Abstract
Cathepsin X is a lysosomal cysteine protease that functions as a carboxypeptidase with broad substrate specificity. Cathepsin X was discovered only recently, and its physiological roles are still not well understood. A number of studies suggest that cathepsin X may be involved in a variety of biological processes, including cancer, aging and degenerative conditions of the brain, inflammation, and cellular communication. Here we present the synthesis and characterization of several activity-based probes (ABPs) that target active cathepsin X. These ABPs were used to label cathepsin X in complex lysates, whole cells, and in vivo. Furthermore, we have developed a method for selectively labeling and visualizing active cathepsin X in vitro and in vivo. Overall, the probes developed in this study are valuable tools for the study of cathepsin X function.
Keywords: Cathepsins, Activity-based probes, Protease inhibitors, Cathepsin X
Introduction
Cathepsin X, also known as cathepsin Z or cathepsin P, is a lysosomal cysteine protease that belongs to the Clan CA/C1 cysteine protease family.(1, 2) Unlike most members of this protease family, cathepsin X functions solely as a carboxypeptidase and exhibits broad substrate specificity.(2, 3) The structure of cathepsin X is similar to that of other papain-like enzymes; however, it contains a distinctive mini-loop that extends into the active site of the enzyme and regulates its activity.(4) Cathepsin X is predominantly expressed in cells of the immune system, including monocytes/macrophages and dendritic cells.(5) This expression pattern suggests that cathepsin X may be involved in inflammatory processes.
Cathepsin X was discovered only recently, and its physiological roles are still not well understood.(6, 7) Cathepsin X may be involved in aging and pathological conditions of the brain, including amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease; increased protein levels and enzyme activity are observed in mouse models of these diseases, as well as in the brains of human patients with Alzheimer’s disease.(8) Cathepsin X may also play a role in cancer processes.(9–13) Both mRNA and protein expression levels of this enzyme are elevated in prostate tumors and gastric carcinomas.(9, 10) Additionally, in the RIP1-Tag2 mouse model of pancreatic cancer, cathepsin X activity is higher in pancreatic tumors than in normal islets.(11) Cathepsin X may also be involved in cellular communication.(2) Both the pro- and mature cathepsin X sequences contain integrin binding motifs, and cathepsin X binds to heparin sulfate proteoglycans.(2, 6, 7, 14) Furthermore, cathepsin X interacts with integrin αvβ3 and the integrin β2 subunit, thus regulating cellular adhesion, phagocytosis, and lymphocyte proliferation.(15–17)
Although cathepsin X seems to have important functions in a variety of normal and disease conditions, the details of cathepsin X function remain elusive. Many of the putative functions of cathepsin X have been proposed based on expression profiles and protein abundance and localization studies. However, because many enzymes are expressed as inactive zymogens or in complex with their endogenous protein inhibitors, protein abundance and gene expression often do not allow the assessment of functional regulation of a protease in a given biological process. In this study, we synthesized fluorescent activity based probes (ABPs) that label active cathepsin X and used these probes to label this protease in complex lysates, whole cells, and in vivo. Additionally, we developed a method for selectively labeling and visualizing active cathepsin X in vitro and in vivo. The probes developed in this study will be useful for future studies of the biological roles of cathepsin X.
RESULTS AND DISCUSSION
Probe Design
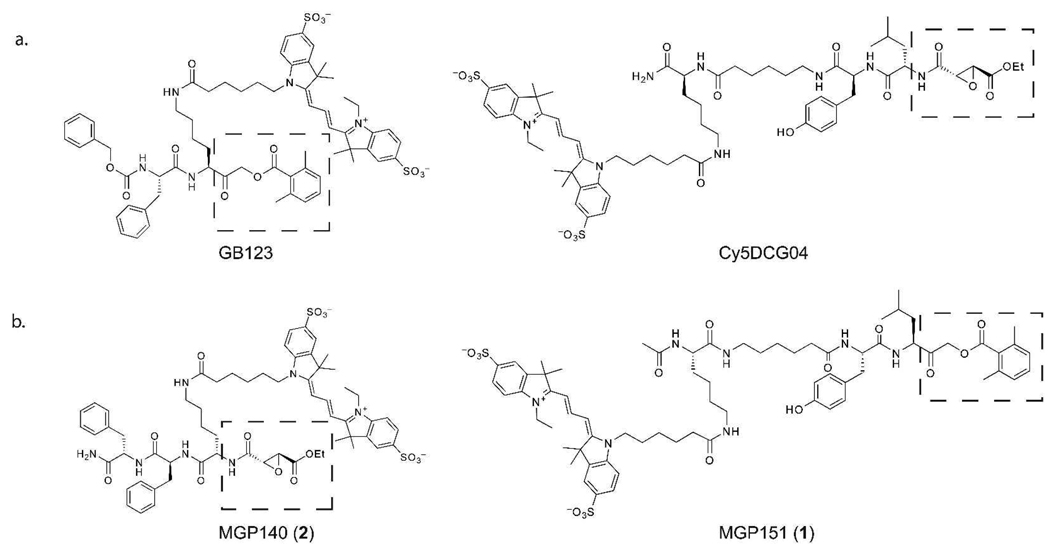
Our group and others have developed ABPs that selectively label the cysteine cathepsins both in vitro and in vivo.(18–24) Two commonly used cysteine cathepsin ABPs are DCG-04 and GB123.(18, 22) DCG-04 was originally designed as a selective activity-based probe of papain family cysteine cathepsins (18). It contains an epoxide warhead that covalently reacts with the active site cysteine residue as well as a biotin tag attached via a lysine sidechain. This biotin group can be easily replaced with a fluorescent tag to yield Cy5DCG04 (Figure 1, panel a). GB123, like DCG-04 is also a broad-spectrum probe for cysteine cathepsins (22). It contains an acyloxymethylketone (AOMK) reactive electrophile and a Cy5 fluorophore as a tag (Figure 1, panel a). DCG-04 has been reported to label numerous cysteine cathepsins in cell and tissue lysates, including cathepsins B, C, H, J, K, L, S, V, and X.(18, 19, 25) GB123, on the other hand, labels only cathepsins B, S, and L.(21, 22) We synthesized Cy5DCG04 via a previously reported solid-phase synthesis technique, and the Cy5 fluorophore was conjugated to the purified peptide at the final step (Supplementary Scheme 1).(18, 19) We then used GB123 and Cy5DCG04 to label proteins in total rat liver extracts and in lysates from NIH-3T3 fibroblasts (Figure 2, panel a). The probes labeled different subsets of the cathepsins, and cathepsin labeling was specifically competed by pretreatment with JPM-OEt, a broad-spectrum cysteine cathepsin inhibitor.(11) Interestingly, Cy5DCG04 labeled cathepsin X, while GB123 did not label this protease, even when used at high concentrations. Cy5DCG04 could also be used at lower concentrations in cell and organ lysates than GB123.
Figure 1. Structures of cysteine cathepsin activity-based probes.
a) Structures of the activity-based probes Cy5DCG04 and GB123 containing epoxide and AOMK reactive electrophiles, respectively (see dashed boxes). b) Structures of MGP151 (1) and MGP140 (2) and, cysteine cathepsin activity-based probes designed based on DCG-04 and GB123. Note these probes have swapped electrophiles (see dashed boxes) relative to the original DCG-04 and GB123 probes.
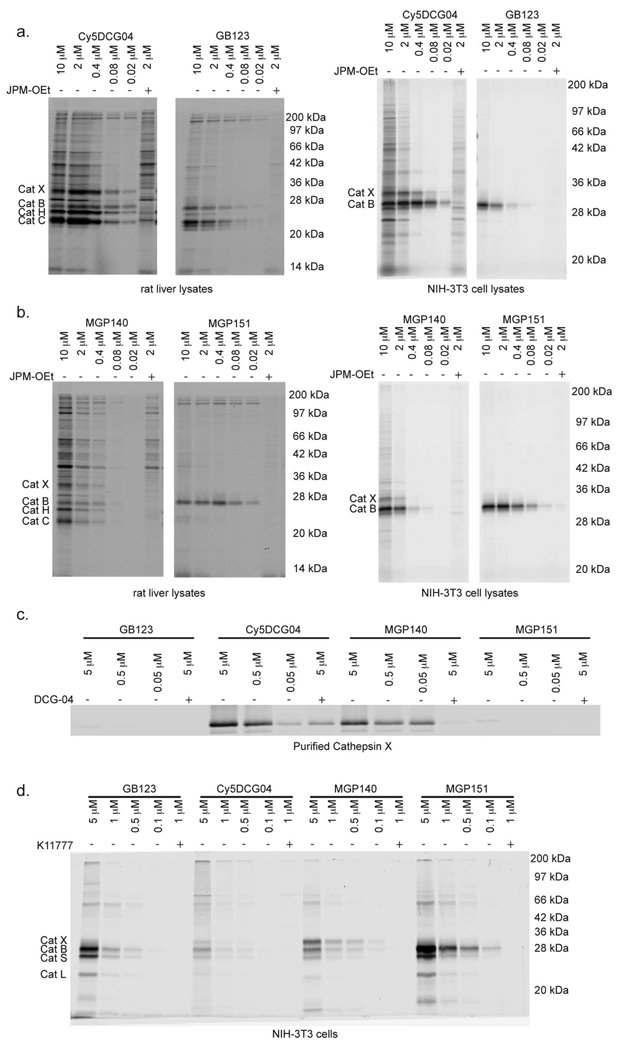
Figure 2. Direct comparison of cysteine cathepsin labeling in intact cells.
a) Where indicated, rat liver lysates or NIH-3T3 lysates were pretreated with the cysteine cathepsin inhibitor JPM-OEt and labeled by addition of Cy5DCG04 or GB123 at the indicated concentrations. Labeled proteins were analyzed by SDS-PAGE, followed by scanning of the gel using a flatbed laser scanner. b) Where indicated, rat liver lysates or NIH-3T3 lysates were pretreated with JPM-OEt and labeled by MGP140 or MGP151 at the indicated concentrations and labeled proteins analyzed as in (a). c) Labeling of purified cathepsin X by GB123, Cy5DCG04, MGP140, and MGP151. Where indicated, cathepsin X was pretreated with the cysteine cathepsin inhibitor DCG-04. d) Direct labeling of cysteine cathepsins in intact NIH-3T3 fibroblasts by GB123, Cy5DCG04, MGP140, and MGP151. Where indicated, cells were pretreated with the cell-permeable cysteine cathepsin inhibitor K11777 before probe labeling. Labeled proteins were analyzed as in (a).
Cy5DCG04 and GB123 use different reactive functional groups to covalently modify target proteases. Cy5DCG04 contains an epoxide electrophile, whereas GB123 contains an AOMK electrophile. In order to determine if the reactive electrophile was responsible for the differences in cathepsin labeling, we designed and synthesized ABPs: MGP151 (1), which contains the primary peptide sequence of DCG-04, but uses an AOMK electrophile and MGP140 (2), which contains the epoxide electrophile attached to the lysine and phenylalanine peptide from GB123 (Figure 1). It should also be noted that, while the peptide scaffolds of MGP151 and MGP140 contain the same amino acids found in the parent compounds, they have reverse polarity relative to their original counterparts due to the placement of the reactive electrophile. We synthesized these two probes via previously reported solid-phase synthesis techniques (Supplementary Schemes 2 and 3).(18, 22)
We first used the probe to label total rat liver extracts and lysates from NIH-3T3 cells at a variety of concentrations (Figure 2, panel b). MGP140 and MGP151 labeled two different subsets of cathepsins in these lysates. Cathepsin labeling could be specifically competed by pretreatment of the lysates with JPM-OEt suggesting that both retain specificity for cysteine cathepsins. Similar to Cy5DCG04, MGP140 labeled multiple cysteine cathepsins, including cathepsins X, B, H, and C. MGP151, on the other hand, was much more selective and labeled only cathepsin B in both lysates. MGP151 was also a more potent label than MGP140.
To confirm that only the epoxide-containing probes were capable of labeling cathepsin X, we labeled purified enzyme with all four probes (Figure 2, panel c). As expected, both of the epoxide-containing probes, Cy5DCG04 and MGP140, labeled cathepsin X in a dose-dependent manner, while the AOMK-containing probes, GB123 and MGP151, failed to label this protease. These results suggest that labeling of cathepsin X by ABPs is due to the epoxide electrophile and not due to the peptide binding region. The crystal structure of human cathepsin X offers some insight into these results.(4) Cathepsin X exhibits an S1 substrate binding site that is somewhat restrictive towards substrates and therefore may not accommodate the steric bulk of the AOMK electrophile.(4)
We subsequently wanted to verify that these probes label active cathepsin X, as well as other cathepsins, in intact NIH-3T3 cells (Figure 2, panel d). All of the probes labeled the cysteine cathepsins, and labeling could be blocked by pretreatment of the cells with the pan-cathepsin inhibitor K11777.(26) As previously observed in lysates, the epoxide-containing probes, Cy5DCG04 and MGP140, labeled cathepsin X in addition to other cathepsins, including cathepsins B, S, and L, whereas the AOMK-containing probes did not label cathepsin X. Interestingly, MGP140 was a more potent cathepsin probe in intact cells than Cy5DCG04. This is in direct contrast to what we observed in lysates, where Cy5DCG04 was more potent than MGP140 (Figures 2, panels a and b). This result could be due to differential uptake of the two probes. MGP140 is more hydrophobic than Cy5DCG04; therefore it is likely that MGP140 more effectively enters the lysosomes in intact cells and labels the lysosomal cathepsins. Of all the probes tested in intact NIH-3T3 cells, MGP151 was the most selective for cathepsin B. Additionally, all the probes labeled cathepsins S and L in intact cells, but not in lysates. It is possible that the preparation of cellular lysates may cause release of endogenous protein inhibitors that block the activities of multiple cathepsins. Nevertheless, these data indicate that epoxide-containing probes, such as Cy5DCG04 and MGP140, are useful compounds for labeling active cathepsin X in whole cells. Furthermore, these results demonstrate that MGP151 is an effective cathepsin B-selective probe in both lysates and in intact cells.
Epoxide-containing ABPs label cathepsin X in vivo
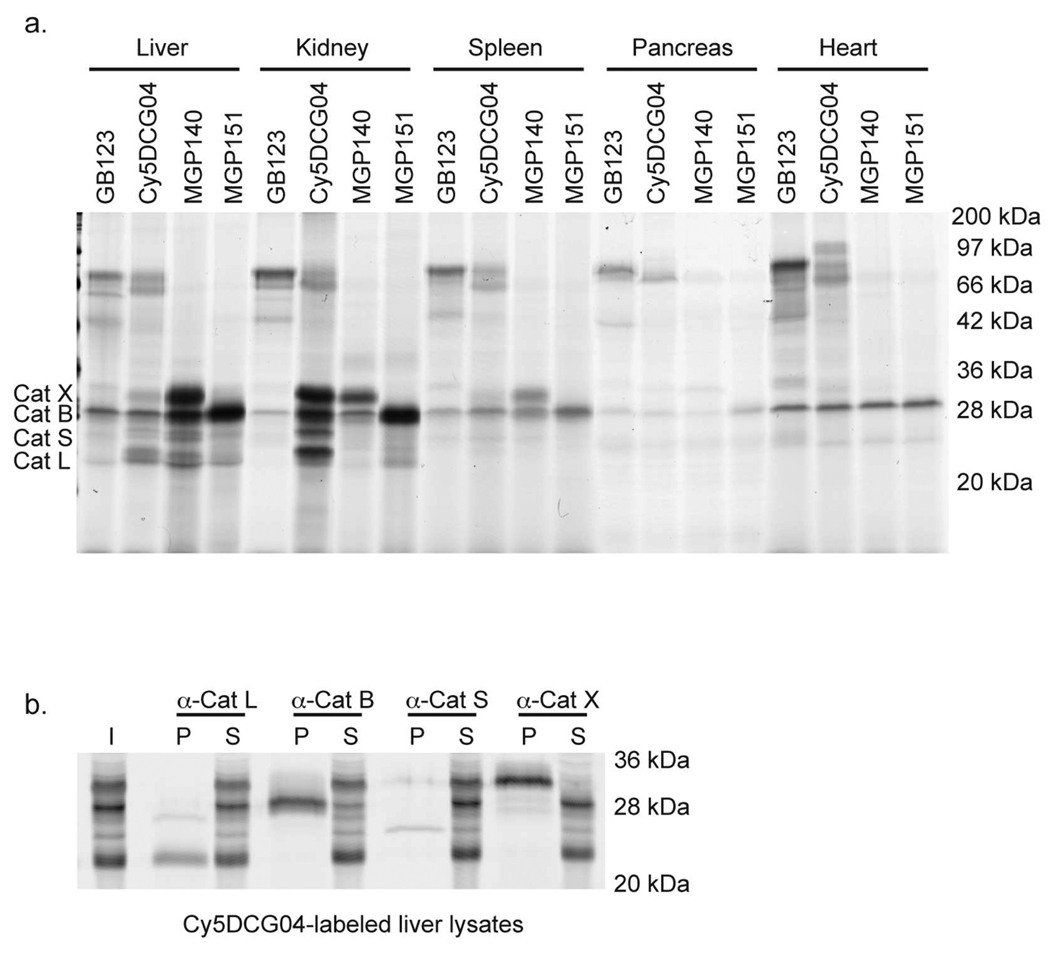
We next evaluated our probes for cathepsin labeling in vivo. All four probes were intravenously (i.v.) injected into mice and allowed to circulate for 2 h, after which the mice were sacrificed, and their organs collected for biochemical analysis (Figure 3, panel a). Similar to our results obtained in intact cells, all of the probes labeled the cathepsins in multiple organs. The identities of the labeled proteins were confirmed by immunoprecipitation with specific antibodies (Figure 3, panel b). As observed in intact cells, the epoxide-containing probes Cy5DCG04 and MGP140 labeled cathepsin X, as well as other cathepsins. Interestingly, MGP151, which contains an AOMK warhead, retained its selectivity for cathepsin B in vivo. Overall, these results demonstrate that the observed specificity of the cathepsin probes in intact cells is retained in vivo. However, the labeling intensities for the probes in intact NIH-3T3 cells differed from those observed in vivo. This result could be due to differential uptake of the probes in intact cells and whole organs. Finally, these data show that MGP151 is a potentially valuable cathepsin B-selective probe that can be used in lysates, intact cells, and in vivo.
Figure 3. Direct comparison of cysteine cathepsin labeling in vivo.
a) Biochemical analysis of tissue extracts from mice injected with GB123, Cy5DCG04, MGP140, or MGP151. The probes were allowed to circulate for 2 h, after which the mice were sacrificed. Tissue lysates from liver, kidney, spleen, pancreas, and heart were analyzed by SDS-PAGE, and labeling of the cysteine cathepsins is indicated. b) Immunoprecipitation of labeled cysteine cathepsins L, S, B, and X from liver lysates from mice injected with Cy5DCG-04. The proteases were immunoprecipitated using specific cathepsin antibodies and analyzed by SDS-PAGE, followed by scanning of the gel for Cy5 fluorescence. Input (I), immunoprecipitated pellet (P), and supernatant (S) samples are indicated.
Design, synthesis, and evaluation of more selective ABPs for cathepsin X
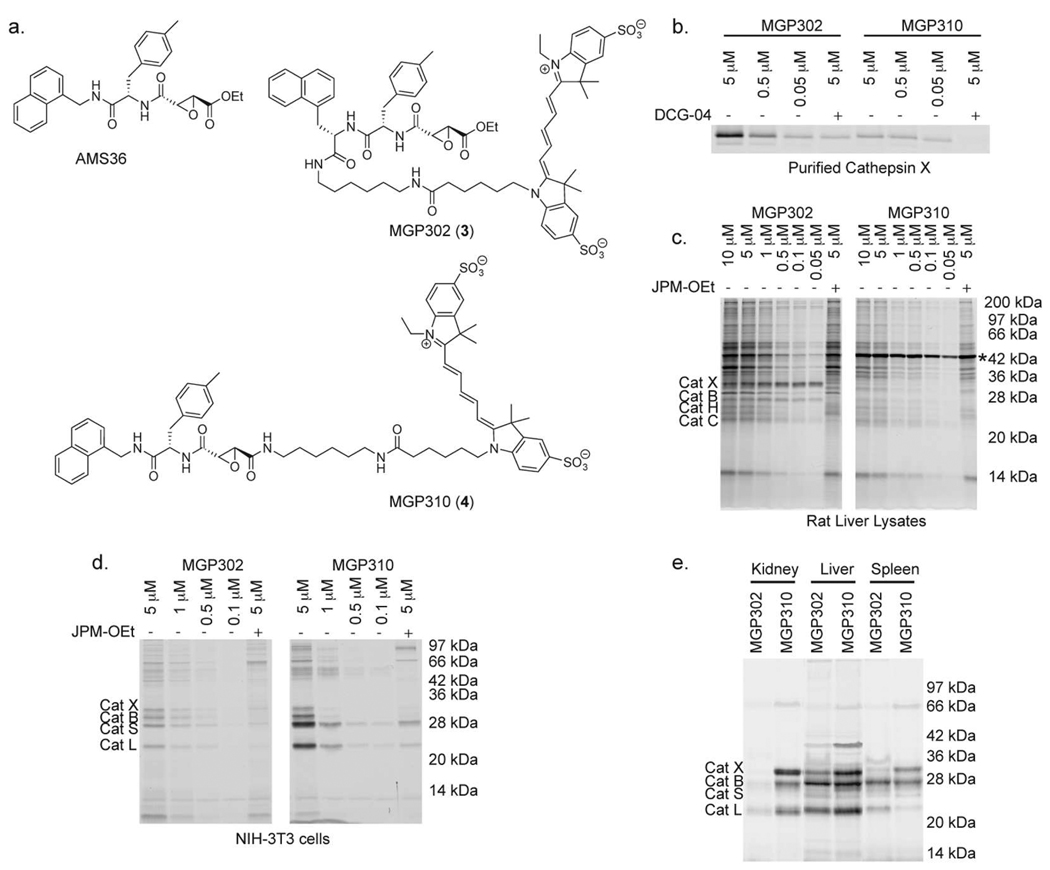
Although both Cy5DCG04 and MGP140 label cathepsin X efficiently in lysates, whole cells, and in vivo, neither are very selective for this protease. Previously, we developed an inhibitor that was selective for cathepsin X (AMS36; Figure 4, panel a).(27) We therefore decided to develop probes based on this general structure. We synthesized two probes, MGP302 (3) and MGP310 (4), that differ in the placement of the Cy5 fluorophore. In MGP302, the Cy5 fluorophore is attached at the N-terminal end of the main peptide sequence, while, in MGP310, the Cy5 fluorophore is attached on the opposite side of the epoxide from the peptide recognition elements. We synthesized both probes via previously reported solid-phase synthesis techniques (Supplementary Schemes 4 and 5).(28)
Figure 4. Structures and labeling selectivity of MGP302 and GP310.
a) Structures of AMS36, a cathepsin X-selective inhibitor, and MGP302 (3) and MGP310 (4). b) Comparison of labeling of purified cathepsin X by MGP302 and MGP310. Where indicated, cathepsin X was pretreated with the cysteine cathepsin inhibitor DCG04. c) Comparison of labeling of rat liver lysates by MGP302 and MGP310. Where indicated, rat liver lysates were pretreated with JPM-OEt and labeled by MGP140 or MGP151 at the indicated concentrations. * indicates a labeled protein that was identified as bile acid-CoA:amino acid N-acyltransferase (BACAT; Supplementary Figure 1). d) Direct labeling of cysteine cathepsins in intact NIH-3T3 cells with MGP302 and MGP310. Where indicated, cells were pretreated with JPM-OEt before probe labeling. Cells were lysed, and the labeled proteins were analyzed by SDS-PAGE, followed by scanning of the gel for Cy5 fluorescence. e) Balb/c mice were injected with MGP302 or MGP310. The probes were allowed to circulate for 2 h, after which the mice were sacrificed. Tissue lysates from kidney, liver, and spleen were analyzed by SDS-PAGE, and labeling of the cysteine cathepsins is indicated.
Both probes labeled cathepsin X in a dose-dependent manner, although MGP302 labeled this protease more strongly than MGP310 (Figure 4, panel b). We next used MGP302 and MGP310 to label rat liver lysates (Figure 4, panel c). MGP302 showed a preference for cathepsin X, especially at low concentrations (i.e., 50 nM). Surprisingly, MGP310 did not label any of the cysteine cathepsins, but labeled a 42 kDa protein (Figure 4, panel c). Isolation of the labeled protein using the biotin-tagged probe bMGP310, identified this protein as bile acid-CoA:amino acid N-acyltransferase (BACAT), an N-acyltransferase found in the liver that is essential for the digestion of dietary cholesterol (Supplementary Figure 1).(29) BACAT contains a cysteine residue in its active site, and this residue is most likely the site of covalent labeling by MGP310. Labeling of BACAT by MGP310 was particularly surprising as we had never observed specific labeling of any non-cathepsin enzymes by analogs of E-64 (a natural product inhibitor of the cathepsins that contains an epoxide warhead like DCG-04). Furthermore, we did not label BACAT with MGP310 in various liver cell lines and other tissues, suggesting that labeling could be due to very high levels of this enzyme in liver tissue extracts (Figure 4, panel d and M.G.P and M.B., unpublished data).
We next evaluated cathepsin labeling in intact NIH-3T3 cells using MGP302 and MGP310 (Figure 4, panel d). Although MGP302 was fairly selective for cathepsin X in lysates, this probe labeled multiple cathepsins in intact cells. Unexpectedly, MGP310, also labeled numerous cathepsins in intact cells. This is likely due to the fact that ABPs such as MGP302 and MGP310 accumulate in lysosomes, leading to labeling of multiple lysosomal cathepsins. To further characterize the properties of these probes, we performed in vivo labeling studies (Figure 4, panel e). As seen in intact cells, MGP310 labeled numerous cathepsins. In fact, MGP310 showed increased labeling of multiple cathepsins compared to MGP302. Overall, the data we obtained in intact cells and in vivo suggest that it may be difficult to obtain ABPs that are absolutely selective for cathepsin X. Even when probes are fairly selective in lysates, they are often non-selective when used in intact cells or in vivo due to accumulation in specific tissues and organelles.
Selective labeling of active cathepsin X in intact cells and in vivo
We found that the epoxide-based probes were efficient labels of cathepsin X, while the AOMK probes were uniformly unable to label this protease. Therefore, we reasoned that it should be possible to treat cells with a previously reported AOMK containing inhibitor, GB111-NH2 (26), to block the active sites of all cysteine cathepsins except for cathepsin X and then specifically label cathepsin X with an epoxide-containing ABP, such as Cy5DCG04 or MGP140. We pretreated NIH-3T3 cells with GB111-NH2 and then labeled with Cy5DCG04, MGP140, and MGP302 (Figure 5, panel a). Using this approach, we were able to selectively label cathepsin X. Importantly, pretreatment of the cells with saturating concentrations of GB111-NH2 produced no ill effects; cells treated with these compounds exhibited similar morphology and growth as cells treated with DMSO as a control (M.G.P. and M.B., unpublished data).
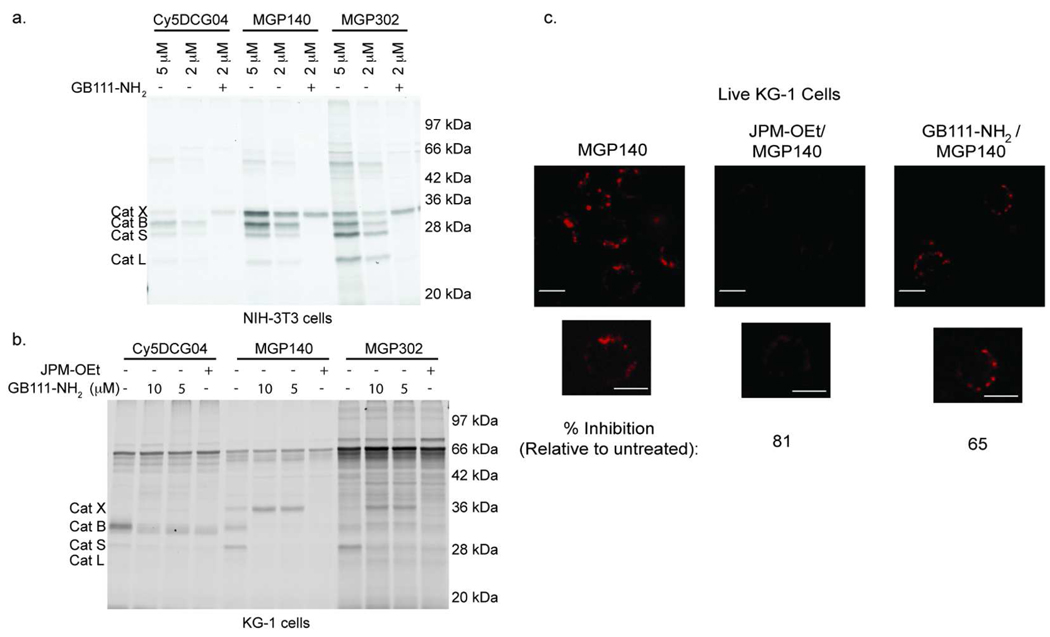
Figure 5. Selective labeling and fluorescent imaging of cathepsin X in lysates and in intact cells.
a) Cells were pretreated with the cysteine cathepsin inhibitor GB111-NH2, followed by labeling with an epoxide-containing cathepsin probe at the indicated concentrations (Cy5DCG04, MGP140, or MGP302). Cells were then lysed, and the labeled proteins were analyzed by SDS-PAGE, followed by scanning of the gel for Cy5 fluorescence. b) Selective labeling of cathepsin X in intact KG-1 cells. Cells were pretreated with the cysteine cathepsin inhibitor GB111-NH2 at the indicated concentrations, followed by labeling with an epoxide-containing cathepsin probe (Cy5DCG04, MGP140, or MGP302). Cells were then lysed, and the labeled proteins were analyzed by SDS-PAGE, followed by scanning of the gel for Cy5 fluorescence. The 66 kDa protein band is serum albumin in the media that non-selectively binds ABPs. c) Live KG-1 cells were treated with MGP140 (left panel), JPM-OEt and then MGP140 (center panel), or GB111-NH2 and then MGP140 (right panel). Images were taken with a 40X objective. Red is Cy5 fluorescence. Scale bar is 10 µm. Quantification of fluorescence intensities was determined using ImageJ software. These data were used to calculate numerical values for percent inhibition as indicated.
We next applied this method to label active cathepsin X in intact KG-1 cells. KG-1 cells are a human myeloblast cell line that differentiates to macrophage-like cells.(30) Notably, KG-1 cells express significant amounts of active cathepsin X and have been used in studies to evaluate the biological roles of cathepsin X.(30) We first pretreated intact KG-1 cells with GB111-NH2, or JPM-OEt as a control, and then incubated the cells with Cy5DCG04, MGP140, or MGP302 (Figure 5, panel b). Both MGP140 and MGP302 effectively labeled cathepsin X, however MGP302 gave significantly more background labeling. Based on these results, we chose to use a combination of GB111-NH2 and MGP140 for all further experiments.
We next used our method to fluorescently image active cathepsin X in intact KG-1 cells. Live KG-1 cells were treated with MGP140 to label all active cathepsins, or with a combination of GB111-NH2 and MGP140 to label only cathepsin X. As a control for non-specific fluorescence, KG-1 cells were also pretreated with JPM-OEt, followed by labeling with MGP140. Treatment of intact KG-1 cells with MGP140 or GB111-NH2/MGP140, followed by imaging of the live cells by fluorescence microscopy gave a distinct punctuate labeling pattern (Figure 5, panel c). This labeling pattern was blocked by pretreatment of the cells with JPM-OEt. Furthermore, treatment of KG-1 cells with GB111-NH2 prior to labeling with MGP140 resulted in a reduction of overall labeling signal compared to the untreated cells. This signal overlapped with the signal from the lysosomal marker LysoTracker (Supplementary Figure 2). Quantification of the total average fluorescence signal per cell after background subtraction indicated that JPM-OEt blocked 81% of the probe signal while GB111-NH2 blocked only 65% (Supplementary Table 1). This partial block by GB111-NH2 suggests that the residual signal is due to active cathepsin X as confirmed by our SDS-PAGE analysis. Furthermore our results are highly consistent with the labeling patterns seen when KG-1 cells are treated with a cathepsin X-specific antibody.(19, 21, 30) Overall, these data confirm that our method for labeling active cathepsin X selectively in intact cells can also be used for imaging active cathepsin X in live cells.
We subsequently tested whether the inhibitor pretreatment method for labeling and imaging cathepsin X selectively would work in vivo (Figure 6, panel a). Overall, we observed selective cathepsin X labeling in both the liver and kidneys from mice treated with a combination of GB111-NH2 and MGP140. In the spleen and heart tissues of these mice, we found only weak labeling of cathepsin X and some indication of weak labeling of other cathepsins even after pre-treatment with GB111-NH2. This result could be due to a combination of poor biodistribution of GB111-NH2 in those tissues coupled with overall low levels of cathepsin X. Regardless, results from these mouse experiments demonstrate that a combination of GB111-NH2 and MGP140 can be used to give selective labeling of cathepsin X in vivo.
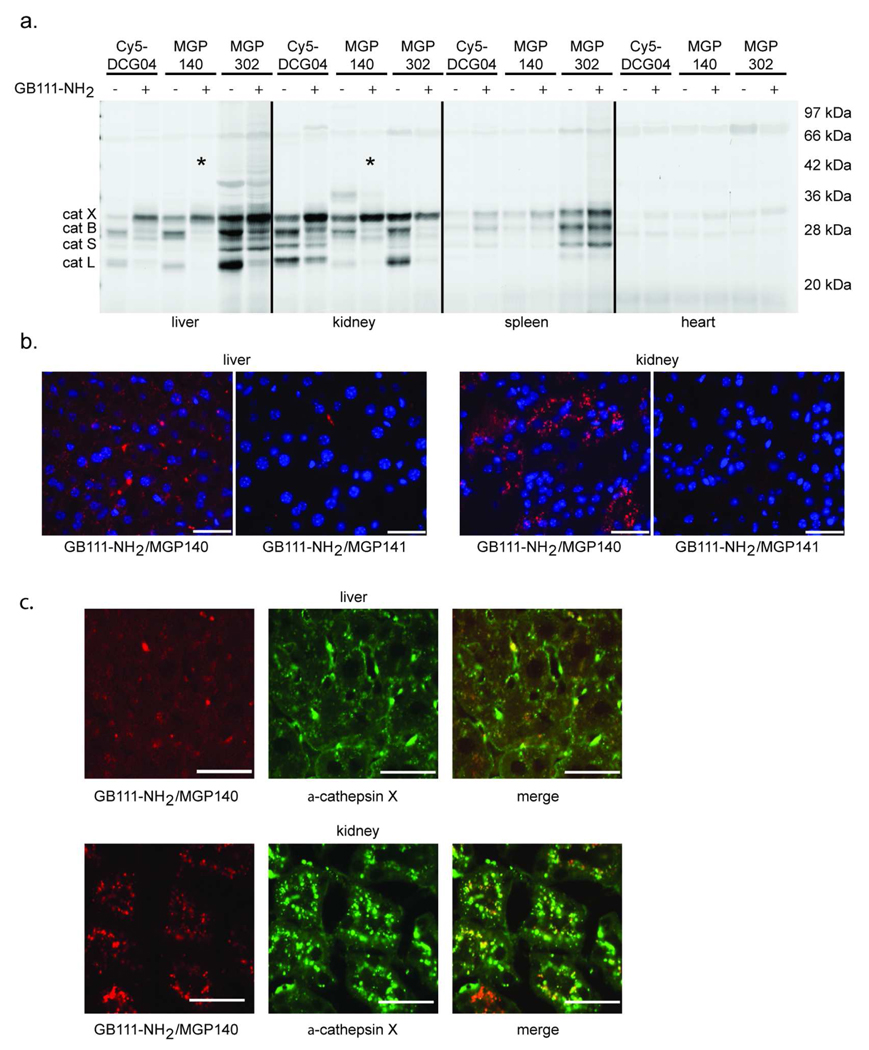
Figure 6. Selective labeling of cathepsin X in vivo.
a) Balb/c mice were injected with GB111-NH2 and then injected with Cy5DCG04, MGP140, or MGP302. The probes were allowed to circulate for 2 h, after which the mice were sacrificed. Tissue lysates from liver, kidney, spleen, and heart were analyzed by SDS-PAGE, and labeling of the cysteine cathepsins is indicated. Selective labeling of cathepsin X is with a *. b) Mice were injected with GB111-NH2, followed by injection with MGP140 or MGP141. MGP141 is a control probe for MGP140 that lacks the epoxide electrophile. Tissues not used in (a) were frozen in OCT medium and sectioned. Tissues were stained with DAPI, and images were taken with a 40X objective. Blue is DAPI fluorescence, and red is Cy5 fluorescence. Scale bar is 30 µm. c) Tissue sections from (b) were frozen in OCT medium, sectioned and then treated with goat anti-cathepsin X, followed by a FITC-anti-goat antibody. Images were taken with a 40X objective. Red is GB111-NH2/MGP140, green is anti-cathepsin X, and yellow is the overlap of green and red signals. Scale bar is 30 µm.
Finally, we examined histological sections of the livers and kidneys from mice treated with GB111-NH2 and either MGP140 or MGP141, a control probe for MGP140 that lacks the epoxide electrophile thatdoes not label any of the cysteine cathepsins in vivo (Supplementary Figure 3). Strong Cy5 fluorescence was observed in tissues from mice treated with GB111-NH2 and MGP140 that was not seen in tissues from mice treated with GB111-NH2 and the control probe MGP141 (Figure 6, panel b). Additionally, in tissues from mice treated with GB111-NH2 and MGP140, the Cy5 fluorescence from ABP labeling overlapped significantly with the FITC fluorescence from antibody staining with a cathepsin X antibody (Figure 6, panel c).
Conclusion
In conclusion, we have synthesized ABPs that label the cysteine cathepsins in lysates, intact cells, and in vivo. Probes containing either an epoxide or an AOMK reactive electrophile can be used to label the cysteine cathepsins; however, probes that contain a bulky AOMK group are unable to label cathepsin X. Using this knowledge, we developed a method to selectively label and visualize active cathepsin X in vitro and in vivo. To accomplish this, intact cells or mice can be first treated with GB111-NH2, an inhibitor that blocks the active sites of all of the cysteine cathepsins except for cathepsin X, followed by labeling with an epoxide-containing ABP. We demonstrated the utility of this method for the labeling and visualization of active cathepsin X in intact cells and in living mice. The chemical tools and methods developed in this study will be valuable to the study of cathepsin X and its biological functions both in vitro and in vivo. We are currently working to apply these tools and methods to mouse models of cancer and other human diseases.
METHODS
General Methods
Purified human cathepsin X was a generous gift from V. Turk (Jozef Stefan Institute, Slovenia). For all lysate and whole cell experiments, the inhibitors and probes were dissolved in DMSO, and the final DMSO concentration of the reactions was maintained at ≤ 0.2%. Fluorescent gels were scanned with a Typhoon 9400 flatbed laser scanner (GE Healthcare). Fluorescent images were acquired on a Zeiss Axiovert 200 M inverted microscope equipped with a 40X objective (Carl Zeiss). Slidebook software was used to control the microscope and camera and for data analysis (Intelligent Imaging Innovations). Quantification of fluorescence intensities was determined using ImageJ software (National Institutes of Health). These data were then used to calculate numerical values for percent inhibition. Male Balb/c nude mice (8 weeks old) were obtained from Charles River, and female Balb/cJ mice (8 weeks old) were obtained from Jackson Labs. All mice were housed in the research animal facility at the Stanford University Department of Comparative Medicine. All animal protocols were approved by the Stanford University Administrative Panel on Laboratory Animal Care, and the procedures were performed in accordance with their guidelines.
Compound synthesis and characterization
Please see the Supporting Information for detailed synthetic procedures and characterization data of final compounds.
Direct labeling of endogenous cathepsin activity in cell and organ lysates
Rat liver lysates (50 µg) or NIH-3T3 cell lysates (25 µg) were diluted to 1 mg mL−1 in reaction buffer (50 mM NaOAc, 2 mM dithiothreitol (DTT), 5 mM MgCl2, pH = 5.5) and subjected to direct labeling. Lysate samples were pretreated with inhibitor (50 µM JPM-OEt) for 30 min at room temperature and labeled with probes for another 30 min at room temperature. The labeled samples were separated by 12% SDS-PAGE and analyzed by scanning the gel with a Typhoon flatbed laser scanner (ex 633nm/em 670 nm).
Direct labeling of purified cathepsin X activity
Purified cathepsin X (75 ng) was diluted to 3 ng µL−1 in reaction buffer (50 mM NaOAc, 2 mM DTT, 5 mM MgCl2, pH = 5.5) and subjected to direct labeling. The samples were pretreated with inhibitor (100 µM DCG-04) for 30 min at room temperature and labeled with probes for another 30 min at room temperature. The labeled samples were separated by 15% SDS-PAGE and analyzed by scanning the gel with a Typhoon flatbed laser scanner.
Direct labeling of endogenous cathepsin activity in intact cells
NIH-3T3 cells (250,000 cells/well) were seeded in a 6-well plate 24 h prior to labeling. Cells were pretreated with inhibitor (100 µM K11777 in complete media) for 1 h at 37 °C, followed by labeling with each probe for 3 h at 37 °C. Cells were rinsed with phosphate-buffered saline (PBS) and pelleted. The cells were lysed in 30 µL of citrate lysis buffer (50 mM sodium citrate, 50 mM sodium phosphate, 1% CHAPS, 0.5% Triton X-100, pH = 4.2), and the lysates were separated by 12% SDS-PAGE and analyzed by scanning the gel with a Typhoon flatbed laser scanner.
Direct labeling of cathepsin activity in vivo
Probes (25 nmol in 10% DMSO in sterile PBS) were i.v. injected into male Balb/c nude mice (15–16 weeks old) and allowed to circulate for 2 h. The mice were sacrificed, and the livers, kidneys, pancreas, hearts, and spleens were removed and flash-frozen in liquid nitrogen. The organs were lysed by a Dounce homogenizer in muscle lysis buffer (PBS, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 0.2% sodium azide, pH = 7.4). Total protein extracts (150 µg) were separated by 12% SDS-PAGE and analyzed by scanning the gel with a Typhoon flatbed laser scanner.
Immunoprecipitation of cathepsins from Cy5DCG04-labeled mouse liver lysates
In vivo-labeled liver lysates (400 µg total protein) were diluted in 500 µL RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, pH = 8.0) and preincubated with the indicated amount of the indicated antibody for 30 min at 4 °C. Antibodies used were as follows: polyclonal anti-mouse cathepsin X antibody (2 µg, R&D Systems, cat. # AF1033), polyclonal anti-mouse cathepsin B antibody (2 µg, R&D Systems, cat. # AF965), polyclonal anti-mouse cathepsin L antibody (2 µg, R&D Systems, cat. # AF1515), and polyclonal anti-mouse cathepsin S antibody (2.5 µg, Abcam, cat. # ab18822). 30 µL of Protein A/G agarose beads were then added to the above solutions, and the reactions were incubated for 20 h at 4 °C. The reactions were then centrifuged (1 min × 13,200 rpm), and the supernatant was removed. After being washed in RIPA buffer (3 × 500 mL), the beads were boiled in 2X SDS sample loading buffer for 5 min. All supernatant samples were acetone-precipitated for 2 h at −80 °C, dried, and resuspended in 1X SDS sample loading buffer. Samples were separated by 12% SDS-PAGE and analyzed by scanning the gel with a Typhoon flatbed laser scanner.
Direct labeling of endogenous cathepsin X activity in intact cells
NIH-3T3 cells (250,000 cells/well) or KG-1 cells (1,000,000 cells/well) were seeded in a 6-well plate 24 h prior to labeling. Cells were pretreated with GB111-NH2 (5–10 µM in complete media) for 1.5 h at 37 °C, followed by labeling with each probe for 2 h at 37 °C. Cells were rinsed with phosphate-buffered saline (PBS) and pelleted. The cells were lysed in 30 µL of citrate lysis buffer, and the lysates were separated by 12% SDS-PAGE and analyzed by scanning the gel with a Typhoon flatbed laser scanner.
Imaging cathepsin X activity in live cells
KG-1 cells (400,000 cells/well) were seeded in a 4-well Lab-Tek chambered coverglass system pretreated with 0.01% poly-L-lysine 24 h prior to labeling. Cells were pretreated with DMSO, GB111-NH2 (10 µM in complete media), or JPM-OEt (100 µM in complete media) for 1 h at 37 °C, followed by labeling with 5 µM MGP140 for 30 min at 37 °C. Cells were rinsed with complete media containing DMSO, 10 µM GB111-NH2, or 100 µM JPM-OEt for 3 h at 37 °C (in order to wash away excess unbound probe). Cells were then subjected to fluorescence microscopy at 25 °C.
Direct labeling of cathepsin X activity in vivo
GB111-NH2 (175 nmol in 40% DMSO in sterile PBS) or DMSO was i.v. injected into female Balb/cJ mice (8–10 weeks old) and allowed to circulate for 2 h. Then, probes (25 nmol in 10% DMSO in sterile PBS) were i.v. injected into the mice and allowed to circulate for 2 h. The mice were sacrificed, and the livers, kidneys, hearts, and spleens were removed and flash-frozen in liquid nitrogen. The organs were lysed by a Dounce homogenizer in muscle lysis buffer, and total protein extracts (150 µg) were separated by 12% SDS-PAGE and analyzed by scanning the gel with a Typhoon flatbed laser scanner.
Histology and immunofluorescent analysis of cathepsin X activity in tissue from mice treated with probes in vivo
Tissues from mice injected with GB111-NH2/MGP140 or GB111-NH2/MGP141 were frozen in OCT medium (an embedding medium for frozen tissue to ensure Optimal Cutting Temperature) and sectioned. Some sections were fixed with acetone, rehydrated, stained with DAPI, and subjected to fluorescence microscopy. Other sections were fixed with acetone, rehydrated, and subjected to immunofluorescence staining with an anti-cathepsin X antibody (R&D Systems, cat. # AF1033). Briefly, after rehydration in PBS, slides were blocked for 1 h at room temperature with 5% rabbit serum in PBS. Then, the slides were treated with anti-cathepsin X (1:200 dilution in 2.5% rabbit serum and 0.3% Triton X-100 in PBS) for 24 h at 4 °C in a humidified chamber. Slides were rinsed with PBS (3 × 8 min) and then treated with FITC-anti-goat (Sigma-Aldrich, 1:400 dilution in 2.5% rabbit serum and 0.3% Triton X-100 in PBS) for 1 h at room temperature in a humidified chamber. The slides were rinsed with PBS (3 × 8 min), stained with DAPI, and subjected to fluorescence microscopy.
Supplementary Material
Acknowledgments
We thank J. Lee and F. Yin for assistance with the mouse injections. We also thank P.W. Bowyer and J.C. Valderramos for mass spectrometry analysis, and V.E. Albrow for helpful synthetic discussions. M.G.P. was supported by an American Lung Association Senior Research Training Fellowship and by Public Health Services grant T32 CA09302, awarded by the US National Cancer Institute, Department of Health and Human Services. This work was supported by grants from the NIH (R01 EB005011) and the American Asthma Foundation (Early Excellence Award).
Footnotes
Supporting Information Available: This material is available free of charge via the Internet.
References
- 1.Deussing J, von Olshausen I, Peters C. Murine and human cathepsin Z: cDNA-cloning, characterization of the genes and chromosomal localization. Biochim Biophys Acta. 2000;1491:93–106. doi: 10.1016/s0167-4781(00)00021-x. [DOI] [PubMed] [Google Scholar]
- 2.Kos J, Jevnikar Z, Obermajer N. The role of cathepsin X in cell signaling. Cell Adh Migr. 2009;3:164–166. doi: 10.4161/cam.3.2.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devanathan G, Turnbull JL, Ziomek E, Purisima EO, Menard R, Sulea T. Carboxy-monopeptidase substrate specificity of human cathepsin X. Biochem Biophys Res Commun. 2005;329:445–452. doi: 10.1016/j.bbrc.2005.01.150. [DOI] [PubMed] [Google Scholar]
- 4.Guncar G, Klemencic I, Turk B, Turk V, Karaoglanovic-Carmona A, Juliano L, Turk D. Crystal structure of cathepsin X: a flip-flop of the ring of His23 allows carboxy-monopeptidase and carboxy-dipeptidase activity of the protease. Structure. 2000;8:305–313. doi: 10.1016/s0969-2126(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 5.Kos J, Sekirnik A, Premzl A, Zavasnik Bergant V, Langerholc T, Turk B, Werle B, Golouh R, Repnik U, Jeras M, Turk V. Carboxypeptidases cathepsins X and B display distinct protein profile in human cells and tissues. Exp Cell Res. 2005;306:103–113. doi: 10.1016/j.yexcr.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Nagler DK, Menard R. Human cathepsin X: a novel cysteine protease of the papain family with a very short proregion and unique insertions. FEBS Lett. 1998;434:135–139. doi: 10.1016/s0014-5793(98)00964-8. [DOI] [PubMed] [Google Scholar]
- 7.Santamaria I, Velasco G, Pendas AM, Fueyo A, Lopez-Otin C. Cathepsin Z, a novel human cysteine proteinase with a short propeptide domain and a unique chromosomal location. J Biol Chem. 1998;273:16816–16823. doi: 10.1074/jbc.273.27.16816. [DOI] [PubMed] [Google Scholar]
- 8.Wendt W, Zhu XR, Lubbert H, Stichel CC. Differential expression of cathepsin X in aging and pathological central nervous system of mice. Exp Neurol. 2007;204:525–540. doi: 10.1016/j.expneurol.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Krueger S, Kalinski T, Hundertmark T, Wex T, Kuster D, Peitz U, Ebert M, Nagler DK, Kellner U, Malfertheiner P, Naumann M, Rocken C, Roessner A. Up-regulation of cathepsin X in Helicobacter pylori gastritis and gastric cancer. J Pathol. 2005;207:32–42. doi: 10.1002/path.1820. [DOI] [PubMed] [Google Scholar]
- 10.Nagler DK, Kruger S, Kellner A, Ziomek E, Menard R, Buhtz P, Krams M, Roessner A, Kellner U. Up-regulation of cathepsin X in prostate cancer and prostatic intraepithelial neoplasia. Prostate. 2004;60:109–119. doi: 10.1002/pros.20046. [DOI] [PubMed] [Google Scholar]
- 11.Joyce JA, Baruch A, Chehade K, Meyer-Morse N, Giraudo E, Tsai FY, Greenbaum DC, Hager JH, Bogyo M, Hanahan D. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5:443–453. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- 12.Decock J, Obermajer N, Vozelj S, Hendrickx W, Paridaens R, Kos J. Cathepsin B, cathepsin H, cathepsin X and cystatin C in sera of patients with early-stage and inflammatory breast cancer. Int J Biol Markers. 2008;23:161–168. doi: 10.1177/172460080802300305. [DOI] [PubMed] [Google Scholar]
- 13.Sevenich L, Schurigt U, Sachse K, Gajda M, Werner F, Muller S, Vasiljeva O, Schwinde A, Klemm N, Deussing J, Peters C, Reinheckel T. Synergistic antitumor effects of combined cathepsin B and cathepsin Z deficiencies on breast cancer progression and metastasis in mice. Proc Natl Acad Sci U S A. 2010;107:2497–2502. doi: 10.1073/pnas.0907240107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nascimento FD, Rizzi CC, Nantes IL, Stefe I, Turk B, Carmona AK, Nader HB, Juliano L, Tersariol IL. Cathepsin X binds to cell surface heparan sulfate proteoglycans. Arch Biochem Biophys. 2005;436:323–332. doi: 10.1016/j.abb.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Lechner AM, Assfalg-Machleidt I, Zahler S, Stoeckelhuber M, Machleidt W, Jochum M, Nagler DK. RGD-dependent binding of procathepsin X to integrin alphavbeta3 mediates cell-adhesive properties. J Biol Chem. 2006;281:39588–39597. doi: 10.1074/jbc.M513439200. [DOI] [PubMed] [Google Scholar]
- 16.Obermajer N, Premzl A, Zavasnik Bergant T, Turk B, Kos J. Carboxypeptidase cathepsin X mediates beta2-integrin-dependent adhesion of differentiated U-937 cells. Exp Cell Res. 2006;312:2515–2527. doi: 10.1016/j.yexcr.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 17.Obermajer N, Repnik U, Jevnikar Z, Turk B, Kreft M, Kos J. Cysteine protease cathepsin X modulates immune response via activation of beta2 integrins. Immunology. 2008;124:76–88. doi: 10.1111/j.1365-2567.2007.02740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenbaum D, Medzihradszky KF, Burlingame A, Bogyo M. Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem Biol. 2000;7:569–581. doi: 10.1016/s1074-5521(00)00014-4. [DOI] [PubMed] [Google Scholar]
- 19.Greenbaum D, Baruch A, Hayrapetian L, Darula Z, Burlingame A, Medzihradszky KF, Bogyo M. Chemical approaches for functionally probing the proteome. Mol Cell Proteomics. 2002;1:60–68. doi: 10.1074/mcp.t100003-mcp200. [DOI] [PubMed] [Google Scholar]
- 20.Verhelst SH, Bogyo M. Solid-phase synthesis of double-headed epoxysuccinyl activity-based probes for selective targeting of papain family cysteine proteases. Chembiochem. 2005;6:824–827. doi: 10.1002/cbic.200400377. [DOI] [PubMed] [Google Scholar]
- 21.Blum G, Mullins SR, Keren K, Fonovic M, Jedeszko C, Rice MJ, Sloane BF, Bogyo M. Dynamic imaging of protease activity with fluorescently quenched activity-based probes. Nat Chem Biol. 2005;1:203–209. doi: 10.1038/nchembio728. [DOI] [PubMed] [Google Scholar]
- 22.Blum G, von Degenfeld G, Merchant MJ, Blau HM, Bogyo M. Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat Chem Biol. 2007;3:668–677. doi: 10.1038/nchembio.2007.26. [DOI] [PubMed] [Google Scholar]
- 23.Hang HC, Loureiro J, Spooner E, van der Velden AW, Kim YM, Pollington AM, Maehr R, Starnbach MN, Ploegh HL. Mechanism-based probe for the analysis of cathepsin cysteine proteases in living cells. ACS Chem Biol. 2006;1:713–723. doi: 10.1021/cb600431a. [DOI] [PubMed] [Google Scholar]
- 24.Watzke A, Kosec G, Kindermann M, Jeske V, Nestler HP, Turk V, Turk B, Wendt KU. Selective activity-based probes for cysteine cathepsins. Angew Chem Int Ed Engl. 2008;47:406–409. doi: 10.1002/anie.200702811. [DOI] [PubMed] [Google Scholar]
- 25.Yasuda Y, Li Z, Greenbaum D, Bogyo M, Weber E, Bromme D. Cathepsin V, a novel and potent elastolytic activity expressed in activated macrophages. J Biol Chem. 2004;279:36761–36770. doi: 10.1074/jbc.M403986200. [DOI] [PubMed] [Google Scholar]
- 26.Abdulla MH, Lim KC, Sajid M, McKerrow JH, Caffrey CR. Schistosomiasis mansoni: novel chemotherapy using a cysteine protease inhibitor. PLoS Med. 2007;4:e14. doi: 10.1371/journal.pmed.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadaghiani AM, Verhelst SH, Gocheva V, Hill K, Majerova E, Stinson S, Joyce JA, Bogyo M. Design, synthesis, and evaluation of in vivo potency and selectivity of epoxysuccinyl-based inhibitors of papain-family cysteine proteases. Chem Biol. 2007;14:499–511. doi: 10.1016/j.chembiol.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Sadaghiani AM, Verhelst SH, Bogyo M. Solid-phase methods for the preparation of epoxysuccinate-based inhibitors of cysteine proteases. J Comb Chem. 2006;8:802–804. doi: 10.1021/cc0601027. [DOI] [PubMed] [Google Scholar]
- 29.O'Byrne J, Hunt MC, Rai DK, Saeki M, Alexson SE. The human bile acid-CoA:amino acid N-acyltransferase functions in the conjugation of fatty acids to glycine. J Biol Chem. 2003;278:34237–34244. doi: 10.1074/jbc.M300987200. [DOI] [PubMed] [Google Scholar]
- 30.Obermajer N, Doljak B, Jamnik P, Fonovic UP, Kos J. Cathepsin X cleaves the C-terminal dipeptide of alpha- and gamma-enolase and impairs survival and neuritogenesis of neuronal cells. Int J Biochem Cell Biol. 2009;41:1685–1696. doi: 10.1016/j.biocel.2009.02.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.