Abstract
The hypoxia-inducible transcription factors (HIF)-1α and -2α play a critical role in cellular response to hypoxia. Elevated HIF-α expression correlates with poor patient survival in a large number of cancers. Recent evidence suggests that HIF-2α appears to be preferentially expressed in neuronal tumor cells that exhibit cancer stem cell characteristics. These observations suggest that expression of HIF-1α and -2α is differentially regulated in the hypoxic tumor microenvironment. However, the underlying mechanisms remain to be fully investigated. In this study, we investigated the transcriptional regulation HIF-1α and -2α under different physiologically relevant hypoxic conditions. We found that transcription of HIF-2α was consistently increased by hypoxia, whereas transcription of HIF-1α showed variable levels of repression. Mechanistically, differential regulation of HIF-α transcription involved hypoxia-induced changes in acetylation of core histones H3 and H4 associated with the proximal promoters of the HIF-1α or HIF-2α gene. We also found that, although highly stable under acute hypoxia, HIF-1α and HIF-2α proteins become destabilized under chronic hypoxia. Our results have thus provided new mechanistic insights into the differential regulation of HIF-1α and -2α by the hypoxic tumor microenvironment. These findings also suggest an important role of HIF-2α in the regulation of tumor progression under chronic hypoxia.
Keywords: hypoxia, hypoxia-inducible factor, HIF-1α and HIF-2α, transcription, promoter
Introduction
The hypoxia-inducible transcription factors (HIF)-1α and -2α are the key transcription factors regulating the expression of hypoxia-induced genes critical for a wide range of tumor cell functions from survival, clonal selection to metastasis (1–3). Elevated HIF-α expression correlates with poor patient survival in a large number of cancers. Nonetheless, other evidence indicates a correlation of HIF-1α expression with favorable prognosis in other cases (4, 5). The stability of the HIF-α proteins is post-translationally regulated by prolyl-4-hydroxylase (PHD)-mediated hydroxylation of two proline residues located in the oxygen-dependent degradation domain, which leads to degradation of the hydroxylated HIF-α via interaction with the von Hippel Lindau (VHL) protein (6, 7). In addition to their non-overlapping transcription activities (8, 9), HIF-1α and -2α are expressed in different tissues with HIF-1α being more widely expressed (10). Recent evidence suggests that HIF-2α appears to be preferentially expressed in neuronal tumor cells with cancer stem cell characteristics (11–13). Furthermore, elevated HIF-2α expression is co-localized in vivo with expression of neural crest progenitor markers, suggesting a preferential association of HIF-2α expression with the immature stem cell-like neuroblastoma cells (12). These observations suggest that expression of HIF-1α and -2α is differentially regulated in the hypoxic tumor microenvironment.
Oxygenation in solid tumors varies from physiological levels of approximately 5–8% O2 to near anoxia (14, 15). Tumor hypoxia is also highly heterogeneous with both chronic and acute hypoxia (16). In this study, we investigated the transcriptional regulation of HIF-1α and -2α under different hypoxic conditions. In addition to conventional hypoxia treatment, we developed an adaptive chronic hypoxia approach by preconditioning tumor cells at 5% O2 before reducing pO2 to hypoxia levels (≤2% O2) to mimic in vivo tumor hypoxia. We found that transcription of HIF-2α was consistently increased by hypoxia in a panel of neuroblastoma cell lines, whereas transcription of HIF-1α showed variable levels of repression. Mechanistically, differential regulation of HIF-α transcription involved hypoxia-induced changes in actetylation of core histones H3 and H4 associated with the proximal promoters of the HIF-1α or HIF-2α gene. We also found that, although highly stable under acute hypoxia, HIF-1α and HIF-2α proteins become destabilized under chronic hypoxia. Our results have thus provided new mechanistic insights into the differential expression and localization of HIF-1α and -2α proteins within the hypoxic tumor microenvironment. These findings further underscore the importance of HIF-2α in the regulation of tumor progression, especially in the regulation of the stem cell-like tumor cell population as observed in neuronal tumors.
Materials and Methods
Cell culture under normoxia or hypoxia
SK-N-BE(2)C, SK-N-ER, and SH-SY5Y cells were maintained in Minimum Essential Medium and F12 (1:1) and IMR-32 cells, in Minimum Essential Medium. The media were supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate, 25 mM HEPES at pH 7.4 to maintain pH stability. The medium was replaced every other day.
For experiments at reduced pO2 (5% O2) or hypoxia (≤2% O2), cells were incubated in a hypoxia chamber (Invivo2 400, Ruskinn Technology). Anoxia experiments were performed in a Bactron Anaerobic Chamber (Sheldon MFG Inc.). Deferoxamine mesylate (DFO, Sigma-Aldrich) was used to mimic hypoxia effects at 21% O2 (17, 18). Culture media were replaced every other day inside the hypoxia chamber. During long-term hypoxic incubation, cells were trypsinized and culture passages were done inside the chamber to prevent reoxygenation.
Western blot
Nuclear extracts were used for Western blots as described previously (17, 19) with antibodies to the following antigens: HIF-1α (1:2000), and HIF-2α (1:1000), HIF-1β (1:500), and DEC1 (1:2,000), all of which were purchased from Novus Biologicals.
Firefly luciferase reporter constructs
All constructs were validated by DNA sequencing. The HIF-1α promoter/enhancer region from +122 to −4,871 relative to the transcription start site (NT_026437.11) was PCR-amplified using the following primers: 5’-TATTC TTGCC TTGGC TGTATC C-3’ (forward) and 5’-ACTGT GCACT GAGGA GCTGA G-3’ (reverse), and then inserted between Mlu 1 and Nhe 1 sites of the pGL3 basic vector. The 2.6-kbp construct was generated by restriction digestion of the 5-kbp construct using Mlu 1 and Pst 1, followed by ligation of the remaining construct. The 0.6-kbp construct was generated by restriction digestion of the 2.6-kbp construct using Kpn 1 and Pml 1, followed by ligation of the remaining construct.
The HIF-2α promoter/enhancer region from +116 to −4,883 relative to the transcription start site (NT_022184.14) was PCR-amplified using the following primers: 5’-AGTCC CATTT TAACA CTTTG CTACA-3’ (forward) and 5’-AGCTG ACCAT ACAGT CTCAG GAC-3’ (reverse), and then inserted between Mlu 1 and Nhe 1 sites of the pGL3 basic vector. The 3.3-kbp construct was generated by deletion of the 5’ sequence from the 5-kbp construct using Mlu 1 and Agl 1. The 1.0-kbp construct was generated by deletion of the 5’ sequence from the 3.3-kbp construct using Kpn 1 and Stu 1. The 0.8-kbp construct was generated by deletion of the 5’ sequence from the 3.3-kbp construct using Kpn 1 and Pvu 1.
Real-time RT-PCR
First-strand cDNA was synthesized from total RNA. Real-time PCR was performed on StepOne Plus (Applied Biosystems) using Power SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer’s recommended protocol. The primer sequences can be found in Supplemental Materials and Methods (Table 1).
Chromosome immunoprecipitation (ChIP)
SK-N-BE(2)C cells were incubated at 1% O2 for 24 hr and were used for ChIP according to our previously published protocol (19, 20). The ChIP primer sequences can be found in Supplemental Materials and Methods (Table 2).
Statistical analysis
The statistical difference between two groups was analyzed by the two-tailed, unpaired Student’s t-test using Prizm 3.0 (GraphPad Software Inc.). Significant difference between two groups was declared if p < 0.05.
Results
Differential regulation of transcription of HIF-1α and HIF-2α by hypoxia
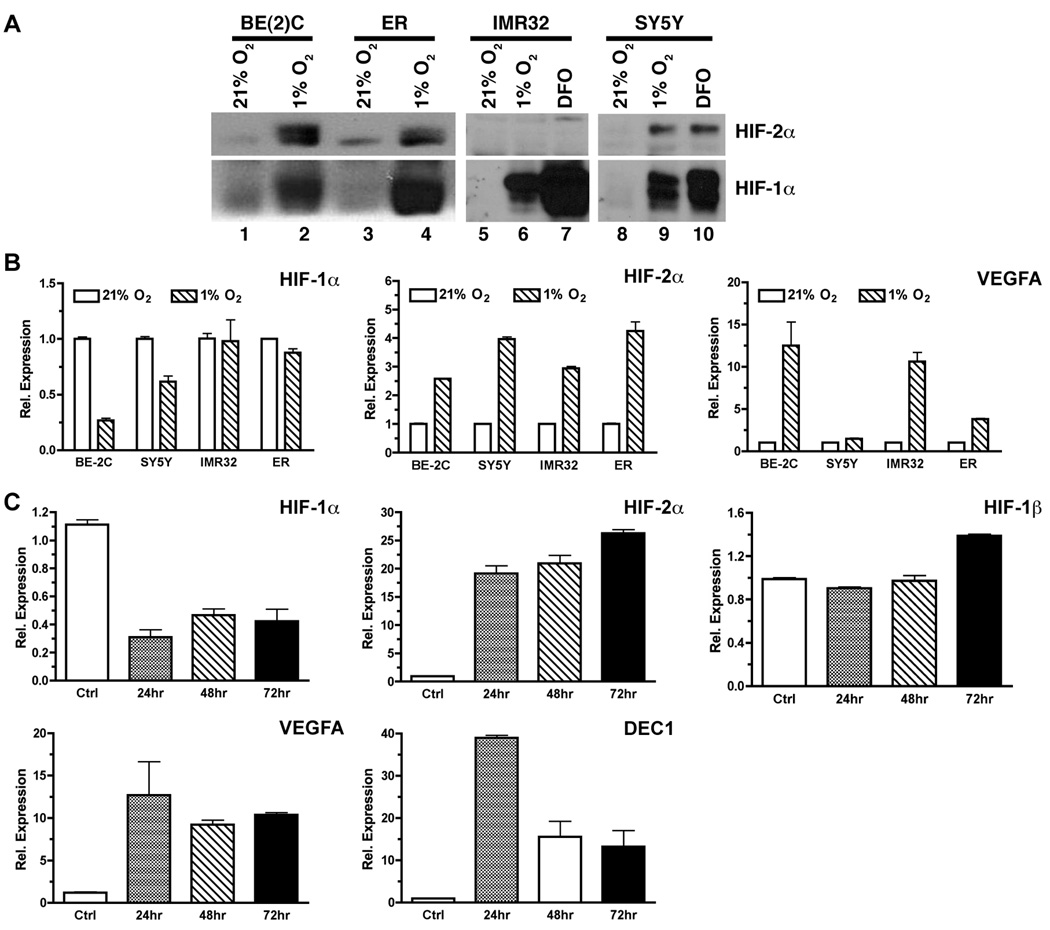
We investigated the transcription of HIF-1α and HIF-2α under hypoxia using a selected panel of neuroblastoma (NB) cell lines either with N-myc amplification [SK-N-BE(2)C and IMR32] or without N-myc amplification (SK-N-ER and SH-SY5Y). Hypoxia induced robust accumulation of HIF-1α protein in all the four cell lines (Figure 1A). In comparison, accumulation of HIF-2α protein was readily detected in hypoxia-treated BE(2)C, ER, and SY5Y cells, but not in IMR32 cells (Figure 1A). A lack of detectable HIF-2α protein in hypoxia-treated IMR32 cells was also observed by others (21). This is likely due to the low HIF-2α mRNA expression in IMR32 cells (>40 fold less than in BE(2)C cells based on our quantitative RT-PCR analysis).
Figure 1. Differential regulation of HIF-1α and HIF-2α transcription by hypoxia.
(A) Hypoxic induction of HIF-1α and HIF-2α protein in neuroblastoma cell lines: BE(2)C = SK-N-BE(2)C, ER = SK-N-ER, IMR32, SY5Y = SH-SY5Y. Cells were incubated for 20–24 hr at 1% O2 or in the presence of 50 µM Deferoxamine (DFO). HIF-1α and HIF-2α proteins in nuclear extracts were detected by Western blot.
(B) Neuroblastoma cell cultures were incubated for 20–24 hr at 1% O2. Total RNA was prepared and subjected to qRT-PCR for quantitative analysis of gene expression. Data are shown as mean relative expression ± sem (n = 4).
(C) Expression of HIF-α and related genes in BE(2)C cells incubated at 1% O2 for 24, 48, or 72 hr. Total RNA was used for analysis of gene expression by qRT-PCR (mean ± sd). Cells cultured at 21% O2 was used as the normoxia control.
Interestingly, transcription of HIF-1α showed a bias toward downregulation by hypoxia, whereas transcription of HIF-2α was increased by hypoxia, in the above four representative NB cell lines (Figure 1B). In addition to NB cell lines, we found similar differential regulation of HIF-1α and HIF-2α in the glioblastoma cell line U373 (Supplementary Figure 1), suggesting a common regulatory mechanism of hypoxia-regulated HIF-α expression in neuronal tumors.
Since tumor hypoxia is heterogeneous and dynamic (16), we further examined the effect of acute (≤24 hr) and chronic (>24 hr) hypoxia on transcription of HIF-1α and HIF-2α. Using the BE(2)C cell line expressing high levels of both HIF-1α and HIF-2α proteins as a model, we found that transcription of HIF-1α was consistently repressed by both acute and chronic hypoxia, whereas transcription of HIF-2α was consistently upregulated under the same hypoxic condition (Figure 1C). Transcription of the classical hypoxia-induced genes VEGFA and DEC1/BHLHE40 (DEC1 is used herein) was increased under both acute and chronic conditions although more robust induction appeared to occur under acute hypoxia (Figure 1C and Figure 2). In contrast, transcription of HIF-1α was not significantly affected by either acute or chronic hypoxia (Figure 1C). These observations indicate that transcription of HIF-1α and HIF-2α is differentially regulated under hypoxia.
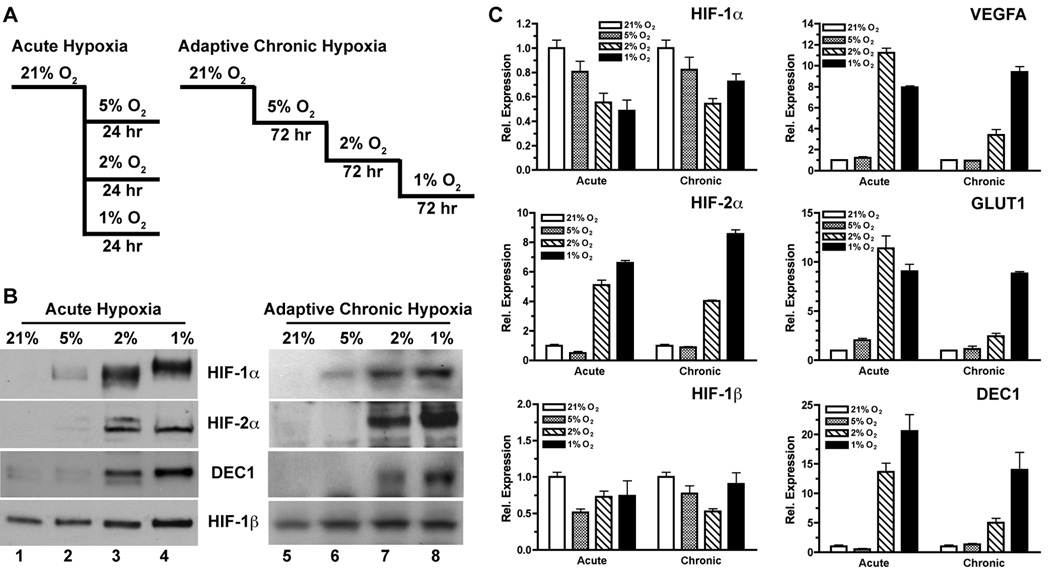
Figure 2. Effect of acute and adaptive chronic hypoxia on the expression of HIF-α and related genes.
(A) Schematics of acute hypoxia and adaptive chronic hypoxia in vitro. The latter serves as a physiologically relevant model for in vivo hypoxia.
(B) Western blots of HIF-α and related proteins in nuclear extracts of BE(2)C cells treated by acute and adaptive chronic hypoxia.
(C) Quantitative analysis of HIF-α and related genes in BE(2)C cells treated by acute and adaptive chronic hypoxia by qRT-PCR (mean ± sd).
The most commonly used approach to in vitro hypoxia studies involves transferring cells from the hyperoxic atmosphere (21% O2) to a hypoxic condition (e.g. ≤2% O2). However, pO2 levels in physiological normal tissues are mostly found to be around 5–8% O2, with intratumoral pO2 levels often found to be <10 mmHg (<1.3% O2) (14, 15). In order to gain insight into the transcriptional regulation of HIF-1α and HIF-2α by tumor hypoxia in vivo, we designed a stepwise adaptive hypoxia model to mimic tumor hypoxia. BE(2)C cells were first allowed to adapt to 5% O2 (tissue-level normoxia) and then to hypoxia (≤2% O2) on a chronic (72 hr exposure) basis (Figure 2A). Decrease of pO2 from 21% to 5% resulted in subtle accumulation of HIF-1α, but did not induce significant stabilization of HIF-2α proteins (Figure 2B). Only minor changes occurred in the transcription of HIF-1α, HIF-2α, and the three classical hypoxia-induced genes when the environmental pO2 changed from 21% to 5% (Figure 2C). In contrast, both acute (24 hr) and adaptive chronic (72 hr) hypoxia of ≤2% O2 reduced the transcription of HIF-1α but strongly increased HIF-2α transcription (Figure 2C). Interestingly, VEGFA, GLUT1 and DEC1 experienced more robust induction by acute hypoxia (≤2% O2) than by adaptive chronic hypoxia (Figure 2C), suggesting a possible involvement of HIF-independent mechanisms of hypoxia-regulated gene expression.
Differential regulation of HIF-1α and HIF-2α protein stabilities by acute and chronic hypoxia
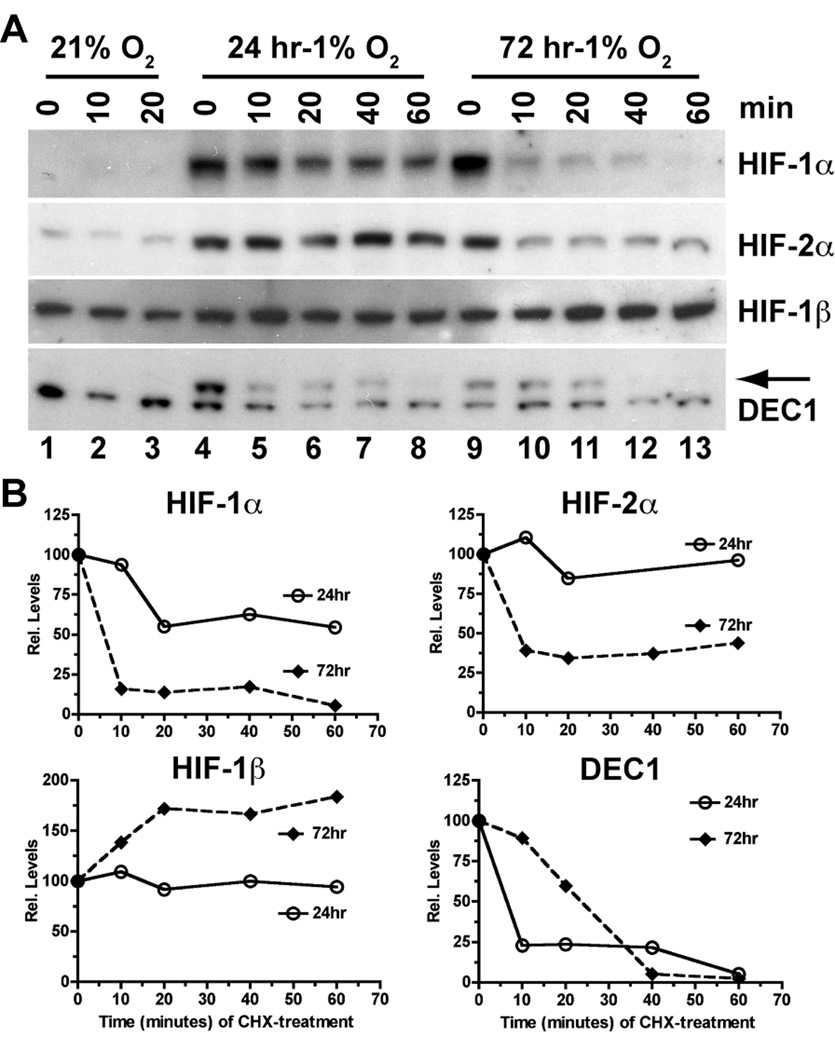
Although it is generally believed that hypoxic induction of HIF-1α and HIF-2α proteins results from increased protein stability or decreased prolylhydroxylase (PHD)-dependent degradation, it is not clear whether HIF-α protein stabilities are regulated differently under acute or chronic hypoxia. As shown in Figure 3, both HIF-1α and HIF-2α proteins were strongly induced by both acute (24 hr at 1% O2, lane 4 vs. lane 1) and chronic (72 hr at 1% O2, lane 9 vs. lane 1) hypoxia. However, the stabilities of both HIF-1α and HIF-2α proteins were much higher under acute hypoxia than under chronic hypoxia. In contrast, HIF-1β protein appears to become even more stable under chronic than under acute hypoxia. These results suggest that stabilities of HIF-1α, HIF-2α, and HIF-1β proteins are differentially regulated under chronic hypoxia.
Figure 3. Impact of acute and chronic hypoxia on the stabilities of HIF-1α and HIF-2α proteins.
(A) BE(2)C cells were either maintained at 21% O2 (normoxia control) or preconditioned at 1% O2 for 24 and 72 hr, respectively. Nuclear extracts were harvested at the indicated time point after the treatment with cycloheximide (CHX, 25 µg/ml) to inhibit protein synthesis. Levels of HIF-1α, HIF-2α, HIF-1β and DEC1 (arrow) proteins were detected by Western blot analysis.
(B) Band intensities were analyzed using NIH Image J and were plotted against time (minutes) after addition of CHX.
Hypoxic regulation of HIF-1α and HIF-2α mRNA stabilities
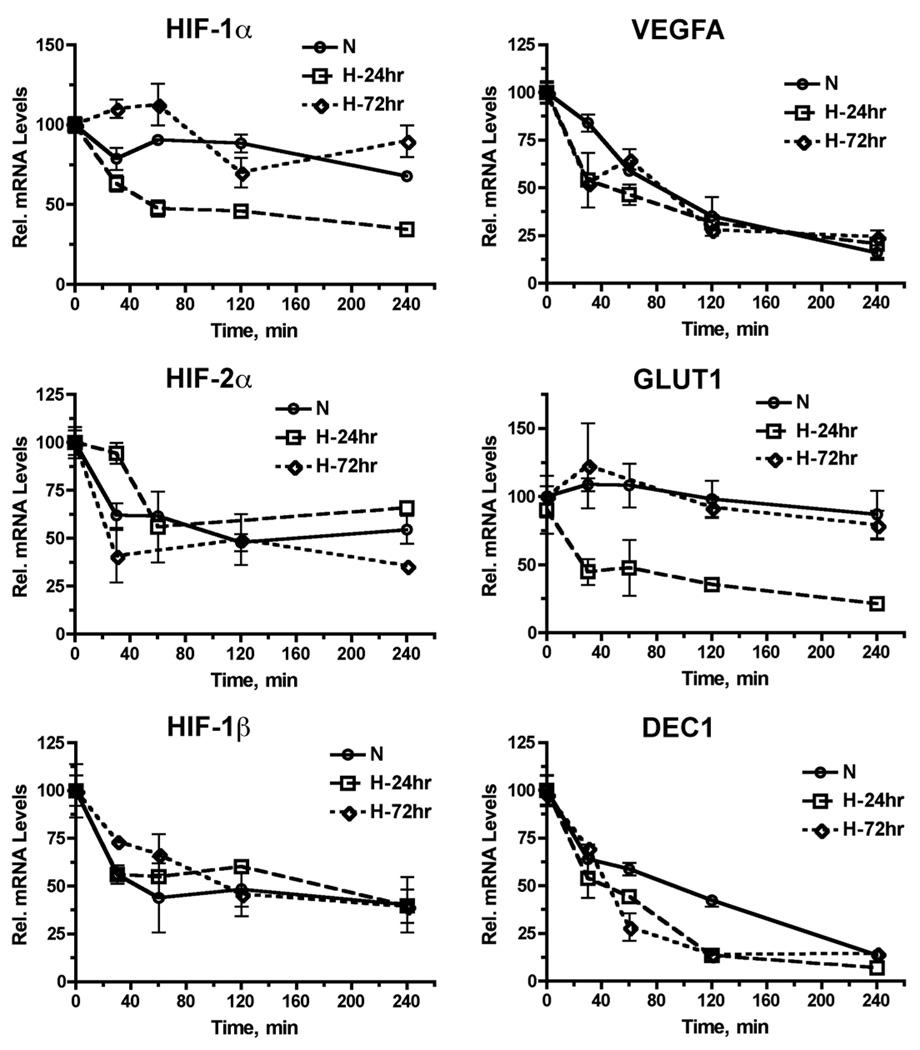
We determined the mRNA stabilities of HIF-1α, HIF-2α, HIF-1β as well as three HIF-regulated genes (VEGFA, GLUT1 and DEC1) using the actinomycin D approach. We found that the stabilities of HIF-2α and HIF-1β mRNA did not change significantly under either acute (24 hr) or chronic (72 hr) hypoxia at 1% O2, as compared to that at 21% O2 (Figure 4). In contrast, HIF-1α mRNA appeared to be less stable under acute hypoxia than under chronic hypoxia or at 21% O2. Among the three HIF-target genes, GLUT1 mRNA experienced the most dramatic change in mRNA stability among the three experimental conditions with the lowest mRNA stability found under acute hypoxia, whereas stabilities of VEGFA and DEC1 mRNA were similar under both acute and chronic hypoxia (Figure 4). These observations demonstrate that the stability of HIF-1α mRNA is more sensitive to regulation by the duration of hypoxia, as compared to that of HIF-2α mRNA.
Figure 4. Impact of acute and chronic hypoxia on the stabilities of HIF-1α and HIF-2α mRNA.
BE(2)C cells were either maintained at 21% O2 (normoxia control) or preconditioned at 1% O2 for 24 and 72 hr, respectively. Cellular RNA was harvested at the indicated time point after the treatment with 5 µg/ml actinomycin D to inhibit RNA synthesis. Levels of HIF-1α, HIF-2α, HIF-1β and hypoxia-induced genes were determined by qRT-PCR (mean ± sd).
Differential regulation of HIF-1α and HIF-2α promoters by acute and chronic hypoxia
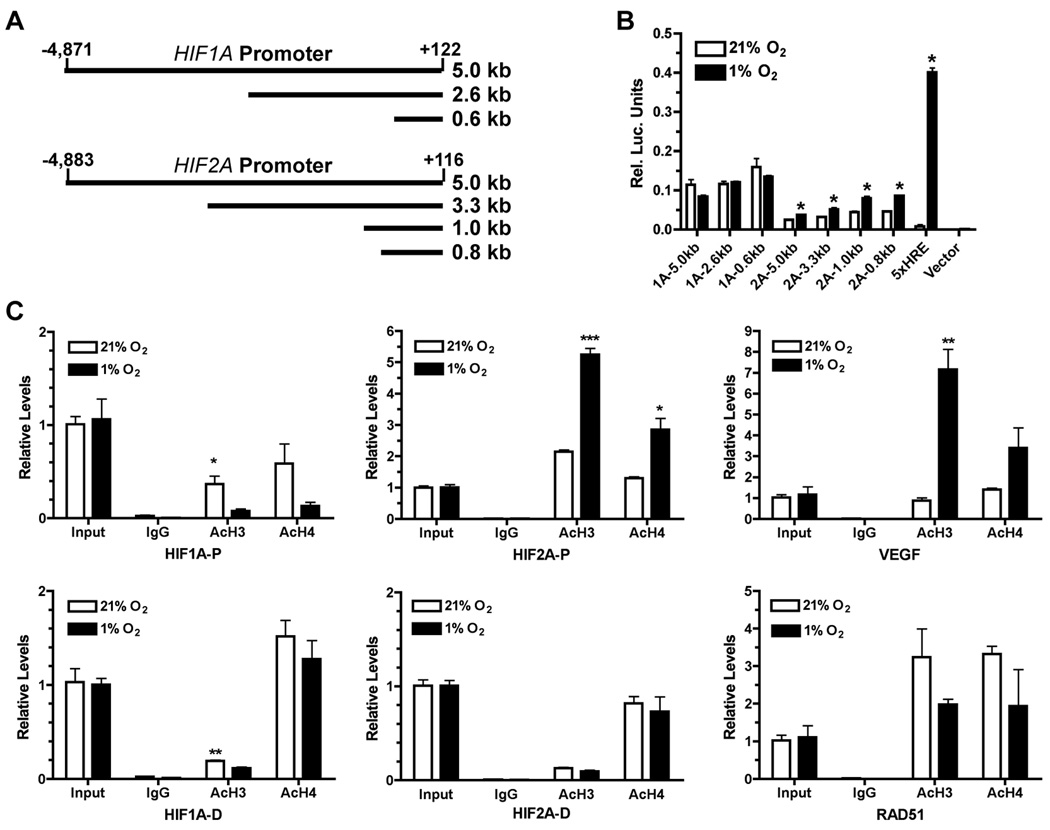
As shown by our results, HIF-2α expression is transcriptionally increased by acute and chronic hypoxia whereas HIF-1α mRNA levels were either decreased or little changed under the same conditions. Based on sequence comparison, the proximal region of the HIF-1α gene (NT_026437.11) is relatively GC-rich and lacks a TATA box whereas the proximal region of the HIF-2α gene (NT_022184.14) contains a putative TATA box (TTTAAA) located around −25-bp from the transcription start site. In order to understand how HIF-1α and HIF-2α are differentially regulated by hypoxia at the transcriptional level, we investigated the transcriptional activities of their respective gene promoters under hypoxia. As shown in Figure 5A, we cloned an approximately 5-kb upstream promoter/enhancer fragment from either the HIF-1α or the HIF-2α genomic sequence. Shorter promoter/enhancer fragments were further generated by restriction digest. As indicated by the luciferase reporter activities (Figure 5B), transcriptional activities of the three HIF-1α promoter/enhancer fragments showed a slight decrease under hypoxia. In contrast, all four HIF-2α promoter/enhancer fragments exhibited higher transcriptional activities under hypoxia than under normoxia. Because all the shorter promoter/enhancer fragments within the 5-kbp region of either gene showed similar transcription activities under hypoxia, specific promoter or enhancer elements were not likely to be significantly involved in the transcriptional regulation by hypoxia. Consistent with this notion, we did not find any plausible conserved transcription sites using the transcription-factor binding-site prediction algorithms (22). These observations suggest that hypoxia-mediated transcriptional regulation of HIF-1α and HIF-2α expression likely involves chromatin modifications in their promoter/enhancer regions.
Figure 5. Differential regulation of HIF-1α and HIF-2α promoter/enhancer by hypoxia.
(A) Schematics of firefly lucerifase reporter constructs driven by different regions of the HIF-1α and HIF-2α promoter/enhancer located upstream to the transcription start site (TSS).
(B) Each indicated firefly luciferase construct was co-transfected with a Renilla luciferase reporter construct into ER cells. After 24 hr incubation, transfected cells were either maintained at 21% O2 or in a hypoxia chamber at 1% O2 for another 24 hr. Firefly luciferase activity was normalized to that of Renilla luciferase. Data shown were average of three independent experiments (p < 0.02). 1A = HIF-1α, 2A = HIF-2α. 5XHRE = a firefly luciferase reporter containing five tandem repeats of a hypoxia-responsive element (HRE). Vector = pGL3 Basic.
(C) Chromatin immunoprecipitation was performed in normoxic (21% O2) or hypoxic (1% O2, 24 hr) BE(2)C cells using specific antibodies against acetylated histone H3 (AcH3) or acetylated histone H4 (AcH4) with a naÔve antibody (IgG) as control. The immunoprecipated promoter fragments were quantitatively analyzed by qPCR. For the HIF-1α promoter/enhancer, HIF1A-P = proximal region (−176 to −30); HIF1A-D = distal region (approximately −8.5 kb); for the HIF-2α promoter/enhancer, HIF2A-P = proximal region (−22 to +76); HIF2A-D = distal region (approximately −8.0 kb). Data shown were average of three independent experiments. *p < 0.05, **p < 0.005, ***p < 0.0005.
Because changes in transcriptional activities are often accompanied by changes in acetylation of core histones near the transcription start site (TSS) (23, 24), we therefore examined the effects of hypoxia on histone acetylation in the HIF-1α and HIF-2α promoter/enhancer regions using chromatin immunoprecipitation. We found (Figure 5C) that hypoxia decreased acetylation of histone H3 and H4 in the HIF-1α promoter/enhancer region with stronger decreases near TSS (HIF1A-P: −30 to −176 bp) than around the −8.5 kbp distal region (HIF1A-D), which correlates well with the downregulation of HIF-1α expression by hypoxia (Figures 1 & 2). On the other hand, acetylation of histone H3 and H4 in the HIF-2α proximal promoter/enhancer region (HIF2A-P: +84 to −59 bp) was significantly increased by hypoxia, whereas there was little change in H3 and H4 acetylation in the distal region around −8.0 kbp (HIF2A-D). As expected, acetylation of histone H3 and H4 in the promoter/enhancer region of the hypoxia-induced gene VEGFA was increased, whereas histone acetylation was decreased in the promoter of RAD51, a hypoxia-repressed gene (25). Our results suggest that differential regulation of HIF-1α and HIF-2α expression is likely due to hypoxia-induced changes in acetylation of histones associated with their respective promoter/enhancers. Specifically, increased acetylation of histones H3 and H4 around the TSS of HIF-2α promoter can potentially facilitate the recruitment of transcription co-activators and formation of the RNA polymerase complex for efficient transcription under hypoxia. In contrast, decreased acetylation of histones H3 and H4 is likely to render the promoter of HIF-1α less accessible to transcription co-factors and to result in suppressed HIF-1α expression under hypoxia.
Discussion
Post-translational modifications are thought to be the key mechanisms of regulation both HIF-1α and HIF-2α proteins in response to changes of environmental pO2. As shown by our current study and other reports (26, 27), the transcription of HIF-1α and HIF-2α genes can be differentially regulated by hypoxia. We reasoned that the structural basis for such differential transcriptional regulation would lie in the different DNA sequences of the upstream promoter/enhancer regions between the HIF-1α and HIF-2α genes. By examining the transcription activities of the approximately 5-kilobase promoter/enhancer regions of HIF-1α and HIF-2α, respectively, we have found that specific hypoxia-responsive motifs are unlikely to be responsible for the differential transcription of HIF-1α and HIF-2α genes under hypoxia. In contrast, our data have shown that hypoxia specifically increases acetylation of the core histones H3 and H4 within the proximal (around TSS), but not the distal (approximately −8.0 kb), promoter region of HIF-2α, coinciding with increased HIF-2α transcription. In contrast, hypoxia decreases histone acetylation of the HIF-1α proximal (near TSS), but not the distal (approximately −8.5 kb) promoter/enhancer region. These findings indicate that chromatin remodeling is likely a key mechanism for transcriptional regulation of HIF-1α and HIF-2α expression under hypoxia.
However, the exact mechanisms of transcriptional regulation are like to be quite complex. Consistent with our findings that HIF-2α transcription likely involves chromatin-level regulation, Johnson et al. have found that hypoxia induces a wide range of histone modifications associated with both transcriptional activation and repression (28). Our earlier work (20) showed that differential histone modifications was involved in adaptive gene expression under chronic hypoxia. It has been reported that hypoxia can regulate expression and activities of several histone-modifying enzymes. Kim et al. have shown that hypoxia can enhance HDAC function to promote angiogenesis (29). Recent studies have shown that members of Jumonji C-domain-containing histone demethylases are also involved in epigenetic regulation of hypoxia-dependent gene transcription (30–33). It is likely that chromatin-level regulation may underline one of the fundamental mechanisms about hypoxia-modulated global gene expression, especially the expression of HIF-independent genes.
It is worth noting that several other mechanisms may also be involved in the transcriptional regulation of HIF-1α and HIF-2α genes. In A549 human lung adenocarcinoma cells, HIF-2α transcription can be increased via an unknown auto-feedback mechanism (34). Other evidence suggests possible trans-regulation between HIF-1α and HIF-2α in human renal cell carcinoma cells (35). Because our promoter analysis does not reveal a likelihood of a functional hypoxia-responsive element (HRE) in either HIF-α gene, chromatin modifications could potentially play a role in either auto-feedback or trans-regulation of HIF-1α and/or HIF-2α genes albeit the exact molecular mechanisms remain to be delineated.
Another interesting observation of the current study is that stabilities of HIF-1α and HIF-2α proteins are also differentially regulated by acute and chronic hypoxia. As widely reported, acute hypoxia (≤24 hr) results in stabilization of HIF-α proteins mainly due to inhibition of PHD-mediated proline hydroxylation (36). In this study, we have found that both HIF-1α and HIF-2α proteins become destabilized under chronic hypoxia (72 hr). It is likely that hypoxia-induced expression of PHDs (37) could restore the negative regulation of HIF-α protein under chronic hypoxia at 1% O2. It is also likely that effective O2-affinity of PHDs might be increased under chronic hypoxia due to potential changes in other co-factors including Fe2+, 2-oxoglutarate and/or ascorbate, or due to yet uncharacterized post-translational modifications of PHDs. Furthermore, other regulators of HIF protein stabilities (reviewed in 38) may also play a role under chronic hypoxia. The increased rates of degradation of HIF-α proteins by chronic hypoxia may constitute a mechanism to fine tune hypoxic responses.
Taken together, our data suggest the HIF-1 and HIF-2 have the potential to play different roles under acute and chronic hypoxia. HIF-1 is likely to be more involved in response to acute hypoxia via hypoxia-induced stabilization of HIF-1α protein. In contrast, HIF-2 appears to play a more important role in cellular adaptation to chronic hypoxia via increased HIF-2α transcription, which may offer growth and/or survival advantages under chronic hypoxia. Our results have thus provided new mechanistic insights into the differential expression of HIF-1α and -2α proteins within the hypoxic tumor microenvironment. In light of recent findings that HIF-2α appears to be preferentially expressed in stem cell-like tumor cells in vivo (11–13) and has the potential to facilitate cell growth by enhancing c-myc transcriptional activities (39), our findings further underscore the importance of HIF-2α in the regulation of malignant tumor progression, especially in the regulation of stem cell-like tumor cells.
Supplementary Material
Acknowledgements
Financial Support: This work was supported by a grant from the National Institutes of Health to ZY (R01CA125021).
We thank Dr. Nai-Kong V. Cheung of Memorial Sloan-Kettering Cancer Center for SK-N-ER cells, Dr. Robert Ross of Fordham University for BE(2)C cells, and Lisa Cabral for her excellent editorial assistance.
Footnotes
Disclosure of Potential Conflicts of Interests: The authors claim no potential conflicts of interests.
References
- 1.Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 2.Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008;15:678–685. doi: 10.1038/cdd.2008.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 4.Lidgren A, Hedberg Y, Grankvist K, Rasmuson T, Vasko J, Ljungberg B. The expression of hypoxia-inducible factor 1alpha is a favorable independent prognostic factor in renal cell carcinoma. Clin Cancer Res. 2005;11:1129–1135. [PubMed] [Google Scholar]
- 5.Vleugel MM, Greijer AE, Shvarts A, et al. Differential prognostic impact of hypoxia induced and diffuse HIF-1α expression in invasive breast cancer. J Clin Pathol. 2005;58:172–177. doi: 10.1136/jcp.2004.019885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivan M, Kondo K, Yang H, et al. HIFa targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 7.Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 8.Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–7934. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang V, Davis DA, Haque M, Huang LE, Yarchoan R. Differential gene up-regulation by hypoxia-inducible factor-1α and hypoxia-inducible factor-2α in HEK293T cells. Cancer Res. 2005;65:3299–3306. doi: 10.1158/0008-5472.CAN-04-4130. [DOI] [PubMed] [Google Scholar]
- 10.Wiesener MS, Jurgensen JS, Rosenberger C, et al. Widespread hypoxia-inducible expression of HIF-2α in distinct cell populations of different organs. FASEB J. 2003;17:271–273. doi: 10.1096/fj.02-0445fje. [DOI] [PubMed] [Google Scholar]
- 11.Li Z, Bao S, Wu Q, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pietras A, Gisselsson D, Ora I, et al. High levels of HIF-2α highlight an immature neural crest-like neuroblastoma cell cohort located in a perivascular niche. J Pathol. 2008;214:482–488. doi: 10.1002/path.2304. [DOI] [PubMed] [Google Scholar]
- 13.Seidel S, Garvalov BK, Wirta V, et al. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2α. Brain. 2010;133:983–995. doi: 10.1093/brain/awq042. [DOI] [PubMed] [Google Scholar]
- 14.Brown JM. Tumor hypoxia in cancer therapy. Methods Enzymol. 2007;435:297–321. doi: 10.1016/S0076-6879(07)35015-5. [DOI] [PubMed] [Google Scholar]
- 15.Vaupel P, Hockel M, Mayer A. Detection and characterization of tumor hypoxia using pO2 histography. Antioxid Redox Signal. 2007;9:1221–1235. doi: 10.1089/ars.2007.1628. [DOI] [PubMed] [Google Scholar]
- 16.Dewhirst MW, Cao Y, Moeller B. Cycling hypoxia and free radicals regulate angiogenesis and radiotherapy response. Nat Rev Cancer. 2008;8:425–437. doi: 10.1038/nrc2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Q, Lee YJ, Yun Z. Differentiation arrest by hypoxia. J Biol Chem. 2006;281:30678–30683. doi: 10.1074/jbc.C600120200. [DOI] [PubMed] [Google Scholar]
- 18.Yun Z, Maecker HL, Johnson RS, Giaccia AJ. Inhibition of PPARγ2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev Cell. 2002;2:331–341. doi: 10.1016/s1534-5807(02)00131-4. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y, Lin Q, Glazer PM, Yun Z. Hypoxic tumor microenvironment and cancer cell differentiation. Curr Mol Med. 2009;9:425–434. doi: 10.2174/156652409788167113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yun Z, Lin Q, Giaccia AJ. Adaptive myogenesis under hypoxia. Mol Cell Biol. 2005;25:3040–3055. doi: 10.1128/MCB.25.8.3040-3055.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jogi A, Ora I, Nilsson H, et al. Hypoxia alters gene expression in human neuroblastoma cells toward an immature and neural crest-like phenotype. Proc Natl Acad Sci U S A. 2002;99:7021–7026. doi: 10.1073/pnas.102660199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 23.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Bindra RS, Glazer PM. Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene. 2007;26:2048–2057. doi: 10.1038/sj.onc.1210001. [DOI] [PubMed] [Google Scholar]
- 26.Holmquist-Mengelbier L, Fredlund E, Lofstedt T, et al. Recruitment of HIF-1α and HIF-2α to common target genes is differentially regulated in neuroblastoma: HIF-2α promotes an aggressive phenotype. Cancer Cell. 2006;10:413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 27.Uchida T, Rossignol F, Matthay MA, et al. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1α and HIF-2α expression in lung epithelial cells: implication of natural antisense HIF-1α. J Biol Chem. 2004;279:14871–14878. doi: 10.1074/jbc.M400461200. [DOI] [PubMed] [Google Scholar]
- 28.Johnson AB, Denko N, Barton MC. Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutat Res. 2008;640:174–179. doi: 10.1016/j.mrfmmm.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim MS, Kwon HJ, Lee YM, et al. Histone deacetylases induce angiogenesis by negative regulation of tumor suppressor genes. Nat Med. 2001;7:437–443. doi: 10.1038/86507. [DOI] [PubMed] [Google Scholar]
- 30.Beyer S, Kristensen MM, Jensen KS, Johansen JV, Staller P. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J Biol Chem. 2008;283:36542–36552. doi: 10.1074/jbc.M804578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollard PJ, Loenarz C, Mole DR, et al. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1α. Biochem J. 2008;416:387–394. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- 32.Xia X, Lemieux ME, Li W, et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci U S A. 2009;106:4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krieg AJ, Rankin EB, Chan D, Razorenova O, Fernandez S, Giaccia AJ. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1α enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30:344–353. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato M, Tanaka T, Maeno T, et al. Inducible expression of endothelial PAS domain protein-1 by hypoxia in human lung adenocarcinoma A549 cells. Role of Src family kinases-dependent pathway. Am J Respir Cell Mol Biol. 2002;26:127–134. doi: 10.1165/ajrcmb.26.1.4319. [DOI] [PubMed] [Google Scholar]
- 35.Krieg M, Haas R, Brauch H, Acker T, Flamme I, Plate KH. Up-regulation of hypoxia-inducible factors HIF-1α and HIF-2α under normoxic conditions in renal carcinoma cells by von Hippel-Lindau tumor suppressor gene loss of function. Oncogene. 2000;19:5435–5443. doi: 10.1038/sj.onc.1203938. [DOI] [PubMed] [Google Scholar]
- 36.Quaegebeur A, Carmeliet P. Oxygen sensing: a common crossroad in cancer and neurodegeneration. Curr Top Microbiol Immunol. 2010;345:71–103. doi: 10.1007/82_2010_83. [DOI] [PubMed] [Google Scholar]
- 37.Henze AT, Riedel J, Diem T, et al. Prolyl hydroxylases 2 and 3 act in gliomas as protective negative feedback regulators of hypoxia-inducible factors. Cancer Res. 2010;70:357–366. doi: 10.1158/0008-5472.CAN-09-1876. [DOI] [PubMed] [Google Scholar]
- 38.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2α promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.