulfan clearance was associated with polymorphisms in the genes regulating the predominant metabolizing enzymes involved in busulfan conjugation, specifically glutathione S-transferase (GST) isoenzymes A1 (GSTA1) and M1 (GSTM1). Busulfan clearance was estimated after the morning dose on days 1, 2, and 3; each patient’s average clearance was used for analyses. The average (± standard deviation) busulfan clearance was 3.2 ± 0.56 ml/min/kg in the separate population of 95 patients who received oral busulfan and 103 ± 24 ml/min/m2 in the 57 patients who received IV busulfan. Oral busulfan clearance was associated with GSTA1 (p=0.008) but not GSTM1 (p=0.57) genotypes. However, among the GSTA1 haplotypes (i.e., *A*A, *A*B, *B*B), there was significant overlap in the observed oral busulfan clearance and similar rates of achieving the target busulfan exposure. Clearance of IV busulfan was not associated with GSTA1 (p=0.21) or GSTM1 (p=0.99). These data suggest that personalizing either IV or oral busulfan dosing cannot be simplified on the basis of GSTA1 or GSTM1 genotype.
Keywords: Busulfan, glutathione S-transferase, hematopoietic cell transplant, pharmacogenetic, therapeutic drug monitoring
INTRODUCTION
The alkylating agent busulfan (BU) is an integral part of many hematopoietic cell trasnplantation (HCT) conditioning regimens.1 Dosing busulfan based on body weight or body surface area (BSA) is associated with considerable interpatient variability in the efficacy and toxicity of busulfan-containing conditioning regimens. This variability in clinical outcomes is due, in part, to interpatient differences in busulfan clearance which result in variable systemic exposure, expressed as area under the plasma concentration-time curve (AUC) or concentration at steady state (Css=AUC/dosing interval).2 Pharmacodynamic relationships of busulfan systemic exposure and hepatic veno-occlusive disease (VOD- also referred to as sinusoidal obstruction syndrome), rejection, and disease relapse have been observed in patients receiving BU/cyclophosphamide (CY) conditioning and more recently BU/fludarabine conditioning.2–3 These observations have led many HCT centers to use therapeutic drug monitoring, which is also termed kinetics-based4 dose targeting, in patients receiving either intravenous (IV) or oral busulfan.
Kinetics-based dose targeting of busulfan involves the collection of several (usually 4–7) blood samples at known time points after busulfan administration, which are subsequently used to calculate an individual’s busulfan clearance. Effective kinetics-based busulfan dose targeting has been shown to lower rates of rejection, nonrelapse mortality, and relapse in select HCT recipients.5–6 However, the resource intensity of pharmacokinetic sampling has been a barrier to universal acceptance of busulfan dose targeting. Although the recent increase in kinetics-based dose targeting of busulfan shows that this strategy is feasible, more efficient methods to estimate busulfan systemic exposure and clearance (as clearance = dose/AUC) are desirable. In addition, targeting busulfan doses based on genetic polymorphisms may decrease the need for the resource intensive kinetics-based dose targeting that requires quantitation of busulfan concentrations in plasma with subsequent pharmacokinetic modeling.
After IV administration of radiolabeled busulfan, less than 50% of the administered dose is recovered in the urine.7–8 Approximately one-third (i.e., 32.8±2.2%) of busulfan is irreversibly bound to plasma proteins, primarily albumin.9 Only a small fraction (<3%) of a busulfan dose is excreted unchanged in the urine, with neglible amounts in the feces.10–12 The primary elimination route for busulfan is glutathione conjugation, resulting in formation of gamma -glutamyl-beta -(S-tetrahydrothiophenium)-alanyl-glycine (THT+).13–15 The initial step in hepatic metabolism of busulfan is conjugation, catalyzed by glutathione S-transferase (GST).16–18 The enzymatic process is predominantly conjugated by GST isoenzyme A1-1 based on hepatic protein expression of GSTA1.18 Other GST isoenzymes, GSTM1 and GSTP1, contribute to busulfan conjugation at ~5% and 0.2%, respectively, after accounting for their lower activity for busulfan conjugation and lower hepatic expression relative to GSTA1.19 Therefore, genetic polymorphisms regulating GSTA1 and GSTM1 hepatic protein expression are likely to be of most importance to busulfan conjugation and thus, clearance.19–21 Conflicting in vitro data have been presented regarding the relationship between hepatic expression of GSTA1 and single nucleotide polymorphisms (SNPs) in the proximal promoter region of the GSTA1 gene (i.e., GSTA1*A, *B).19–20 The GSTM1 null genotype is associated with lower hepatic protein expression, and an increased risk of VOD in β-thalassemia patients receiving BU/CY with antithymocyte globulin (ATG) conditioning.22
Thus, we sought to evaluate the association of GSTA1 haplotype and GSTM1 genotype with IV or oral busulfan clearance in two separate HCT populations.
MATERIALS AND METHODS
Study populations
This was a retrospective study in two patient cohorts who received HCT conditioning with either IV or oral busulfan. The cohorts were separate and no patients received both IV and oral busulfan. All patients had their busulfan dose personalized to a target busulfan Css using kinetics-based busulfan dosing. Patients were enrolled in Fred Hutchinson Cancer Research Center (FHCRC) clinical treatment protocols from June 2002 to November 2006 (IV busulfan) and from July 2004 to January 2006 (oral busulfan). Approval of the FHCRC Investigational Review Board was obtained prior to data analysis. Inclusion criteria were as follows: patients receiving their first HCT, receiving a conditioning regimen containing busulfan, and being enrolled in protocols that stipulated busulfan kinetics-based dose targeting. For oral busulfan only, children less than 10 years of age were excluded from this analysis because of previously documented age-dependent clearance.13 For IV busulfan, 67 patients met the inclusion criteria, but genomic DNA was not available from 10 participants; thus, the final database contained information on 57 patients who received IV busulfan. The criteria were met in 131 patients receiving oral busulfan; however, GSTA1 and GSTM1 genotypes could not be obtained for 34 and 2 patients, respectively. Thus, the final database contained 95 patients who received oral busulfan.
Records were examined for demographic data (i.e., age, sex, height, weight, body surface area), and clinical data (i.e., disease, conditioning regimen). The ideal body weight in adults was calculated as follows: for males = 50 kg + (2.3 kg for each inch over 5 feet); for females = 45.5 kg + (2.3 kg for each inch over 5 feet). The adjusted ideal body weight (AIBW = ideal body weight + 0.25 (actual body weight - ideal body weight) was determined.23 Body surface area (BSA) was calculated using height and AIBW; BSA was calculated by taking the square root of actual weight × height, divided by 60.
Standard practice for prophylaxis to busulfan-induced seizures was phenytoin.
Intravenous busulfan dosing and pharmacokinetic sampling
Intravenous busulfan was administered every six hours (Q6hr) to 18 patients and every 24 hours (i.e., daily) to 39 patients. All patients received busulfan for 4 days for a total of 16 doses (if administered Q6hr) or 4 doses (if administered daily). The first busulfan dose and the target busulfan Css were based upon the FHCRC treatment protocol and could be adjusted by the attending physician. In those patients receiving Q6hr IV busulfan, the mean (± standard deviation) first busulfan dose was 0.92 ± 0.15mg/kg and the cumulative busulfan dose ranged from 11–23 mg/kg. For 22 of the patients receiving daily IV busulfan, the first IV busulfan dose was 3.2 mg/kg for a total IV busulfan dose of 9.7–20 mg/kg. Based upon the average clearance after daily IV busulfan in this cohort, the first IV busulfan dose was increased to 4 mg/kg in the subsequent 17 patients. These patients received a total dose of 10–19 mg/kg.
A total of 21 pharmacokinetic blood samples were obtained from each patient receiving IV busulfan. Patients receiving IV busulfan Q6hr had pharmacokinetic sampling after the morning dose on days 1 through 3 at the following times relative to the start of the 2 hour infusion: end of infusion, and at 2.25, 2.5, 3, 4, 5, and 6 hours (i.e., prior to subsequent dose). Those patients receiving daily IV busulfan had pharmacokinetic sampling on days 1 through 3 at the following time points relative to the start of the 3 hour infusion: end of infusion, and at 3.25, 4.5, 6, 8, 11 and 24 hours (i.e., prior to subsequent dose).
Oral busulfan dosing and pharmacokinetic sampling
Oral busulfan was administered every six hours (Q6hr) for 4 days for a total of 16 doses. The first busulfan dose and the target busulfan Css were based upon the FHCRC treatment protocol and could be adjusted by the attending physician. For all patients receiving oral busulfan, the mean first oral busulfan dose was 1 ± 0.01 mg/kg and a total oral busulfan dose ranging from 9.3–24 mg/kg.
For each patient receiving oral busulfan, a total of 19 pharmacokinetic blood samples were obtained. Pharmacokinetic sampling was conducted after the morning dose on days 1 through 3. Samples were obtained immediately before and 30, 60, 90, 120, 180, 240, 300, and 360 minutes after oral administration on day 1. On days 2 and 3, samples were obtained immediately before and 60, 120, 240, and 360 minutes after oral busulfan administration.
Quantitation and Pharmacokinetic Modeling of Busulfan Samples
Busulfan concentrations were determined by gas chromatography with mass spectrometry detection as previously described.24 Dynamic range was from 25 to 4500 ng/mL, and the inter-day CV was less than 8%. After quantitation of busulfan samples, the concentration-time data were fit using WinNonlin (version 5.0.1) via noncompartmental (IV or oral) or compartmental (IV only) modeling, determined by visual inspection of the model fit to the individual concentration-time data. The AUC from time 0 to infinity (∞) was calculated after the first dose. The AUC from 0 to the end of the dosing interval (abbreviated tau [τ]) was calculated after the day 2 and day 3 doses in patients receiving busulfan Q6hrs (IV and oral) and the AUC from time 0 to infinity (∞) was calculated after the day 2 and day 3 doses in patients receiving busulfan daily (IV only). The AUC0 to ∞ after the first dose equals the AUC0 to τ at steady state (i.e., days 2 and 3).25 Clearance was calculated by dividing the dose by the AUC. After calculation of the patient’s clearance, the target dose for subsequent doses was calculated linearly to achieve the target Css, which was chosen by the attending physician. Successful targeting is confirmed by determining the busulfan clearance after the morning dose on days 2 and 3, with further dose adjustments as needed.
Preparation of DNA
Peripheral blood leukocytes were collected from freshly obtained samples and stored at the FHCRC DNA repository (Clinical Research Division). Genomic DNA was extracted using a QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA) from peripheral blood leukocytes. The quality of DNA samples was tested by spectrophotometric analysis at 260 and 280 nm before genotyping.
Determination of GSTA1 and GSTM1 Genotypes
Genotyping was performed at the DNA Sequencing and Gene Analysis Center (Department of Pharmaceutics, University of Washington, Seattle, WA). The four sequence variants of the human GSTA1 proximal promoter that define the *A and *B alleles (−631T or G, −567T, −69C, −52G and −631G, −567G, −69T, −52A, respectively) were determined by direct sequencing of a 780 bp amplicon.20 The cycling conditions for PCR were as follows: 95 °C for 2 min; 40 cycles of 95 °C for 30 sec, 60 °C for 30 sec, 72 °C for 2 min; 72 °C for 7 min; hold at 4 °C. The PCR reaction contained 1.25 U PfuTurbo Hotstart DNA polymerase (Stratagene, La Jolla, CA) in 1X reaction buffer with 200 μM dNTPs and 500 nM oligonucleotides with ~50–150 ng genomic DNA template in a total volume of 25 μl. The PCR amplicons were purified on QIAquik PCR purification columns (Qiagen) according to manufacturer’s instruction. The primer sequences for PCR (AF7 and AR1) were taken from the original citation20 and BigDye Terminator V 3.1 (Applied Biosystems, Foster City, CA) sequencing of the purified amplicon was performed using both primer AF7 and a second internal primer (hGSTA1.seq.for 5′CCAGCTATGCTCACAGTAGAG3′) to identify the SNPs at −631/−567 and −69/−52, respectively.
GSTM1 gene deletion genotyping was performed using the Applied Biosystems “TaqmanR Gene Copy Number Assay” reagents according to the manufacturer’s protocol. In brief, DNA (~25 ng) was combined with 5 μL Taqman 2X MasterMix, 1 μL 20X GSTM1 6-FAM-labeled assay (PN4331182) and 1 μL 20X RnaseP VIC-labeled assay (PN4316844) in a total volume of 10 μL. Each sample was run in triplicate in a 96 well PCR plate on a ABI 7900HT (Applied Biosystems) instrument with the following thermal cycling conditions: 95 °C for 10 min and 40 cycles at 95 °C for 15 sec, 60 °C for 1 min. The real-time PCR data were collected using ABI sequence detection system software (SDS V2.1) (Applied Biosystem) and the cycle threshold (Ct) values for GSTM1 and RnaseP triplicates calculated. The Ct values for the RnaseP gene served as a reference when analyzing the samples for the presence or absence of GSTM1 sequences. Each assay included controls known to contain (or not to contain) the GSTM1 gene, and patient genotypes were called as either being GSTM1 positive or GSTM1 null; no attempt was made to determine zygosity.
Statistical Methods
An a priori power analysis was conducted using our clinical database to assist with kinetics-based dosing of IV or oral busulfan (reported in part in McCune and Holmberg26). Eight patients per GSTA1 genotype group would provide 80% power to observe a 20% difference in IV busulfan clearance. For oral busulfan clearance, six patients per GSTA1 genotype group would provide 80% power to observe a 20% difference. All statistical analyses were performed using SAS version 9 (SAS, Inc., Cary, NC).
Each participant’s average busulfan clearance was calculated from busulfan clearance on days 1, 2 and 3. Log transformation of the average busulfan clearance was performed to normalize the distribution of data before inferential analysis, as specified a priori. Categorical patient variables included frequency of administration (Q6hr vs. daily, IV clearance only), GSTA1 genotype (categorized as *A*A, *A*B, or *B*B) and GSTM1 genotype (categorized as null or present). Continuous patient variables included age at the time of HCT, BSA, and AIBW, which was done for the IV busulfan dataset only as this population had greater heterogeneity.
A linear regression analysis was conducted on log average busulfan clearance as the dependent variable to identify which variables were significant predictors of clearance. All predictors with a p<0.05 were considered significant.
Genotype frequencies of GSTA1 and GSTM1 were determined to be in Hardy-Weinberg equilibrium using χ2 analysis.
RESULTS
Patient characteristics
Patient pre-transplant demographics and HCT characteristics are described in Table 1.
Table 1.
Description of patient populationa
| Group | IV Busulfan | Oral Busulfan |
|---|---|---|
| N | 57 | 95 |
| Age (years) | ||
| Mean ± standard deviation (SD) | 38 ± 21 | 45 ± 12 |
| N < 18 years | 13 | 1 |
| AIBW (kg) | ||
| Mean ± SD | 58 ± 25 | 66 ± 10 |
| N < 12 kgb | 8b | 0 |
| Body mass indexc (kg/m2) | ||
| Mean | 27 ± 6.9 | 27 ± 5.7 |
| Underweight | 8 (14%) | 0 |
| Normal (18.5–24.9) | 22 (39%) | 34 (36%) |
| Overweight (25.0–29.9) | 18 (32%) | 30 (32%) |
| Obesity I (30.0–34.9) | 5 (9%) | 21 (22%) |
| Obesity II (35.0–39.9) | 1 (2%) | 7 (7%) |
| Extreme Obesity (40.0+) | 3 (5%) | 3 (3%) |
| Ethnic background | ||
| Caucasian | 46 (81%) | 76 (80%) |
| Asian | 2 (4%) | 6 (6%) |
| African American | 1 (2%) | 5 (5%) |
| East Indian | 1 (2%) | 3 (3%) |
| Hispanic | 1 (2%) | 2 (2%) |
| Native American | 1 (2%) | 2 (2%) |
| Unknown | 5 (9%) | 1 (1%) |
| Gender | ||
| Male | 34 (60%) | 48 (51%) |
| Female | 23 (40%) | 47 (49%) |
| Conditioning Regimen | ||
| Busulfan + Flu ± THY | 39 (68%) | 0 |
| Busulfan + CY ± THY ± etoposide | 15 (26%) | 94 (98%) |
| Busulfan + Flu + CY | 2 (4%) | 1 |
| Busulfan + melphalan | 1 (2%) | 0 |
| Diagnosis | ||
| Myelodysplastic syndrome | 17 (30%) | 27 (28.4%) |
| Acute myeloid leukemia | 13 (23%) | 29 (30.5%) |
| Acute myeloid leukemia/myelodysplastic syndrome | 0 | 6 (6.3%) |
| Myelofibrosis | 6 (11%) | 9 (9.5%) |
| Chronic myelomonocytic leukemia | 5 (9%) | 0 |
| Wiskott-Aldrich syndrome | 3 (5%) | 0 |
| Osteopetrosis | 3 (5%) | 0 |
| Chronic myeloid leukemia | 2 (4%) | 16 (16.9%) |
| Hemophagocytic lymphohistiocytosis | 2 (4%) | 0 |
| Agnogenic myeloid metaplasia | 1 (2%) | 0 |
| Multiple myeloma | 1 (2%) | 0 |
| Blackfan-Diamond anemia | 1 (2%) | 0 |
| Congenital dyserythropoietic | 1 (2%) | 0 |
| Sickle cell anemia | 1 (2%) | 0 |
| β-Thalassemia | 1 (2%) | 0 |
Data presented as N or mean ± standard deviation;
within the package insert, children less than 12 kg should receive a higher initial busulfan dose than heavier children and adults;
categorized per National Heart Lung and Blood Institute
For patients receiving IV busulfan, the median age was 43 years (range: 0.5–66). Thirteen of the patients (23%) were less than 18 years of age, and eight patients (14%) weighed less than 12 kg, at which weight higher initial IV busulfan doses are necessary per the package insert.12 Sixty percent (34 of 57) of the patients receiving IV busulfan were male. The majority of patients receiving IV busulfan (41 of 57, 71%) received busulfan plus fludarabine conditioning. Forty-six patients (81%) received ATG as part of the conditioning regimen. Additionally, etoposide and melphalan were given to a limited number of patients (4% and 2%, respectively). No other antineoplastic agents or irradiation were given immediately before or concomitantly with busulfan.
Of those patients receiving oral busulfan, the median age was 48 years (range: 16–63); only one patient (1%) was less than 18 years of age. Fifty percent (48 of 95) of the patients were male. The majority of patients receiving oral busulfan received busulfan plus cyclophosphamide (60 mg/kg/dose, once daily for two days) conditioning. Three patients (3%) received ATG as part of the conditioning regimen. Additionally, fludarabine was given to one patient (1%). No other antineoplastic agents or irradiation were given immediately before or concomitantly with busulfan.
IV busulfan pharmacokinetics
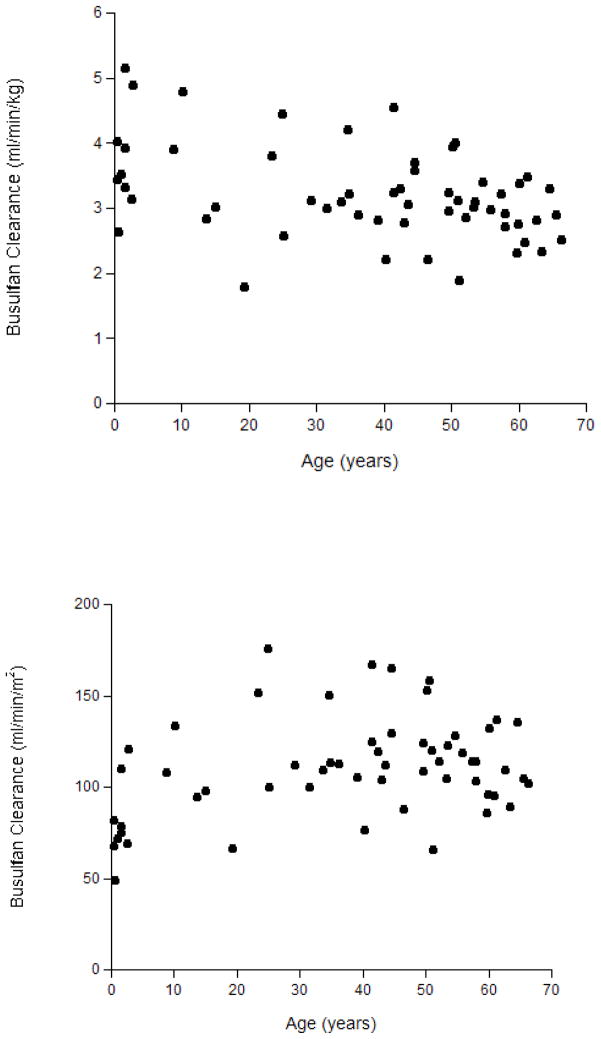
The average (± standard deviation) IV busulfan clearance was 103 ± 23.6 ml/min/m2, consistent with busulfan clearance in other HCT populations receiving concomitant phenytoin.27 Busulfan clearance did not differ by age (Figure 1) or by administration frequency (i.e., Q6 hours compared to daily, data not shown). Thus, subsequent statistical analyses were conducted using busulfan clearance standardized by BSA (i.e., ml/min/m2) in all 57 patients. Of note, IV busulfan clearance standardized to BSA (i.e., ml/min/m2) is optimal because liver weight expressed relative to BSA (g/m2) is similar for children and adults.13, 28 Liver weight expressed relative to body weight (g/kg) is higher in young children than in older children and adults.13, 28
Figure 1.
Figure 1a (top) and 1b (bottom)
Association of Age with IV Busulfan Clearance Standardized by Body Weight (panel A) or by Body Surface Area (panel B)
Oral busulfan pharmacokinetics
The average (± standard deviation) busulfan clearance was 3.2 ± 0.56 ml/min/kg (119 ± 20 ml/min/m2), consistent with busulfan clearance in other HCT populations receiving concomitant phenytoin.26
Genotype Frequencies
The frequencies of GSTA1 and GSTM1 genotypes observed in the IV and oral busulfan cohort are displayed in Table 2. All genotypes were within the Hardy-Weinberg equilibrium (P>0.05).
Table 2.
GSTA1 and GSTM1 Frequencies
Pharmacogenetic Associations with IV Busulfan Clearance
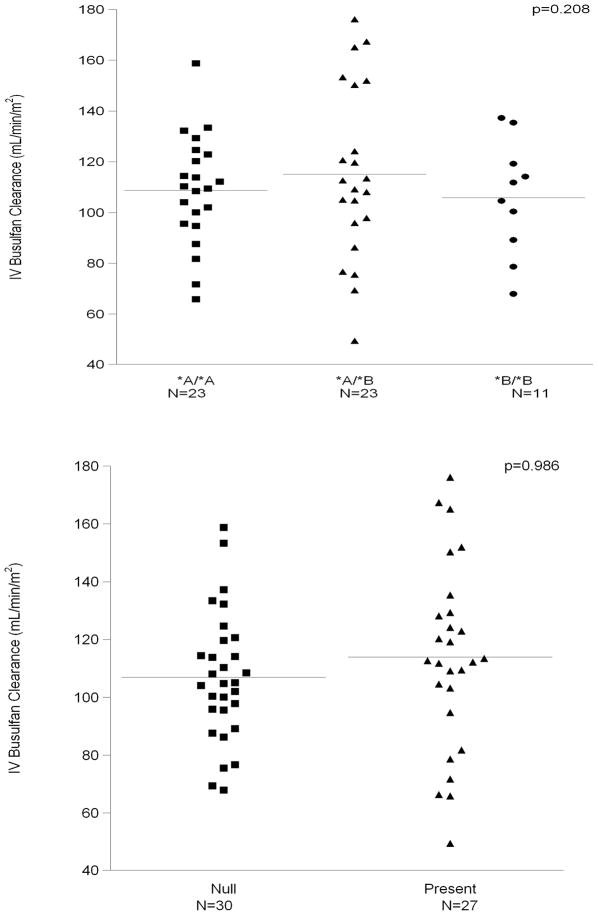
We analyzed the association between IV busulfan clearance and the genetic variants of GSTA1 and GSTM1. Figure 2 shows the average and the range of IV busulfan clearance by genotype. Neither genotype was statistically associated with busulfan clearance [GSTA1 (p=0.21, panel A) and GSTM1 (p=0.99, panel B)].
Figure 2.
Figure 2a (top) & 2b (bottom)
Lack of Association of IV Busulfan Clearance with GSTA1 (panel 2A, p=0.208) and GSTM1 (panel 2B, p=0.986). Bars indicate mean values for each category.
Pharmacogenetic Associations with Oral Busulfan Clearance
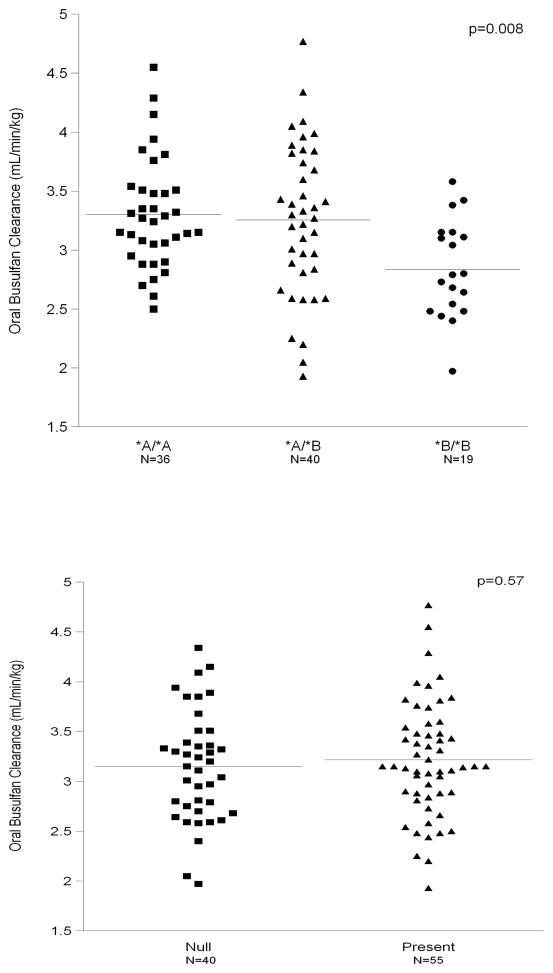
We analyzed the associations between oral busulfan clearance and the GSTA1 and GSTM1 genotypes in this study population. Figure 3 shows the average and the range of oral busulfan clearance by genotype, specifically GSTA1 (p=0.008, panel A) and GSTM1 (p=0.57, panel B). The GSTA1 haplotype was statistically significantly associated with oral busulfan clearance, with those patients with germline GSTA1*A*A or *A*B haplotype having a higher oral busulfan clearance than those carrying the GSTA1*B*B haplotype. Those patients with the GSTA1*A*A haplotype had an oral busulfan clearance that was 0.45 ml/min/kg (16%) higher, on average, than those with GSTA1*B*B genotype. Similarly, the oral busulfan clearance of patients with the GSTA1*A*B genotype was, on average, 0.41 ml/min/kg (14%) higher than patients with GSTA1*B*B. GSTM1 genotype was not associated with oral busulfan clearance (Figure 3B).
Figure 3.
Figure 3a (top) & 3b (bottom)
Association of Oral Busulfan Clearance with GSTA1 (panel 3A, P=0.008) but not GSTM1 (panel 3B, p=0.57). Bars indicate mean values for each category.
We then sought to evaluate the impact of GSTA1 haplotype upon kinetics-based dosing of oral busulfan to a target busulfan Css. As shown in Table 3, those patients with the GSTA1*B*B haplotype often had their oral busulfan dose decreased from the starting weight-based dose (i.e., dose #1), as evidenced by the mean ratio of dose 5/dose 1 of 0.88. There is considerable interpatient variability in the range of the dose adjustments. The most common busulfan Css target in the oral busulfan cohort was 800 to 900 ng/ml, which was the target Css for 76 of 95 patients (80%). Across all three haplotypes, the percentage of patients achieving the target Css after dose 1 was low (i.e., 20–32%). Specifically, the target Css wasachieved after dose 1 in 32% of the 28 GSTA1*A*A carriers, 20% of the 30 GSTA1*A*B carriers and 28% of the 18 GSTA1*B*B carriers. The target Css after all the busulfan doses was achieved among almost all the patients (i.e., 93–94%).
Table 3.
Oral busulfan dose adjustments to achieve target busulfan Css according to GSTA1 haplotype
| Number of patients (Total = 95) | GSTA1 Haplotype | ||
|---|---|---|---|
| *A/*A | *A/*B | *B/*B | |
| 36 | 40 | 19 | |
| Dose 5 | 1.04 ± 0.15 | 1.01 ± 0.20 | 0.88 ± 0.13 |
| Dose 1 | (0.79–1.52) | (0.46–1.41) | (0.60–1.16) |
| Dose 9 | 1.02 ± 0.09 | 1.02 ± 0.09 | 1.00 ± 0.07 |
| Dose 5 | (0.88–1.24) | (0.80–1.23) | (0.91–1.14) |
Data are presented as mean ± standard deviation (range). Busulfan dose 1 was 1 mg/kg for all patients; doses 5 and 9 were adjusted to achieve target Css.
DISCUSSION
We sought to gain a better understanding of the genetic covariates associated with busulfan clearance with the long-range goal of discovering less resource-intensive methods to target both IV and oral busulfan doses in HCT patients. The key findings of this analysis were: 1. Using the candidate gene approach, no significant pharmacogenetic associations were observed between IV busulfan clearance and reported genetic variants of GSTA1 or GSTM1 (Figure 2); 2. Although oral busulfan clearance was associated with GSTA1 haplotype, targeting oral busulfan doses appeared unlikely to be simplified by genetics-based dosing due to the considerable interpatient variability amongst the three GSTA1 haplotypes (Figure 3A).
Use of the established GSTA1 and GSTM1 genotypes to predict IV and oral busulfan clearance cannot be recommended at this time. The genotypes evaluated were chosen based on their association with hepatic protein expression.19–21 Our dataset was adequately powered to reveal a 20% difference in busulfan clearance between GSTA1 haplotypes. The data regarding the importance of these GST polymorphisms to HCT patients receiving busulfan are conflicting. In vitro, GSTA1*A/*A is associated with higher hepatic protein expression of this most active GST enzyme but not busulfan conjugation. Hepatic expression of GSTA1 protein is associated with GSTA1 haplotype, with the GSTA1*A*A haplotype having the highest protein expression.20 However, a separate in vitro study did not find an association between GSTA1 or GSTM1 genotype and busulfan conjugation.19
The data have been conflicting regarding the association of GSTA1 and GSTM1 genotypes with the pharmacokinetics of busulfan. In a small population of twelve Japanese patients receiving oral busulfan, Kusuma et al observed that the apparent oral clearance of busulfan was lower in patients with the GSTA1*A*B haplotype than with the GSTA1*A*A haplotype.29 Notably, no patients were homozygotes for the GSTA1*B*B variant. With IV busulfan, the data have been conflicting with reports indicating that GSTA1 is30 or is not associated31–32 with IV busulfan clearance. The majority of the data with GSTM1 suggests that this genotype is not associated with IV busulfan clearance,31 although contradictory data do exist.32 Not surprisingly, the majority of these studies were from small patient populations (numbers accrued ranging from 1229 to 7731). Similarly, our study was from a single institution; however, we powered our sample size a priori to observe a 20% difference in busulfan clearance. Two separate studies in pediatric populations noted that GSTA1 haplotype was30 or was not31 associated with IV busulfan clearance. Our results suggest that GSTA1 and GSTM1 are not associated with IV busulfan clearance.
To date, the association between GST genotypes and circulating busulfan metabolites –such as the thiophenium ion13 – in blood has not been evaluated in patients receiving busulfan. However, such studies may elucidate a physiologic rationale, potentially regulation of intestinal GSTA1 expression, for the association of GSTA1 haplotype with oral (Figure 3A) but not IV (Figure 2A) busulfan clearance.
Consideration should be given to factors which may influence the busulfan clearance phenotype. Concomitant medications, specifically phenytoin, may have altered busulfan clearance and obscured a genotype-phenotype relationship. Phenytoin is commonly used to prevent busulfan-induced seizures, and its effects upon busulfan clearance are difficult to assess since the majority of busulfan pharmacokinetic data has been obtained in patients also receiving phenytoin. However, phenytoin has been reported to increase busulfan clearance33–34 and may alter busulfan clearance such that an association between GST genotype and busulfan clearance phenotype cannot be discerned under these conditions. Unfortunately, no metabolic study data are available that compare busulfan metabolism in patients receiving phenytoin to that in patients not receiving phenytoin. The potential impact of phenytoin upon busulfan metabolism and thus, genotype – phenotype relationships, remains to be evaluated.
The GST genotypes have also been associated with clinical outcomes in HCT recipients receiving BU/CY, although a mechanistic rationale for their association remains elusive. Because pharmacokinetics-based busulfan dose targeting was performed in our dataset, we did not evaluate the association of these GST genotypes with clinical outcomes. In Korean patients receiving BU/CY conditioning prior to an allogeneic graft, GSTA1*A*A is associated with a lower incidence of acute graft versus host disease (GVHD) but not hepatic VOD.35 The authors hypothesized that the underlying mechanism is that GSTA1*A*A metabolizes busulfan and CY more rapidly, thus decreasing tissue injury which has been implicated as a key event in the etiology of GVHD.1 While busulfan Css is associated with VOD, it is not consistently associated with acute GVHD.6, 36 Thus, the mechanistic rationale for this observation is more likely to be due to altered metabolism of CY.6, 36 GSTM1-1 also mediates busulfan conjugation, accounting for ~5% of total GST activity.18 The GSTM1 null genotype is associated with increased risk of VOD, lower busulfan Css, and faster oral busulfan clearance in β-thalassemia patients receiving BU/CY ± ATG regimen.22, 37 These results led to the hypothesis that VOD was caused by liver damage due to a metabolite of busulfan but not busulfan itself, potentially due to depletion of the glutathione pool. Our results suggest that these pharmacogenetic associations between clinical outcomes and GST genotypes are multifactorial, since we did not observe an association between GST genotypes and IV busulfan clearance.
After IV administration, the busulfan clearance was similar between those receiving busulfan Q6hr and daily, confirming the results of Madden et al.27 The increasing popularity of daily administration of busulfan makes it imperative that more efficient tools are developed to personalize busulfan doses. Daily administration of busulfan leads to fewer busulfan doses being administered in HCT conditioning (i.e., 4 days of busulfan conditioning, formerly Q6 hour × 16 doses to now Q24 hour × 4 doses).27, 38 For example, with the current practice of weight-based dosing of daily IV busulfan in conjunction with kinetics-based dose adjustments, patients will receive at least one (i.e., 25% of the total busulfan therapy) to three (i.e., 75% of the total busulfan therapy) doses before information on the individual’s busulfan clearance is available. In addition, shorter courses (e.g., 2 days) of IV or oral busulfan are being evaluated in reduced-intensity conditioning regimens and prior to infusion of genetically modified cells.38–41 With these latter regimens, there is inadequate time to target busulfan doses without an on-site laboratory quantitating busulfan concentrations. Current approaches to targeting require extended and intensive blood sampling schedules to characterize an individual’s busulfan clearance. Alternative methods to target IV busulfan doses are needed due to the continuing trend for shorter busulfan courses.27, 38–41 We have created a population pharmacokinetic model for daily IV busulfan42 with the intent of addressing both major busulfan dose targeting hurdles: modeling of variability and mitigation of resource intensity of kinetics-based targeting. Population pharmacokinetic modeling can also identify genetic and non-genetic covariates of busulfan conjugation and elimination. These models, coupled with further in vitro studies, may provide insight regarding why GST polymorphisms are associated with hepatic protein expression20 but not busulfan conjugation in vitro.19 Population pharmacokinetic models also facilitate development of limited blood sampling schedules, which require fewer blood samples to characterize an individual patient’s busulfan clearance. Such population pharmacokinetic models and limited sampling schedules could lead to less resource-intensive methods to target busulfan doses and have been utilized with oral busulfan dosing.43
In summary, IV busulfan clearance is not associated with the genetic variants of the most promising genes related to its metabolism: GSTA1 and GSTM1. Similarly, GSTM1 genotype was not associated with oral busulfan clearance. Although oral busulfan clearance was associated with GSTA1 haplotype, the considerable interpatient variability within each haplotype would not provide sufficient confidence in prediction of oral busulfan clearance, and therefore the dose needed to achieve target busulfan exposure. Thus, intensive pharmacokinetic sampling remains the standard for targeting busulfan doses in HCT recipients.
Acknowledgments
The authors would like to acknowledge the contributions of the technical staff in the Seattle Cancer Care Alliance Pharmacokinetics Laboratory with busulfan dose targeting; Mr. Eric Mickelson in the FHCRC Hansen Laboratory for DNA isolation; and Ms. Tot Bui Nguyen in the University of Washington the DNA Sequencing and Gene Analysis Center for GST sequencing.
Financial Support: Supported by in part by PDL Biopharma (JSM), UW School of Pharmacy DMTPR Funds (JSM), and the Milo Gibaldi Endowment Fund (RJYH). The clinical hematopoietic cell transplant protocols were supported by HL036444, CA 18029. The DNA repository at FHCRC is supported by NIH PO1-AI33484 and R01 HL087690 (Principal Investigator: John Hansen, MD).
Abbreviations
- AIBW
adjusted ideal body weight
- ALT
alanine transaminase
- AST
aspartate transaminase
- ATG
antithymocyte globulin
- AUC
area under the plasma concentration-time curve
- BSA
body surface area
- BU
busulfan
- Css
concentration at steady state
- CY
cyclophosphamide
- FHCRC
Fred Hutchinson Cancer Research Center
- GST
glutathione S-transferase
- GVHD
graft versus host disease
- HCT
hematopoietic cell transplant
- VOD
veno-occlusive disease
- IV
intravenous
- SNPs
single nucleotide polymorphisms
References
- 1.Deeg HJ, Maris MB, Scott BL, Warren EH. Optimization of allogeneic transplant conditioning: not the time for dogma. Leukemia. 2006 Oct;20(10):1701–1705. doi: 10.1038/sj.leu.2404327. [DOI] [PubMed] [Google Scholar]
- 2.McCune JS, Gibbs JP, Slattery JT. Plasma concentration monitoring of busulfan: does it improve clinical outcome? Clin Pharmacokinet. 2000;39(2):155–165. doi: 10.2165/00003088-200039020-00005. [DOI] [PubMed] [Google Scholar]
- 3.Field T, Perkins J, Alsina M, et al. Busulfan Area-under-the-Curve Finding Study within a Busulfan/Fludarabine (BuFlu) Conditioning Regimen before Allogeneic Hematopoietic Cell Transplantation (HCT) Blood. 2006 November 16;108(11) abstract #2939. [Google Scholar]
- 4.Undevia SD, Gomez-Abuin G, Ratain MJ. Pharmacokinetic variability of anticancer agents. Nat Rev Cancer. 2005 Jun;5(6):447–458. doi: 10.1038/nrc1629. [DOI] [PubMed] [Google Scholar]
- 5.Deeg HJ, Storer BE, Boeckh M, et al. Reduced incidence of acute and chronic graft-versus-host disease with the addition of thymoglobulin to a targeted busulfan/cyclophosphamide regimen. Biol Blood Marrow Transplant. 2006 May;12(5):573–584. doi: 10.1016/j.bbmt.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 6.Radich JP, Gooley T, Bensinger W, et al. HLA-matched related hematopoietic cell transplantation for chronic-phase CML using a targeted busulfan and cyclophosphamide preparative regimen. Blood. 2003 July 1;102(1):31–35. doi: 10.1182/blood-2002-08-2619. [DOI] [PubMed] [Google Scholar]
- 7.Nadkarni MV, Trams EG, Smith PK. Preliminary Studies on the Distribution and Fate of TEM, TEPA, and Myeleran in the Human. Cancer Res. 1959;19:713–718. [PubMed] [Google Scholar]
- 8.Vodopick H, Hamilton HE, Jackson HL, Peng CT, Sheets RF. Metabolic fate of tritiated busulfan in man. J Lab Clin Med. 1969;73(2):266–276. [PubMed] [Google Scholar]
- 9.Ehrsson H, Hassan M. Binding of busulfan to plasma proteins and blood cells. J Pharm Pharmacol. 1984;36(10):694–696. doi: 10.1111/j.2042-7158.1984.tb04847.x. [DOI] [PubMed] [Google Scholar]
- 10.Hassan M, Oberg G, Ehrsson H, et al. Pharmacokinetic and metabolic studies of high-dose busulphan in adults. Eur J Clin Pharmacol. 1989;36(5):525–530. doi: 10.1007/BF00558081. [DOI] [PubMed] [Google Scholar]
- 11.Ehrsson H, Hassan M, Ehrnebo M, Beran M. Busulfan Kinetics. Clin Pharmacol Ther. 1983;34:86–89. doi: 10.1038/clpt.1983.134. [DOI] [PubMed] [Google Scholar]
- 12.Package Insert. Busulfex (busulfan) Otsuka America Pharmaceutical, Inc; Tokyo, Japan: [accessed May 30, 2008]. (Revision Date: February 2008): http://www.ivbusulfex.com/Otsuka_IVBusulfex_v2AA.pdf. [Google Scholar]
- 13.Gibbs JP, Murray G, Risler L, Chien JY, Dev R, Slattery JT. Age-dependent tetrahydrothiophenium ion formation in young children and adults receiving high-dose busulfan. Cancer Res. 1997;57(24):5509–5516. [PubMed] [Google Scholar]
- 14.Hassan M, Ljungman P, Bolme P, et al. Busulfan bioavailability. Blood. 1994 Oct 1;84(7):2144–2150. [PubMed] [Google Scholar]
- 15.Hassan M, Ehrsson H, Ljungman P. Aspects concerning busulfan pharmacokinetics and bioavailability. Leuk Lymphoma. 1996;22(5–6):395–407. doi: 10.3109/10428199609054777. [DOI] [PubMed] [Google Scholar]
- 16.Hassan M, Ehrsson H. Metabolism of 14C-busulfan in isolated perfused rat liver. Eur J Drug Metab Pharmacokinet. 1987 Jan-Mar;12(1):71–76. doi: 10.1007/BF03189864. [DOI] [PubMed] [Google Scholar]
- 17.Gibbs JP, Czerwinski M, Slattery JT. Busulfan-glutathione conjugation catalyzed by human liver cytosolic glutathione S-transferases. Cancer Res. 1996;56(16):3678–3681. [PubMed] [Google Scholar]
- 18.Czerwinski M, Gibbs JP, Slattery JT. Busulfan conjugation by glutathione S-transferases alpha, mu, and pi. Drug Metab Dispos. 1996;24(9):1015–1019. [PubMed] [Google Scholar]
- 19.Bredschneider M, Klein K, Murdter TE, et al. Genetic polymorphisms of glutathione S-transferase A1, the major glutathione S-transferase in human liver: consequences for enzyme expression and busulfan conjugation. Clin Pharmacol Ther. 2002 Jun;71(6):479–487. doi: 10.1067/mcp.2002.124518. [DOI] [PubMed] [Google Scholar]
- 20.Coles BF, Morel F, Rauch C, et al. Effect of polymorphism in the human glutathione S-transferase A1 promoter on hepatic GSTA1 and GSTA2 expression. Pharmacogenetics. 2001 Nov;11(8):663–669. doi: 10.1097/00008571-200111000-00004. [DOI] [PubMed] [Google Scholar]
- 21.Bolt HM, Thier R. Relevance of the deletion polymorphisms of the glutathione S-transferases GSTT1 and GSTM1 in pharmacology and toxicology. Curr Drug Metab. 2006 Aug;7(6):613–628. doi: 10.2174/138920006778017786. [DOI] [PubMed] [Google Scholar]
- 22.Srivastava A, Poonkuzhali B, Shaji RV, et al. Glutathione S-transferase M1 polymorphism: a risk factor for hepatic venoocclusive disease in bone marrow transplantation. Blood. 2004 September 1;104(5):1574–1577. doi: 10.1182/blood-2003-11-3778. [DOI] [PubMed] [Google Scholar]
- 23.Pai MP, Paloucek FP. The origin of the “ideal” body weight equations. Ann Pharmacother. 2000 Sep;34(9):1066–1069. doi: 10.1345/aph.19381. [DOI] [PubMed] [Google Scholar]
- 24.Slattery JT, Risler LJ. Therapeutic monitoring of busulfan in hematopoietic stem cell transplantation. Ther Drug Monit. 1998;20(5):543–549. doi: 10.1097/00007691-199810000-00017. [DOI] [PubMed] [Google Scholar]
- 25.Jusko WJ. In: Guidelines for Collection and Analysis of Pharmacokinetic Data. 4. Burton ME, editor. Philadelphia: Lippincott Williams & Wilkins; 2006. Applied Pharmacokinetics & Pharmacodynamics. [Google Scholar]
- 26.McCune JS, Holmberg LA. Busulfan in hematopoietic stem cell transplant setting. Expert Opin Drug Metab Toxicol. 2009 Aug;5(8):957–969. doi: 10.1517/17425250903107764. [DOI] [PubMed] [Google Scholar]
- 27.Madden T, de Lima M, Thapar N, et al. Pharmacokinetics of once-daily IV busulfan as part of pretransplantation preparative regimens: a comparison with an every 6-hour dosing schedule. Biol Blood Marrow Transplant. 2007 Jan;13(1):56–64. doi: 10.1016/j.bbmt.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 28.Murry DJ, Crom WR, Reddick WE, Bhargava R, Evans WE. Liver volume as a determinant of drug clearance in children and adolescents. Drug Metab Dispos. 1995;23(10):1110–1116. [PubMed] [Google Scholar]
- 29.Kusama M, Kubota T, Matsukura Y, et al. Influence of glutathione S-transferase A1 polymorphism on the pharmacokinetics of busulfan. Clinica chimica acta; international journal of clinical chemistry. 2006 Jun;368(1–2):93–98. doi: 10.1016/j.cca.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 30.Johnson L, Orchard PJ, Baker KS, et al. Glutathione S-Transferase A1 Genetic Variants Reduce Busulfan Clearance in Children Undergoing Hematopoietic Cell Transplantation. J Clin Pharmacol. 2008 Jul 17; doi: 10.1177/0091270008321940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zwaveling J, Press RR, Bredius RG, et al. Glutathione S-transferase polymorphisms are not associated with population pharmacokinetic parameters of busulfan in pediatric patients. Ther Drug Monit. 2008 Aug;30(4):504–510. doi: 10.1097/FTD.0b013e3181817428. [DOI] [PubMed] [Google Scholar]
- 32.Ansari M, Lauzon-Joset JF, Vachon MF, et al. Influence of GST gene polymorphisms on busulfan pharmacokinetics in children. Bone Marrow Transplant. 2010 Feb;45(2):261–267. doi: 10.1038/bmt.2009.143. [DOI] [PubMed] [Google Scholar]
- 33.Hassan M, Oberg G, Bjorkholm M, Wallin I, Lindgren M. Influence of prophylactic anticonvulsant therapy on high-dose busulphan kinetics. Cancer Chemother Pharmacol. 1993;33(3):181–186. doi: 10.1007/BF00686213. [DOI] [PubMed] [Google Scholar]
- 34.Sandstrom M, Karlsson MO, Ljungman P, et al. Population pharmacokinetic analysis resulting in a tool for dose individualization of busulphan in bone marrow transplantation recipients. Bone Marrow Transplant. 2001 Oct;28(7):657–664. doi: 10.1038/sj.bmt.1703229. [DOI] [PubMed] [Google Scholar]
- 35.Kim I, Keam B, Lee KH, et al. Glutathione S-transferase A1 polymorphisms and acute graft-vs.-host disease in HLA-matched sibling allogeneic hematopoietic stem cell transplantation. Clin Transplant. 2007 Mar-Apr;21(2):207–213. doi: 10.1111/j.1399-0012.2006.00624.x. [DOI] [PubMed] [Google Scholar]
- 36.Slattery JT, Clift RA, Buckner CD, et al. Marrow transplantation for chronic myeloid leukemia: the influence of plasma busulfan levels on the outcome of transplantation. Blood. 1997 Apr 15;89(8):3055–3060. [PubMed] [Google Scholar]
- 37.Terakura S, Murata M, Nishida T, et al. Increased risk for treatment-related mortality after bone marrow transplantation in GSTM1-positive recipients. Bone Marrow Transplant. 2006 Feb;37(4):381–386. doi: 10.1038/sj.bmt.1705257. [DOI] [PubMed] [Google Scholar]
- 38.Kletzel M, Jacobsohn D, Duerst R. Pharmacokinetics of a test dose of intravenous busulfan guide dose modifications to achieve an optimal area under the curve of a single daily dose of intravenous busulfan in children undergoing a reduced-intensity conditioning regimen with hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2006 Apr;12(4):472–479. doi: 10.1016/j.bbmt.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 39.Ott MG, Schmidt M, Schwarzwaelder K, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006 Apr;12(4):401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 40.Aiuti A, Slavin S, Aker M, et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002 Jun 28;296(5577):2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- 41.Kahl CA, Tarantal AF, Lee CI, et al. Effects of busulfan dose escalation on engraftment of infant rhesus monkey hematopoietic stem cells after gene marking by a lentiviral vector. Exp Hematol. 2006 Mar;34(3):369–381. doi: 10.1016/j.exphem.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Salinger DH, Vicini P, Blough DK, O’Donnell PV, Pawlikowski MA, McCune JS. Development of a Population Pharmacokinetics-Based Sampling Schedule to Target Daily Intravenous Busulfan for Outpatient Clinic Administration. J Clin Pharmacol. 2010 Jan 14; doi: 10.1177/0091270009357430. [DOI] [PubMed] [Google Scholar]
- 43.Bleyzac N, Souillet G, Magron P, et al. Improved clinical outcome of paediatric bone marrow recipients using a test dose and Bayesian pharmacokinetic individualization of busulfan dosage regimens. Bone Marrow Transplant. 2001 Oct;28(8):743–751. doi: 10.1038/sj.bmt.1703207. [DOI] [PubMed] [Google Scholar]