Abstract
Type II polyketides include antibiotics such as tetracycline, and chemotherapeutics such as daunorubicin. Type II polyketides are biosynthesized by the type II polyketide synthase (PKS) that consists of 5 – 10 stand-alone domains. In many type II PKSs, the type II ketoreductase (KR) specifically reduce the C9-carbonyl group. How the type II KR achieves such a high regio-specificity, and the nature of stereo-specificity, are not well understood. Sequence alignment of KRs led to a hypothesis that a well-conserved 94-XGG-96 motif may be involved in controlling the stereochemistry. The stereo-specificity of single, double and triple mutant combinations of P94L, G95D and G96D were analyzed in vitro and in vivo for the actinorhodin KR (actKR). The P94L mutation is sufficient to change the stereospecificity of actKR. Binary and ternary crystal structures of both wild type and P94L actKR were solved. Together with assay results, docking simulations, and co-crystal structures, a model for stereochemical control is presented herein that elucidates how type II polyketides are introduced into the substrate pocket such that the C9-carbonyl can be reduced with high regio- and stereo-specificities. The molecular features of actKR important for regio- and stereo-specificities can potentially be applied to biosynthesize new polyketides via protein engineering that rationally controls polyketide ketoreduction.
Streptomycete soil bacteria are one of nature’s largest producers of clinically relevant pharmaceuticals (1, 2). A majority of these products belong to the polyketide family, which are used as antibiotic (actinorhodin), anti-cancer (daunorubicin), and immunosuppressive (FK506) agents (3). The wide range of bioactivities of polyketide natural products is due in part to the presence of multiple chiral centers that contribute to their structural diversity. Recently, the importance of developing chiral drugs and their effect on pharmacokinetics have been reviewed (4). Often, the specific bioactivity and biosynthesis of these natural products are dependent on the presence of chiral centers introduced in a controlled and systematic manner. In fact, many natural products lack biological activity in the absence of modifications such as hydroxylations and glycosylations that create chiral centers (5). The importance of chirality for the action and biosynthesis of polyketides therefore presents a new facet that can be controlled to develop new, therapeutic polyketide products.
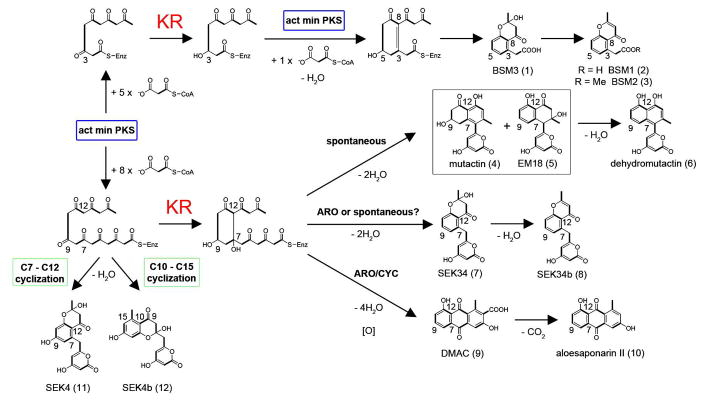
The chemical diversity of aromatic polyketides arises as a result of biosynthesis by polyketide synthase (PKS), which is closely related to fatty acid synthase (FAS). Both FAS and PKS can be classified as type I or type II enzymes. The type I FAS and PKS are megasynthases, in which the domains are covalently linked together, whereas the type II FAS and PKS contain stand-alone domains. The high sequence conservation between FAS and PKS domains suggests that a general rule for stereochemical control may be broadly applied to both systems. For example, aromatic polyketides such as the antibiotic actinorhodin (6) (Figure 1) are produced from acyl thioesters by a type II PKS that is structurally and functionally related to the bacterial type II FAS (2). The type II PKS is comprised of 5–10 stand-alone proteins that act iteratively during the repeated condensation of malonyl thioesters to produce aromatic polyketides (Figure 2) (7). In the bacterial type II FAS, the iterative, concerted action of ketoreductase (KR), dehydratase (DH), and enoyl reductase (ER) leads to a fully reduced aliphatic fatty acid, in which the KR reduces every carbonyl group of the growing fatty acyl chain. In contrast, the type II PKS utilizes a highly conserved KR that specifically reduces only one carbonyl group (typically the C9) of a full-length polyketide chain (Figure 1). Subsequently, the presence of multiple carbonyl groups on the polyketide chain provides reactive α-carbon centers that lead to intra-molecular cyclizations (Figure 1, the formation of 1–6). Further diversity can be introduced by altering the chain length of the starting polyketide or varying the presence of other modification enzymes such as aromatase/cyclase (ARO/CYC) (Figure 1, the formation of 7–10). Because KR has been well recognized as a versatile oxidoreductase that can be applied to both directing biosynthesis and organic synthesis (8–11), there is a need to understand how KR directs the formation of chiral centers in polyketides. Compared to the type I KRs, whose stereospecificity has been investigated in depth (12–15), little is known about the stereospecificity of the type II polyketide KR. In other words, how the type II polyketide KR specifically reduces the C9-carbonyl group, as well as the resulting stereospecificity, have remained mysteries (16).
Figure 1.
Structures of the proposed and isolated KR intermediates. Multiple products have been reported from genetically engineered strains of Streptomyces that express the act minimal PKS (actKS, CLF and ACP) in the presence or absence of actKR. The position and timing of ketoreduction is shown in red. Polyketides BSM1–3 are truncated hexaketide products.
Figure 2.
Naturally occurring polyketides and substrate analogs. (A) Type II polyketides that are reduced at the C9-position by a ketoreductase (KR). (B) Substrate analogs used in this study.
The goal of this study is to elucidate the stereospecificity of KR from the actinorhodin type II PKS (referred to as actKR throughout the text). Earlier, we solved the crystal structures of wild type actKR in complex with NADP+ or NADPH, both in the presence or absence of the inhibitor emodin (17, 18). The cocrystal structures, combined with the development of an in vitro assay, enabled us to identify residues that are important for substrate binding and orientation. Significantly, the in vitro assay results show that actKR can only reduce the C9-carbonyl group if the substrate is pre-cyclized between C7 and C12 (Figure 1, leading to 4–6) (17, 18). Building on these results, in this work we aim to elucidate the stereospecificity of actKR. Although little is known about the stereospecificity of type II KRs, much research has been conducted to determine the reduction stereochemistry of isolated KR domains from the type I PKSs (14, 15, 19). For these type I KRs, the presence or absence of an “LDD motif” determines the reduction stereochemistry (detailed below) (20–22). The type I KRs have low sequence homology to the type II polyketide KRs, suggesting that actKR follows a different set of rules that govern the stereospecific C9-reduction. We hypothesize that, similar to type I polyketide KRs, the type II polyketide KRs also control their stereospecificity through distinct structural motifs located in the substrate binding pocket.
To test this hypothesis, we generated a sequence alignment of actKR with other short-chain dehydrogenase/reductase (SDR) proteins, specifically the type I KRs, and we identified the 94-PGG-96 region of actKR as a potential motif that is important for stereospecificity. The PGG region of actKR aligns with the LDD motif of the type I KRs (Figure 3B). Here, we present the mutations, in vitro assays, crystal structures, and docking simulations of actKR that strongly support the importance of the PGG motif in determining actKR stereospecificity. The above results allow us to propose a model that interprets the observed stereospecificity of type II KRs.
Figure 3.
Structural comparison among solved SDR proteins. (A) The location of the “LDD motif” (ball and stick representations) relative to the lid region in SDR proteins. Top: A comparison of actKR (blue ribbon), DEBS KR1 (green ribbon), and amphotericin KR2 (black ribbon) highlights major differences in the lid region (red triangles connected by dashes). Bottom: A comparison of actKR (blue ribbon), E. coli FabG (green ribbon), and TR-I (black ribbon). Helix α6 is conserved in these structures and α7 overlays well (a ribbon comparison between actKR and FabG is shown). (B) Sequence alignment shows that the PGG region (green highlight) is conserved among type II KRs. Sequences included: actinorhodin KR, frenolicin KR, granaticin KR, griseucin KR, hedamycin KR, nogalamycin KR, oxytetracycline KR, urdamycin KR, E. coli FabG, TR-I, TR-II, TH3N reductase, amphotericin KR2, DEBS KR1, and tylosin KR1.
MATERIALS AND METHODS
Materials
trans-1-Decalone, S-(+)-tetralol, R-(−)-tetralol, cofactor NADPH, and emodin were purchased from Sigma and were the highest grade available. Restriction endonucleases XbaI, EcoRI and SpeI, as well as Antarctic Phosphatase and T4 DNA Ligase, were purchased from New England Biolabs. Custom oligonucleotides were purchased from Operon Biotechnologies. All organic solvents including DMSO were HPLC grade or better and purchased from Fisher.
Gene Manipulation and Protein Purification in E. coli
Plasmid pYT284 contains the actIII gene cloned into the pET28b(+) expression vector (Novagen) (17). Single mutants G95D and G96D were generated by PCR using the Quick-Change Site Directed Mutagenesis Kit (Stratagene) with WT plasmid pYT284 as the template. The P94L point mutation was generated previously (18). Double- and triple-mutants were constructed using single- or double-mutant DNA as template. E. coli strain XL1-Blue (Stratagene) was used for plasmid amplification. All mutations were confirmed by sequencing.
ActKR Protein Expression and Purification
Recombinant WT or mutant actKR protein was expressed in E. coli strain BL21(DE3) as described previously (17, 18). Briefly, following transformation, cells were grown by shaking at 37 °C in 2 × 1 L Luria-Bertani media supplemented with 50 μg/mL kanamycin to an OD600 of 0.6. At that point, protein expression was induced by the addition of 0.1 mM IPTG at 18 °C overnight. Cells were harvested by centrifugation (3500 rpm × 30 min) at 4 °C, resuspended in 100 mL buffer A (50 mM Tris-Cl pH 7.5, 300 mM NaCl, 10 % glycerol), and lysed on ice by sonication (5 × 30 sec pulses). Cell debris were removed by centrifugation (14000 rpm × 45 min) followed by binding of recombinant protein to 5 mL Ni-IMAC resin (Bio-Rad) in batch mode at 4 °C. Bound actKR was eluted at 40, 60, 80, 100, 150, and 250 mM imidazole in buffer A. At this point, the WT or mutant proteins were approximately 95% pure. Imidazole was removed by dialysis of the pooled protein fractions overnight at 4 °C against 4 L buffer containing 50 mM Tris-Cl pH 7.5, 300 mM NaCl, 10% glycerol. The protein was concentrated to ~10 mg/mL with Pierce iCON 9000 MWCO protein concentrators.
In Vitro Assay for ActKR Mutant Activity
To monitor the effect of mutation on the stereospecificity of the reaction, steady-state kinetic parameters for WT and mutant complexes were determined by monitoring the reverse reaction of actKR in the presence of S- or R-tetralol and NADP+. The change in absorbance from the conversion of NADP+ to NADPH was monitored at 340 nm (ε340 = 6220 M−1 cm−1) on a Cary 3E UV-Vis Spectrophotometer (Varian) over 10 minutes. All assays were performed in 400 mM KPi buffer, pH 7.4 and were initiated with the addition of the enzyme at a concentration of 1.8 μM. The Michaelis-Menten constants Km and kcat for each substrate were obtained by varying the substrate concentration in the presence of 250μM NADPH. Data were fitted directly to the Michaelis–Menten equation using the program Kaleidagraph (Synergy).
Protein Crystallization and Data Collection
Crystallization conditions for the P94L actKR single mutant with NADPH in the absence or presence of emodin were described previously (18). In order to obtain full occupancy of both emodin molecules in the mutant active site, a solution of P94L actKR at 9 mg/mL in the presence of 2 mM NADPH was treated with 500 μM emodin dissolved in DMSO (5% v/v final) for at least 1 hour before setting up drops. Crystals grew within one week. Prior to flash-freezing, the crystals were soaked with an extra 500 μM emodin followed by back-soaking in 30% glycerol. Extensive screening of all other mutant complexes did not yield crystals. Data for the binary structure were collected on beamline BL9-1 at the Stanford Synchrotron Radiation Lightsource (SSRL) and data for the ternary structure were collected on beamline 8.2.2 at the Advanced Light Source (ALS) to a resolution of 2.3 Å. Diffraction intensities were indexed, integrated and scaled using HKL2000 (23). The data statistics are listed in Table 2.
Table 2.
Crystallization, data collection, and refinement statistics.
| P94L-NADPH | P94L-NADPH-emodin | |
|---|---|---|
| A. Crystallization | 4 M sodium formate | 4 M sodium formate |
| B. Crystallographic Data | ||
| Space group | P3221 | P3221 |
| Cell dimension (Å) | 104.58 104.58 123.59 | 105.09 105.09 123.65 |
| α = β =90 °, γ = 120 ° | α= β =90 °, γ = 120 ° | |
| Resolution (Å) | 90.57-2.79 | 50-2.29 |
| Mosaicity | 0.3 | 0.3 |
| No. of observations | 214691 | 392747 |
| No. of unique reflections | 37315 | 35974 |
| Completeness % (last shell) | 99.6 (99.2) | 100.0 (100.0) |
| I/σ(I) (last shell) | 29.4 (5.9) | 25.0 (5.5) |
| Rmerge % (last shell) | 9.0 (46.4) | 11.0 (49.5) |
| C. Refinement | ||
| Resolution (Å) | 2.79 | 2.29 |
| No. of reflections | 19609 | 35919 |
| No. of protein atoms | 3799 | 3752 |
| No. of cofactor atoms | 96 | 96 |
| No. of ligand atoms | 3 | 80 |
| No. of water atoms | 41 | 107 |
| Rfree % | 22.6 | 20.8 |
| Rcrys % | 16.4 | 18.3 |
| D. Geometry | ||
| RMS bonds (Å) | 0.008 | 0.008 |
| RMS angles (°) | 1.20 | 1.41 |
| RMS B main chain | 0.904 | 1.32 |
| RMS B side chain | 2.90 | 3.31 |
| Ramachandran plot (%) | ||
| Most favored | 89.1 | 90.3 |
| Favored | 10.9 | 9.7 |
| Generously allowed | 0 | 0.8 |
Phasing by Molecular Replacement and Refinement
The binary structure of the P94L actKR mutant was solved by molecular replacement using PHASER in CCP4i (24, 25). The previously solved WT actKR structure was used as the search model with waters removed. Following an initial round of refinement with REFMAC (26), manual rebuilding was performed and waters were added in COOT (27). For the ternary structure, emodin was generated using SKETCHER (25) and added to the 2fo-fc density contoured to 1 σ. Subsequent rounds of rebuilding and refinement in REFMAC were done for the binary and ternary structures. Further refinement of both structures was conducted in PHENIX (47) until the final R and Rfree were 0.164 and 0.226 for the binary structure and 0.183 and 0.208 for the ternary structure. The quality of the final structures was analyzed by PROCHECK (28). The refinement and model statistics are listed in Table 2.
DNA Manipulation and Generation of In Vivo Expression Vectors
Shuttle vectors containing the act min PKS (actKS, CLF and ACP) and actIII (KR) genes were derived from pBR1 (29). The actKR gene was amplified via PCR from pYT284 or mutant derivatives with engineered primers containing unique XbaI and SpeI restriction sites (underlined) and a ribosomal binding site (parentheses) with the following sequences: 5′-GCTCTAGA(AGGAGG)AGCCCATATGGCCACGCAGGACTCCGAAGT -3′- and 5′-GCGACTAGTTCAGTAGTTCCCCAGCCCGCCGCA -3′. To facilitate analysis and sequencing, the resulting ~0.8 kb fragment was digested with XbaI and SpeI and ligated into pBR1 that had been linearized with XbaI and treated with Antarctic Phosphatase for 2 hours at 37 °C. After DNA sequencing, a 1.7 kb fragment containing the actKR and actACP genes plus a 0.6 kb silent fragment was then retrieved from pBR1 through restriction digestion with XbaI and EcoRI, and purified by electrophoresis on a 1% agarose gel. This 1.7 kb fragment was then cloned into pYT84 after the vector was digested with XbaI and EcoRI to create pTK1 through pTK8, which contain actKS, actCLF, actACP, and actKR. The actKR in pTK1, 2, 3, 4, 5, 6, 7, and 8 is: wild type, P94L, G95D, G96D, P94L:G95D, G95D:G96D, P94L:G96D, and P94L:G95D:G96D.
Polyketide Production in Streptomycete Hosts
The heterologous in vivo production of polyketides was initiated by transformation of plasmids pTK1 through pTK8 independently into either S. lividans K4-114 (30) or S. coelicolor CH999 (31) on R5 plates supplemented with thiostrepton (50μg/mL) as a selectivity marker, as described in (32). Prior to transformation into S. coelicolor CH999, the plasmids were transformed into E. coli K12 ER2925 (New England Biolabs, dam dcm endA1 hsdR2 StrR CamR) to generate unmethylated DNA. For production and analysis of polyketides, transformed S. lividans K4-114 colonies were plated onto R5 agar plates supplemented with 50 μg/mL thiostrepton and incubated at 30 °C for 10 days. For product analysis, a single plate was finely chopped and extracted once with 30 mL of 94% ethyl acetate, 5% methanol, and 1% acetic acid. The extract was filtered and solvent removed under vacuum. The resulting resin was resuspended in 200 μL of 100% DMSO and stored at −20 °C prior to analysis. For large scale preparation of polyketides, 30 plates were extracted 3 times with 300 mL of 94% ethyl acetate, 5% methanol, and 1% acetic acid, dried under vacuum, and the resulting resin resuspended in 1.5 mL of 100% DMSO. Polyketide product analysis was performed by reverse phase HPLC on a C6-phenyl analytical column (Phenomenex, 5 μ, 250 × 4.6 mm). 10–20 μL of extract sample were loaded and separated with a 30–80% acetonitrile plus 0.1% formic acid gradient over 20 minutes, with absorbance monitored by a UV/vis diode array displayed at 254 nm.
Purification of Mutactin
Large-scale extracts were separately prepared from plates of S. lividans K4-114 expressing pTK1 (wild type actKR) or pTK2 (P94L actKR). Sample was run on a reverse phase C6-phenyl preparative column (Phenomenex, 5 μ, 250 × 21.2 mm) with a 15–60% acetonitrile plus 0.1% formic acid gradient over 65 minutes. The peak corresponding to mutactin, based on the UV profile (33), was collected along with the immediately preceding peak, which corresponds to EM18. Acetonitrile was removed from the collected eluent under vacuum and water removed through lyophilization. The remaining yellowish solid was dissolved in 1 mL of DMSO and run on a reverse phase C18 semi-preparative column (Beckman Coulter, 5 μ, 150 × 10 mm) with a 20–22.5% acetonitrile plus 0.1% formic acid gradient over 25 minutes. Both peaks were separately collected, and acetonitrile and water removed as before. Negative ion mode ESI-MS (Figure S2) and NMR analysis (Table S1 and Figure S3) confirmed that the peaks correspond to EM18 and mutactin.
Chiral HPLC Analysis of Mutactin
Mutactin and EM18 were collected together during the first semi-preparative HPLC step and lyophilized. Approximately 1 mg of solid was dissolved in 750 μL of DMSO, and 20 μL of this sample was injected onto a Chiralcel OD-R column equilibrated with 20% acetonitrile plus 0.1% formic acid and run at an isocratic gradient over 20 minutes. The HPLC chromatograms are shown in Figure S1.
RESULTS AND DISCUSSION
Rationale for Targeting the PGG Motif of ActKR
We propose to target the 94-PGG-96 motif of actKR based on a critical comparison between available crystal structures of type I versus type II polyketide KRs. Both types of KRs possess the short-chain dehydrogenase/reductase (SDR) fold (Figure 3A), which consists of a Rossman fold with the active site tetrad S144-Y157-K161-N114 (actKR numbering) at the core interacting with the cofactor NADPH, and two flexible loop regions (β4-α4 and α6-α7) co-defining the substrate binding pocket. The major difference between type I and type II polyketide KRs lies in their oligomeric state: while the type I polyketide KR is typically a monomer containing two SDR domains (one for catalysis and the other for structural support), the type II polyketide KR is tetrameric, and each monomer contains only one SDR domain. Another difference between the type II polyketide KRs and other SDR proteins is that the former contain a 10-residue insertion between α6 and α7, which is also the least conserved region among SDR proteins.
For type I polyketide KRs, the relationship between protein sequence and ketoreduction stereospecificity was first described by Celmer in 1965 through the analysis of reduced polyketide stereochemistry (34). Subsequent studies by Leadlay (20, 22, 35–38), Weissman (39, 40), and Khosla and Cane (15, 41, 42) determined sequence elements that are important for the type I KR stereospecificity. Type I KRs can be further classified as A-type or B-type, which respectively lead to S or R stereochemistry for the product alcohol (43). The A-type KR has a Trp at position 149, while the B-type has Leu-Asp-Asp residues at positions 94–96 (actKR numbering) located along the β4-α4 loop that defines part of the substrate binding pocket (Figure 3A). The molecular basis of A- and B-type KRs has been investigated in-depth by Keatinge-Clay et al. recently (12–15, 44). The major determinant of β-hydroxyl stereochemistry has been attributed to the second aspartic acid, which is invariant in KRs that produce the R isomer and absent in those that produce the S isomer (14). Similar motifs that align with the LDD motif are also important for the stereospecificity of related SDR proteins, such as the fatty acid KR FabG (45) and tropinone reductases (TR-I and TR-II) (46). The LDD motif of type I KR overlaps with 94-PGG-96 of actKR (Figure 3B). A structural overlay of actKR with the above homologous SDR structures (Figure 3A) shows that: (1) the position and orientation of the loop containing the PGG or LDD motif is similar in all of these structures and (2) the main difference is the location of helix α6 that forms the lid region of the substrate pocket. For the type I KR (14), Keatinge-Clay et al. hypothesized that the presence of the LDD motif influences the orientation of the polyketide substrate by interacting with the hydrophobic portion of the ACP-bound phosphopantetheine (PPT) group. Based on the above structural comparison between type I and II KRs, we hypothesize that the 94-PGG-96 motif may serve as a determinant of stereospecificity in type II KRs, similar to the LDD motif in type I KRs.
In Vitro Assay of Wild Type ActKR: The Ability to Catalyze the Oxidation of Tetralol
The evaluation of actKR stereospecificity requires a versatile enzymatic assay with easily accessible chiral substrates. We previously demonstrated that pure actKR is active in vitro in the presence of the bicyclic substrates trans-1-decalone and α-tetralone (Figure 2) (18). These previous results showed that actKR accepts a cyclic substrate (Figure 1), but not a linear one; however, we did not probe the stereochemistry of ketoreduction. To determine the stereospecificity of actKR, we measured the Michaelis-Menten parameters for the oxidation of enantiopure R-(−)-tetralol (22) or S-(+)-tetralol (23) by actKR in the presence of NADP+ (Figure 2). When wild type actKR was assayed with S-(+)-tetralol or R-(−)-tetralol, it displayed a threefold preference for S-(+)-tetralol over R-(−)-tetralol (Table 1). By inference, other type II KRs that are highly homologous to actKR (Figure 3B) should also exhibit a preference for S stereochemistry. By the rule of microscopic reversibility, the fact that actKR accepts both tetralol enantiomers seems to agree with a previous report that the reduced polyketide mutactin (4) is present as a racemic mixture (Figure 1, more details below) (16). The ability to accept both tetralol enantiomers suggests that the actKR active site can bind the in vitro substrate in two orientations, albeit with a preference for one orientation (S) over the other (R).
Table 1.
Comparison of actKR activity in the presence of S-(+)-tetralol or R-(−)-tetralol. The activity of actKR mutants shows that the P94L mutation is sufficient to alter the stereospecificity such that the S stereomer is preferred over the R stereomer.
| Kinetic Parameters for the Oxidation of S-(+)-Tetralol and R-(−)-Tetralol by Wild Type and Mutants of Actinorhodin Ketoreductase (ActKR) | ||||||
|---|---|---|---|---|---|---|
 |
 |
|||||
| kcat (s−1) | Km (mM) | kcat/Km (s−1 mM−1) | kcat (s−1) | Km (mM) | kcat/Km (s−1 mM−1) | |
| WT | 0.24 ±0.01 | 6.91 ±1.2 | 0.035 ±0.006 | 0.06 ±0.004 | 6.47 ±0.90 | 0.010 ±0.001 |
| P94L | 0.24 ±0.03 | 6.57 ±1.8 | 0.036 ±0.011 | ND | ≫40 | ND |
| G95D | inactive | inactive | inactive | inactive | inactive | inactive |
| G96D | 0.021 ±0.004 | 33.8 ±9.5 | 0.001 ±0.0002 | ND | ≫40 | ND |
| P94L:G95D | 0.28 ±0.03 | 8.7 ± 2.0 | 0.032 ±0.001 | ND | ≫40 | ND |
| G95D:G96D | inactive | inactive | inactive | inactive | inactive | inactive |
| P94L:G96D | 0.23 ±0.02 | 10.9 ±1.9 | 0.02 ±0.004 | ND | ≫40 | ND |
| P94L:G95D:G96D | inactive | inactive | inactive | inactive | inactive | inactive |
ND: cannot determine the kinetic parameters.
A Single Mutation in ActKR is Sufficient to Change Stereospecificity In Vitro
In order to determine whether the PGG motif is a determinant of stereospecificity in type II KRs, similar to the LDD motif in type I KRs, we generated the single, double and triple mutant combinations of P94L, G95D and G96D and analyzed the respective stereospecificities using the above in vitro assay (Table 1). Significantly, the P94L single mutation is sufficient to alter the stereospecificity of the reaction. In solution, P94L is as stable as the wild type actKR, while all other mutants tend to precipitate after 2–3 days post Ni-column purification, indicating the de-stablization effect caused by mutations other than P94L. While P94L actKR retains wild type activity in the presence of S-(+)-tetralol (kcat/Km = 0.036 ± 0.011 s−1 mM−1 compared to 0.035 ± 0.006 s−1 mM−1 for wild type), it has no detectable activity for R-(−)-tetralol (Table 1). The G96D mutant also shows a dominant preference for S-(+)-tetralol, albeit with 30 times less efficiency (kcat/Km = 0.001 ± 0.0002 s−1 mM−1) than P94L.
The G95D single mutant is inactive toward tetralol (Table 1) or trans-1-decalone (data not shown). This loss of activity is likely due to a change in the loop conformation in this region (Figure 4). Consistent with this hypothesis, we could not crystallize G95D in the crystallization conditions of either wild type or P94L actKR, which is likely caused by a change of protein conformation of G95D. The G95D:G96D and P94L:G95D:G96D mutants are also inactive, as expected from the presence of the G95D mutation (Table 1). Interestingly, P94L “rescues” the G95D mutation in the P94L:G95D double mutant, which retains wild type activity in the presence of S-(+)-tetralol and has no detectable activity toward R-(−)-tetralol. It is possible that the stabilizing effect of P94L exceeds the de-stablization effect of G95D, leading to the observed activity in the P94L:G95D mutant.
Figure 4.
Overlay of the WT and P94L actKR structures. (A) Left panel: Surface representation of the P94L mutant depicting NADPH (ball and stick model), the predicted PPT-binding site, and the direction of polyketide binding into the active site (dashed line). Right panel: Close-up view of the overlaid wild type (grey) and P94L (green) structures. (B) The scheme depicts the different conformations of monomers A and B found in each crystal structure. A green circle corresponds to an open conformation, red triangles correspond to a closed conformation, and the square represents a conformation between fully open and fully closed.
The G96D single mutation results in much reduced activity (Table 1) and the observed S-dominant stereospecificity may be due to our detection limit of this low-activity mutant toward R-(−)-tetralol. Similar to the P94L:G95D double mutant, the activity of the P94L:G96D double mutant is also “rescued” by the presence of the P94L mutation, with kcat and kcat/Km similar to those of wild type and a dominant S-stereospecificity. Presumably, the P94L mutation stabilizes the flexible regions of actKR to such an extent that the de-stabilizing forces originating from the G95D or G96D mutations can be canceled out, resulting in WT-like activity in the double mutants P94L:G95D and P94L:G96D. The above results also showed that the molecular features governing ketoreduction stereochemistry are markedly different between type I and type II polyketide KRs.
Wild Type and P94L ActKR Structures: Binary vs. Ternary
To probe the molecular basis of stereospecificity for type II KRs, we solved a series of actKR crystal structures. We previously solved and analyzed the actKR structures of WT-NADP+, WT-NADPH, WT-NADPH-emodin, and P94L-NADPH-emodin (18). However, previous analyses focused on the emodin binding motif, and little comparative analysis was done to determine the differences between WT and P94L structures (18). In this work, we solved the P94L-NADPH binary crystal structure and also resolved the ternary P94L-NADPH-emodin structure to a higher resolution with full emodin occupancy in monomer A. We then conducted the following structural comparisons among WT-NADPH, WT-NADPH-emodin, P94L-NADPH, and P94L-NADPH-emodin crystal structures, focusing on the conformational changes induced by the P94L mutation and by the presence of emodin (Figure 4).
(1) WT-NADPH vs. P94L-NADPH. Based on the B-factors, the hinge region between helices α6 and α7 is the most flexible region of actKR and it can adopt either an open or closed conformation, subsequently changing the size and shape of the substrate binding pocket. A comparison of this region between monomers A and B in the WT-NADPH and P94L-NADPH binary structures shows that the P94L mutation affects the open versus closed conformations, as well as the shape of the binding pocket (Figure 4). Monomer A of both the WT and P94L binary structures adopts an open conformation. In comparison, for monomer B, WT-NADP+ has a disordered α6-α7 region, WT-NADPH has a partially open conformation, and P94L-NADPH has a completely closed conformation. When overlapping monomers A and B, in the WT-NADPH structure, there is a 3-Å displacement between the α6-α7 regions in the two monomers, as measured by the movement of E207 in helix 6. In comparison, this movement is more than 6 Å in the P94L-NADPH structure, which may be a result of increased favorable interactions between L94 and the V198 and M194 side chains (Figure 4). Such hydrophobic interactions would bring helix α6 closer to the LGG loop of P94L.
(2) WT-NADPH-emodin vs. P94L-NADPH-emodin. The difference between WT and P94L actKR becomes more obvious upon emodin binding: in WT-NADPH-emodin, only one emodin was observed in monomer A and no emodin was bound in monomer B. Binding of emodin locks the enzyme into an open conformation; subsequently, monomers A and B of the WT-NADPH-emodin structure have open and closed conformations, respectively (Figure 4). In comparison, two emodin molecules were observed in both monomers A and B of the P94L-NADPH-emodin structure; subsequently, both monomers have an open conformation. The differences between the monomers in each structure are summarized in Figure 4B. Significantly, docking simulation of the natural actKR substrate, the octaketide, consistently docked the C9-carbonyl group to a position corresponding to the second emodin in the P94L ternary structure, not the emodin position observed in the wild type ternary structure. Therefore, the new P94L-NADPH-emodin structure reported herein helps us locate the binding motif and residues important for enzyme-substrate interaction. The above comparisons between the binary and ternary structures also reveal that P94L is much more sensitive to substrate binding than wild type, an important property linked to its observed increase of stereospecificity.
The Molecular Basis of Stereospecificity of Type II KRs
From the above structural comparisons, the conformation of P94L actKR is much more sensitive to substrate binding, which correlates well with its increased stereospecificity. This is in contrast to previous type I KR mutation results (all resulted in a decrease of stereospecificity), suggesting that the molecular determinants of stereospecificity are different between type I and type II KRs.
In the tylosin (Tyl) or DEBS (Ery) KR structures, the position of the LDD loop is stabilized by a combination of electrostatic interactions between D95 (actKR numbering) and a backbone nitrogen from the “lid helix” (Figure 3), which essentially seals the active site in a zipper-like fashion and prevents entrance of the polyketide chain from the left side (14). In comparison, there are no such stabilizing electrostatic interactions between P94 and helix α7 in type II KRs. Also, when compared to the type I KR structures, the presence of helix α6 in the type II KRs (not found in type I KRs) causes the corresponding “lid helix” α7 to rotate away from the active site (Figure 3A). Helix α6 is present in TR-I, TR-II and TH3N reductase (46, 47) and, along with helix α7, has been shown to play an important role in substrate orientation. This feature suggests that the stereospecificity of actKR is controlled in a fashion more similar to TR-I, TR-II and TH3N reductase than to type I polyketide KRs (Figure 3A). Therefore, the lack of side chains in the PGG motif, in combination with the presence of helix α6, supports the hypothesis that the PPT arm presents the polyketide from the left side (Figure 4) of the pocket at all times and that the high stereospecificity of actKR mutants arises as a result of a change in substrate pocket shape rather than a difference in PPT binding. Consequently, wild type actKR has a preference for S-tetralol, while the more substrate-sensitive P94L actKR is S-dominant. We therefore predict that when holo-PKS is assembled as a complex (KS/CLF + ACP + KR), because of the additional restraint from protein-protein interaction between ACP and KR, the S-dominant stereospecificity should be more prominent.
In Vivo Polyketide Biosynthesis by Type II PKS Complex Supports the Importance of P-G-G Motif
To determine whether the stereospecificity observed from the in vitro assay with S- and R-tetralol could reflect the actual polyketide stereochemistry, and to test the above hypothesis that the holo-PKS complex will further increase the S-stereospecificity, we cloned the actinorhodin KS/CLF, ACP, and WT or mutant actKR into a pRM5-derived vector, and transformed it into the Streptomyces lividans K4-114 host strain. The actinorhodin KS/CLF and ACP constitute the “minimal PKS” (act min PKS), which produces the unreduced polyketide SEK4 (11) (Figure 1). The act min PKS + WT actKR (pTK1) was also transformed into Streptomyces coelicolor CH999 to monitor strain-specific differences. The polyketides produced by each construct were analyzed by HPLC and HRMS.
HPLC analysis of pTK1 shows the presence of at least three major peaks and two minor peaks in the crude extract from both S. lividans K4-114 (Figure 5) and S. coelicolor CH999 host strains (not shown), suggesting that there are no strain-specific differences in polyketide production. The product profile is more complete than in previous studies, which only reported mutactin (4) and dehydromutactin (6) (7, 48). The presence of products 2 and 3 with low molecular weights of 218 and 232 most likely correspond to truncated hexaketides (Figure 1, top), supporting a recent report that the act min PKS can produce truncated polyketides with shorter chain lengths (49). Consistent with the in vitro results, the P94L, P94L:G95D and P94L:G96D product profiles are similar to that of WT (Table 1 and Figure 5). The P94L crystal structures and the observation that the P94L mutant generates the same levels of products in vivo as WT actKR (Figure 5) indicate that P94L retains a reductase-competent active site while maintaining appropriate interactions with ACP and the PPT group. In comparison, when actKR is absent, there is an approximate 1:1 ratio of 11 to 12 produced (Figure 5). The G95D and G96D mutants, which were either inactive or displayed greatly reduced activity in vitro, mainly produced the unreduced octaketides 11 and 12 in vivo. However, these two mutants still produced mutactin (4), albeit at reduced levels (Figure 5), suggesting that in the presence of upstream KS/CLF and ACP, the activities of these two KR mutants are somewhat rescued by appropriate protein-protein interactions between KR, ACP and KS/CLF.
Figure 5.
In vivo analysis of polyketide production. The engineered Streptomyces host expresses the act minimal PKS and WT or mutant actKR genes. HPLC traces of actKR single, double and triple mutants show the effect of mutation on the biosynthesis of mutactin (4), as well as SEK4 (11), SEK4b (12), SEK34 (7), BSM1 (2), BSM2 (3), dehydromutactin (6), and SEK34b (8).
Similarly, the G95D:G96D and P94L:G95D:G96D mutants, inactive in vitro, are also inactive in vivo, producing only the unreduced full-length octaketides 11 and 12 (Figures 1, 5). The fact that no shorter hexaketides are formed with the G95D:G96D and P94L:G95D:G96D mutants supports the hypothesis that the formation of the truncated hexaketide requires the presence of an active actKR, a hypothesis also proposed by Moore et al., that an active actKR may compete with KS/CLF for a penta- or hexa-ketide intermediate, thus derailing the formation of full-length octaketide (49). In the mutants, the KR active site or KR-ACP interactions necessary for such a derailment may have been abolished, resulting in only the full-length octaketide biosynthesized in the upstream KS/CLF active site. In summary, the in vivo heterologous expression results confirm that mutations in the PGG motif affects both enzyme activity and product outcome.
In Silico Analysis of Regio- and Stereo-specific Reduction by ActKR
In the absence of the larger and bulkier leucine, wild type actKR can accommodate both S-(+)- and R-(−)-tetralol. When the proline is mutated to leucine, steric interactions preclude binding of the R-isomer. This observation can be rationalized by computer-simulated docking of S-(+)- and R-(−)-tetralol to the wild type and P94L actKR structures using the program GOLD (50). Docking shows that the presence of the smaller proline provides enough space to accommodate the aromatic ring of either S-(+)- or R-(−)-tetralol. However, in P94L actKR the bulkier leucine blocks this area and allows only S-(+)-tetralol to bind. These results imply that the KR stereospecificity is influenced at least partially by steric constraints imposed by pocket residues near the P-G-G motif (Figure 6).
Figure 6.
Docking of S-(+)- or R-(−)-tetralol to the P94L actKR mutant. Representation of the difference in (A) S-(+)-tetralol and (B) R-(−)-tetralol binding predicted from in vitro assay results and confirmed by in silico docking.
The docking results help explain not only the observed in vitro stereospecificity, but also the in vivo truncated products. Previously, Moore et al. proposed that actKR accepts two possible substrates, the full-length octaketide (16 carbons) or a truncated pentaketide (10 carbons) that undergoes a subsequent round of elongation to produce a reduced hexaketide (12 carbons) (Figure 1, top) (49, 51). We were able to reproduce the Moore result and biosynthesized the truncated hexaketides when we included both the act min PKS and an active actKR (WT or mutants). However, assuming the PPT group is anchored in the arginine patch (R38, R65 and R93), as previously proposed (18), docking of putative actKR substrates with either WT or P94L structures showed that a minimum of 12 carbons is necessary to position the C5-carbonyl in the oxyanion hole (which would result in the formation of 1–3, Figure 1). Therefore, the previous proposal that actKR accepts a 10 carbon substrate would cause physical strain to the enzyme-substrate interactions. Rather, docking indicates the 12-carbon hexaketide as a more likely truncated substrate for actKR. In addition, the arginine patch has been identified as the putative ACP-docking site for both actKR and the homologous SCO1815 KR from S. coelicolor (52) and its presence would restrict the attached polyketide to a single side of the pocket (17, 18). The above analysis leads to the hypothesis that actKR physically “counts” the carbon chain length, starting from PPT binding groove, and that actKR determines which carbonyl group to reduce by the physical distance between the PPT group and the enzyme active site. Consistent with this hypothesis, our in vivo results show that formation of C9-reduced octaketides is in large excess to C5-reduced hexaketide, because the longer octaketide can better reach the actKR active site.
Toward the Absolute Stereochemistry of the C9-Position of Mutactin Produced by WT or P94L ActKR In Vivo
The assignment of absolute stereochemistry for products of actKR is complicated by the fact that water elimination (i.e. aromatization) often removes the C9-hydroxyl group (7, 18, 48, 49) during type II polyketide biosynthesis (Figure 1, formation of 1–3, 5–10), thus rendering an assignment at C9 difficult. The structure of mutactin was characterized by the Floss group in 1990 (16). Circular dichroism analysis of the molecule yielded no signal and NMR analysis using chiral derivatizing or shift reagents was not successful; therefore, it was suggested that mutactin is present as a racemic mixture (16).
The observed S-dominant stereochemistry of the P94L mutant offers a unique opportunity to determine the actKR stereospecificity in vivo. Assuming that the in vivo stereospecificity of actKR correlates to that measured in vitro, the S. lividans K4-114 strain expressing pTK1 (WT actKR) would produce mutactin enantiomers in an approximate 3:1 ratio (S:R), whereas pTK2 (P94L actKR) would lead to the production of S-mutactin alone.
To this end, mutactin was purified from large-scale polyketide extracts from both pTK1- and pTK2-expressing S. lividans K4-114 grown on plates. Different analytical chiral HPLC columns were utilized to resolve the enantiomers of mutactin. To confirm the functionality of the chiral column, mutactin was analyzed along with EM18 (5 in Figure 1, racemic at C6 and C15, detailed in Supporting Information), which elutes immediately before mutactin. Figure S1 shows the pTK1 and pTK2 chiral HPLC chromatograms. For both samples, EM18 resolves into two peaks with the same UV absorbance profile, but neither pTK1- nor pTK2-derived mutactin separates into two peaks, suggesting that mutactin is not present as a mixture of enantiomers as a product of either wild type or P94L actKR. A possible explanation for this result stems from the fact that the in vitro assay utilizes a bicyclic substrate analog that, while it can be oxidized by actKR, does not represent the true in vivo substrate, a polyketide chain cyclized between C7–C12 and linked to ACP by the PPT group. Our docking simulation supports the hypothesis that the PPT arm presents the polyketide from the left side (arginine patch) of the substrate pocket (17, 18) at all times (Fig. 4). If this is the case, the polyketide must appropriately position the C9-carbonyl within the oxyanion hole for ketoreduction only from the left side (17). Subsequently, the hydride from NADPH will consistently be transferred to the same face of the carbonyl group at all times, thus producing only one enantiomer of mutactin.
Although EM18 was effectively resolved into two stereomers through chiral HPLC, while mutactin was not, there does exist a possibility that mutactin is not amenable to this procedure. To address this concern, we sought NMR as a means of determining whether purified pTK1-derived mutactin exists as enantiomers. Derivatization with a reagent such as Mosher’s acid (53) was not successful due to the presence of multiple hydroxyl groups to which the reagent could form an ester. We also conducted proton NMR analysis of mutactin in the presence of Eu(dcm)3, a lanthanide chiral shift reagent (54). While the NMR spectra suggested that the reagent interacts with mutactin, there was no conclusive evidence as to the presence or absence of enantiomers (data not shown). The chiral HPLC results of EM18 co-eluted with mutactin (Fig. S1) did strongly suggest that mutactin produced in vivo through heterologous expression is not enantiomeric, consistent with our structural analysis outlined above. Further analysis is necessary to definitively determine the stereochemistry of mutactin.
Biological Significance
The therapeutic implications of producing stereospecific natural products are deeply appreciated (4). In the case of aromatic polyketide biosynthesis, the potential ability of KR to stereospecifically reduce full-length polyketides presents an additional means of introducing diversity and developing new polyketide products. Building upon previous structural and biochemical analyses (17, 18), the 94-XGG-96 motif in type II KRs was identified to be important for reduction stereospecificity. Mutation of this region in actKR showed that stereospecific reduction could be achieved through a single point mutation (P94L) in vitro. The dominant preference for S-(+)-tetralol of P94L supports the notion that altering native PKS proteins through site-specific mutation is a viable approach to controlling stereospecificity of type II KRs. These results pave the foundation toward the rational control of stereospecificity and design of new polyketides with potential pharmaceutical capabilities.
Supplementary Material
Acknowledgments
This work is supported by the Pew Foundation and National Institute of General Medicinal Sciences (NIGMS R01GM076330).
We thank Prof. Larry E. Overman for use of the Chiralcel OD-R column, as well as Mr. Stephen Canham for his suggestions in regard to chiral HPLC methodology. We also thank Dr. Philip R. Dennison for assistance with NMR data acquisition and processing. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. Portions of this research were carried out at the Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Abbreviations
- KR
ketoreductase
- FabG
β-Ketoacyl [Acyl Carrier Protein] Reductase
- Act
Actinorhodin
- PKS
polyketide synthase
- NADP
nicotinamide adenine dinucleotide phosphate
- NADPH
reduced nicotinamide adenine dinucleotide phosphate
- SDR
short-chain dehydrogenase/reductase
- T3HNR
1,3,8 trihydroxynaphthalene reductase
- T4HNR
1,3,6,8-tetrahydroxynaphthalene
- ACP
acyl carrier protein
Footnotes
The atomic coordinates have been deposited in the Protein Data Bank (accession code 3QRW and 3CSD).
Supporting information of EM18 structural analysis is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Gullo VP, McAlpine J, Lam KS, Baker D, Petersen F. Drug discovery from natural products. J Ind Microbiol Biotechnol. 2006;33:523–531. doi: 10.1007/s10295-006-0107-2. [DOI] [PubMed] [Google Scholar]
- 2.Hopwood DA. Genetic Contributions to Understanding Polyketide Synthases. Chem Rev. 1997;97:2465–2498. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 3.Cane DE, Walsh CT, Khosla C. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 4.McConnell O, Bach A, 2nd, Balibar C, Byrne N, Cai Y, Carter G, Chlenov M, Di L, Fan K, Goljer I, He Y, Herold D, Kagan M, Kerns E, Koehn F, Kraml C, Marathias V, Marquez B, McDonald L, Nogle L, Petucci C, Schlingmann G, Tawa G, Tischler M, Williamson RT, Sutherland A, Watts W, Young M, Zhang MY, Zhang Y, Zhou D, Ho D. Enantiomeric separation and determination of absolute stereochemistry of asymmetric molecules in drug discovery: building chiral technology toolboxes. Chirality. 2007;19:658–682. doi: 10.1002/chir.20399. [DOI] [PubMed] [Google Scholar]
- 5.Hutchinson CR. Combinatorial biosynthesis for new drug discovery. Curr Opin Microbiol. 1998;1:319–329. doi: 10.1016/s1369-5274(98)80036-2. [DOI] [PubMed] [Google Scholar]
- 6.Bartel PL, Zhu CB, Lampel JS, Dosch DC, Connors NC, Strohl WR, Beale JM, Jr, Floss HG. Biosynthesis of anthraquinones by interspecies cloning of actinorhodin biosynthesis genes in streptomycetes: clarification of actinorhodin gene functions. J Bacteriol. 1990;172:4816–4826. doi: 10.1128/jb.172.9.4816-4826.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. Rational design of aromatic polyketide natural products by recombinant assembly of enzymatic subunits. Nature. 1995;375:549–554. doi: 10.1038/375549a0. [DOI] [PubMed] [Google Scholar]
- 8.Jacobsen JR, Keatinge-Clay AT, Cane DE, Khosla C. Precursor-directed biosynthesis of 12-ethyl erythromycin. Bioorg Med Chem. 1998;6:1171–1177. doi: 10.1016/s0968-0896(98)00081-9. [DOI] [PubMed] [Google Scholar]
- 9.Broussy S, Cheloha RW, Berkowitz DB. Enantioselective, ketoreductase-based entry into pharmaceutical building blocks: ethanol as tunable nicotinamide reductant. Org Lett. 2009;11:305–308. doi: 10.1021/ol802464g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Truppo MD, Pollard D, Devine P. Enzyme-catalyzed enantioselective diaryl ketone reductions. Org Lett. 2007;9:335–338. doi: 10.1021/ol0627909. [DOI] [PubMed] [Google Scholar]
- 11.Grau BT, Devine PN, DiMichele LN, Kosjek B. Chemo- and enantioselective routes to chiral fluorinated hydroxyketones using ketoreductases. Org Lett. 2007;9:4951–4954. doi: 10.1021/ol701810v. [DOI] [PubMed] [Google Scholar]
- 12.Zheng J, Taylor CA, Piasecki SK, Keatinge-Clay AT. Structural andfunctional analysis of A-type ketoreductases from the amphotericin modular polyketide synthase. Structure. 18:913–922. doi: 10.1016/j.str.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 13.Keatinge-Clay AT, Stroud RM. The structure of a ketoreductase determines the organization of the beta-carbon processing enzymes of modular polyketide synthases. Structure. 2006;14:737–748. doi: 10.1016/j.str.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Keatinge-Clay AT. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Castonguay R, Valenzano CR, Chen AY, Keatinge-Clay A, Khosla C, Cane DE. Stereospecificity of ketoreductase domains 1 and 2 of the tylactone modular polyketide synthase. J Am Chem Soc. 2008;130:11598–11599. doi: 10.1021/ja804453p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang HL, He XG, Adefarati A, Gallucci J, Cole SP, Beale JM, Keller PJ, Chang CJ, Floss HG. Mutactin, a novel polyketide from Streptomyces coelicolor. Structure and biosynthetic relationship to actinorhodin. The Journal of Organic Chemistry. 1990;55:1682–1684. [Google Scholar]
- 17.Korman TP, Hill JA, Vu TN, Tsai SC. Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity. Biochemistry. 2004;43:14529–14538. doi: 10.1021/bi048133a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korman TP, Tan YH, Wong J, Luo R, Tsai SC. Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase. Biochemistry. 2008;47:1837–1847. doi: 10.1021/bi7016427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castonguay R, He W, Chen AY, Khosla C, Cane DE. Stereospecificity of ketoreductase domains of the 6-deoxyerythronolide B synthase. J Am Chem Soc. 2007;129:13758–13769. doi: 10.1021/ja0753290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holzbaur IE, Harris RC, Bycroft M, Cortes J, Bisang C, Staunton J, Rudd BA, Leadlay PF. Molecular basis of Celmer’s rules: the role of two ketoreductase domains in the control of chirality by the erythromycin modular polyketide synthase. Chem Biol. 1999;6:189–195. doi: 10.1016/S1074-5521(99)80035-0. [DOI] [PubMed] [Google Scholar]
- 21.Holzbaur IE, Ranganathan A, Thomas IP, Kearney DJ, Reather JA, Rudd BA, Staunton J, Leadlay PF. Molecular basis of Celmer’s rules: role of the ketosynthase domain in epimerisation and demonstration that ketoreductase domains can have altered product specificity with unnatural substrates. Chem Biol. 2001;8:329–340. doi: 10.1016/s1074-5521(01)00014-x. [DOI] [PubMed] [Google Scholar]
- 22.Ostergaard LH, Kellenberger L, Cortes J, Roddis MP, Deacon M, Staunton J, Leadlay PF. Stereochemistry of catalysis by the ketoreductase activity in the first extension module of the erythromycin polyketide synthase. Biochemistry. 2002;41:2719–2726. doi: 10.1021/bi0117605. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 29.McDaniel R, Ebert-Khosla S, Fu H, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides: influence of a downstream enzyme on the catalytic specificity of a minimal aromatic polyketide synthase. Proc Natl Acad Sci U S A. 1994;91:11542–11546. doi: 10.1073/pnas.91.24.11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ziermann R, Betlach MC. Recombinant polyketide synthesis in Streptomyces: engineering of improved host strains. BioTechniques. 1999;26:106–110. doi: 10.2144/99261st05. [DOI] [PubMed] [Google Scholar]
- 31.McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides. Science. 1993;262:1546–1550. doi: 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- 32.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomcyes Genetics. The John Innes Foundation; Norwich: 2000. [Google Scholar]
- 33.Ma SM, Zhan J, Xie X, Watanabe K, Tang Y, Zhang W. Redirecting the cyclization steps of fungal polyketide synthase. J Am Chem Soc. 2008;130:38–39. doi: 10.1021/ja078091o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Celmer WD. Biogenetic, constitutional, and stereochemical unitary principles in macrolide antibiotics. Antimicrob Agents Chemother (Bethesda) 1965;5:144–156. [PubMed] [Google Scholar]
- 35.Siskos AP, Baerga-Ortiz A, Bali S, Stein V, Mamdani H, Spiteller D, Popovic B, Spencer JB, Staunton J, Weissman KJ, Leadlay PF. Molecular basis of Celmer’s rules: stereochemistry of catalysis by isolated ketoreductase domains from modular polyketide synthases. Chem Biol. 2005;12:1145–1153. doi: 10.1016/j.chembiol.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 36.Baerga-Ortiz A, Popovic B, Siskos AP, O’Hare HM, Spiteller D, Williams MG, Campillo N, Spencer JB, Leadlay PF. Directed mutagenesis alters the stereochemistry of catalysis by isolated ketoreductase domains from the erythromycin polyketide synthase. Chem Biol. 2006;13:277–285. doi: 10.1016/j.chembiol.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 37.O’Hare HM, Baerga-Ortiz A, Popovic B, Spencer JB, Leadlay PF. High-throughput mutagenesis to evaluate models of stereochemical control in ketoreductase domains from the erythromycin polyketide synthase. Chem Biol. 2006;13:287–296. doi: 10.1016/j.chembiol.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 38.Kellenberger L, Galloway IS, Sauter G, Bohm G, Hanefeld U, Cortes J, Staunton J, Leadlay PF. A polylinker approach to reductive loop swaps in modular polyketide synthases. Chembiochem. 2008;9:2740–2749. doi: 10.1002/cbic.200800332. [DOI] [PubMed] [Google Scholar]
- 39.Bali S, Weissman KJ. Ketoreduction in mycolactone biosynthesis: insight into substrate specificity and stereocontrol from studies of discrete ketoreductase domains in vitro. Chembiochem. 2006;7:1935–1942. doi: 10.1002/cbic.200600285. [DOI] [PubMed] [Google Scholar]
- 40.Bali S, O’Hare HM, Weissman KJ. Broad substrate specificity of ketoreductases derived from modular polyketide synthases. Chembiochem. 2006;7:478–484. doi: 10.1002/cbic.200500430. [DOI] [PubMed] [Google Scholar]
- 41.Valenzano CR, Lawson RJ, Chen AY, Khosla C, Cane DE. The biochemical basis for stereochemical control in polyketide biosynthesis. J Am Chem Soc. 2009;131:18501–18511. doi: 10.1021/ja908296m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin Y, Gokhale R, Khosla C, Cane DE. Erythromycin biosynthesis. The 4-pro-S hydride of NADPH is utilized for ketoreduction by both module 5 and module 6 of the 6-deoxyerythronolide B synthase. Bioorg Med Chem Lett. 2001;11:1477–1479. doi: 10.1016/s0960-894x(00)00529-1. [DOI] [PubMed] [Google Scholar]
- 43.Caffrey P. The stereochemistry of ketoreduction. Chem Biol. 2005;12:1060–1062. doi: 10.1016/j.chembiol.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Caffrey P. Conserved amino acid residues correlating with ketoreductase stereospecificity in modular polyketide synthases. Chembiochem. 2003;4:654–657. doi: 10.1002/cbic.200300581. [DOI] [PubMed] [Google Scholar]
- 45.Price AC, Zhang YM, Rock CO, White SW. Cofactor-induced conformational rearrangements establish a catalytically competent active site and a proton relay conduit in FabG. Structure. 2004;12:417–428. doi: 10.1016/j.str.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 46.Nakajima K, Yamashita A, Akama H, Nakatsu T, Kato H, Hashimoto T, Oda J, Yamada Y. Crystal structures of two tropinone reductases: different reaction stereospecificities in the same protein fold. Proc Natl Acad Sci U S A. 1998;95:4876–4881. doi: 10.1073/pnas.95.9.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersson A, Jordan D, Schneider G, Lindqvist Y. A flexible lid controls access to the active site in 1,3,8-trihydroxynaphthalene reductase. FEBS Lett. 1997;400:173–176. doi: 10.1016/s0014-5793(96)01382-8. [DOI] [PubMed] [Google Scholar]
- 48.McDaniel R, Ebert-Khosla S, Fu H, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides: influence of a downstream enzyme on the catalytic specificity of a minimal aromatic polyketide synthase. Proc Natl Acad Sci U S A. 1994;91:11542–11546. doi: 10.1073/pnas.91.24.11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalaitzis JA, Moore BS. Heterologous biosynthesis of truncated hexaketides derived from the actinorhodin polyketide synthase. J Nat Prod. 2004;67:1419–1422. doi: 10.1021/np0499564. [DOI] [PubMed] [Google Scholar]
- 50.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 51.Zhang W, Watanabe K, Wang CC, Tang Y. Heterologous biosynthesis of amidated polyketides with novel cyclization regioselectivity from oxytetracycline polyketide synthase. J Nat Prod. 2006;69:1633–1636. doi: 10.1021/np060290i. [DOI] [PubMed] [Google Scholar]
- 52.Tang Y, Lee HY, Kim CY, Mathews I, Khosla C. Structural and functional studies on SCO1815: a beta-ketoacyl-acyl carrier protein reductase from Streptomyces coelicolor A3(2) Biochemistry. 2006;45:14085–14093. doi: 10.1021/bi061187v. [DOI] [PubMed] [Google Scholar]
- 53.Vodicka P, Streinz L, Vavra J, Koutek B, Budesinsky M, Ondracek J, Cisarrova I. Synthesis of (R)- and (S)-3,3,3-trifluoro-2-phenylpropanoyl isocyanates and their use as reactive analogues of Mosher’s acid. Chirality. 2005;17:378–387. doi: 10.1002/chir.20176. [DOI] [PubMed] [Google Scholar]
- 54.Smith RV, Erhardt PW, Rusterholz DB, Barfknecht CF. NMR study of amphetamines using europium shift reagents. J Pharm Sci. 1976;65:412–417. doi: 10.1002/jps.2600650324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.