Abstract
African trypanosomes escape the host adaptive immune response by switching their dense protective coat of Variant Surface Glycoprotein (VSG). Each cell expresses only one VSG gene at a time from a telomeric expression site (ES). The ‘pre-genomic’ era saw the identification of the range of pathways involving VSG recombination in the context of mono-telomeric VSG transcription. A prominent feature of the early post-genomic era is the description of the molecular machineries involved in these processes. We describe the factors and sequences recently linked to mutually exclusive transcription and VSG recombination, and how these act in the control of the key virulence mechanism of antigenic variation.
Introduction
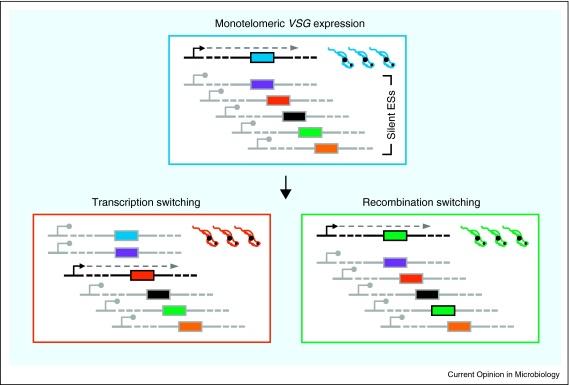
Many pathogens have evolved strategies for phenotypic and clonal variation of surface proteins. This allows for the establishment of a persistent infection in immunocompetent hosts, enhancing transmission. The African trypanosome, Trypanosoma brucei, is one such pathogen; a protozoan of major medical and economic importance. These highly motile cells circulate in the mammalian host bloodstream and are spread by tsetse flies. Evasion of the adaptive host immune response is achieved by changing the composition of a dense Variant Surface Glycoprotein (VSG) coat on bloodstream form cells [1]. The VSG is invariably encoded in a polycistronically transcribed telomeric expression site (ES). Importantly, VSG expression is monoallelic such that only one among 10–20 telomeric ESs is transcribed at a time [2]. Silencing at all other ESs maintains monoallelic expression and the integrity of the evasion strategy while the multiplicity of potential ESs (Figure 1) allows for a co-ordinated switch to transfer active transcription from one telomere to another [3]. Beyond the VSGs found in the ES, T. brucei also possesses a massive archive of ∼1000 silent VSGs and VSG pseudogenes which dominate subtelomeres [27]. Recombination is therefore central to antigenic variation, allowing the parasite to utilise this VSG archive, typically by copying a different gene into the active ES (Figure 1).
Figure 1.
The schematic illustrates mono-telomeric VSG expression and routes of VSG switching. NB: there are >1000 VSG (pseudo)genes available for the exchange or assembly of new VSGs at the active ES; most of these are in long subtelomeric arrays flanked by repetitive elements.
Recent reviews have dealt with a range of topics related to antigenic variation in T. brucei, focusing on DNA breaks as triggers for recombination-based switching [4], expression-site associated genes [5], VSG expression patterns and mechanisms [6] and the trafficking and barrier function of the VSG coat [7,8]. Here, we focus on recent advances in understanding the molecular machineries that maintain VSG allelic exclusion and that execute recombination-based VSG switching.
Control of monoallelic VSG expression site transcription
The epigenetic mechanisms underlying VSG gene silencing and allelic exclusion are of great intrinsic scientific interest and also present potential targets for chemotherapy. Subtelomeric promoters and genes are typically prone to silencing in a range of organisms, a phenomenon first described in yeast [9] and subsequently demonstrated in trypanosomes [10,11•]. Crucially, in T. brucei, only one of the available bloodstream ESs [2] must specifically escape silencing to maintain homogeneity of the VSG coat and the ability to rapidly swap exposed epitopes; the resulting differential in VSG expression between silent and active ESs may be in excess of 10,000-fold. A notable feature of VSG ESs is transcription by RNA polymerase I (RNAPI). Although all VSG ES promoters appear to initiate RNAPI-mediated transcription at a similar rate, transcription is processive only at the single ‘active’ VSG ES [12] and this ES associates with an extranucleolar accumulation of RNAPI known as the ES body (ESB) [13]. It remains unknown whether the ESB self-assembles around the active gene [14], or whether the structure excludes other ESs [13]. Thus, the mechanism allowing one ES to escape silencing remains something of a mystery, and no ESB-specific factor has been identified to date, but there has been some recent progress in understanding the structure and behaviour of the active ES. Nucleosomes are depleted at the active ES [15•,16•], thereby reflecting either transcription-based ejection and/or another form of destabilisation. In addition, sister chromatid cohesion promotes inheritance of the active state; in cells depleted for cohesin components, cohesion at the active ES is compromised leading to an increased rate of transcription switching to alternative telomeres [17•]. These studies, and the DOT1B work described below, may provide some early insight into the elusive mechanism of cross-talk that operates among the active and silent ESs.
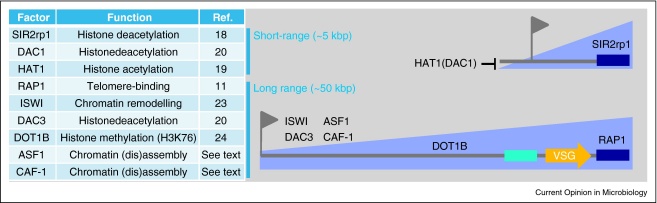
The powerful silencing mechanism itself has been more readily amenable to investigation and at least six factors required to maintain ES silencing have been identified in recent years. It has also become clear that other factors participate in a distinct form of silencing defined by the distance that the effect spreads from the telomere (Figure 2). ‘Short-range’ telomeric silencing is restricted to a region immediately adjacent to the telomeric repeats and in T. brucei, the distal ES promoters and antigenic variation escape this effect. This silencing mechanism requires SIR2rp1, the only nuclear NAD-dependent histone deacetylase in T. brucei [18]. Furthermore, the histone acetyltransferase, HAT1 [19], and a histone deacetylase, DAC1 [20•], appear to regulate SIR2rp1 spreading. These latter findings reinforce the parallels with telomeric silencing in yeast where the putative homologues, Sas2 [21] and Rpd3 [22], control the spreading of Sir2-dependent silencing.
Figure 2.
Factors that impact telomeric and VSG expression site silencing in T. brucei. The role of each factor in the table on the left is illustrated in the schematic on the right. Only the SIR2rp1 and RAP1 effects have been shown to diminish as distance from the telomere increases. The short-range effects have only been shown to affect de novo telomeres but may also impact VSG ES transcription, particularly at the short monocistronic ESs used in the insect mid-gut and during the establishment of a mammalian infection [3]. It is also important to note that these factors could impact recombination. Many of the factors shown were originally named based on phenotypes identified in yeast: ASF, anti-silencing factor; DOT, disruptor of telomeric silencing; SIR, silent information regulator; RAP, repressor/activator protein. The DAC3 homologue in yeast (Hda1p) has also been linked to telomeric exclusion of genes encoding surface-exposed proteins [50,51]. Flags represent promoters and blue boxes represent repetitive sequences; dark, T2AG3, light, 70 bp.
Substantial evidence has emerged recently to also link chromatin structure and modification to the more extensive ‘long-range’ VSG ES silencing (Figure 2). The chromatin chaperones, CAF-1 and ASF1, are required for inheritance of the silent state, presumably through cycles of nucleosome (dis)assembly associated with DNA replication (Alsford et al., unpublished). In addition, several enzymes have been identified that are likely to stabilise the nucleosomes at silent sites thereby repressing transcription. These include a chromatin remodeler, ISWI [23], a histone deacetylase, DAC3 [20•] and a histone (H3K76) methyltransferase, DOT1B [24•]. Most of the factors above are essential for growth and have been depleted using RNA interference. Only the methyltransferase is dispensable and cells lacking this factor display a relatively subtle VSG transcription derepression phenotype [24•]; the essential factors reveal a more pronounced derepression phenotype at the promoter that, nonetheless, due to attenuation, does not lead to detectable VSG expression from the ‘silent’ ESs. This may reflect a more pronounced role at the promoter or simply stasis associated with only partial alteration of the chromatin through the long polycistronic ES. Thus, current results suggest that chromatin modifiers and remodelers cooperate to reinforce and propagate the silent state. Specifically, the silent sites are likely to comprise hypoacetylated, hypomethylated (H3K76, the DOT1B methyltransferase effect is thought to be indirect) and ordered chromatin. The viable dot1b methyltransferase mutants also presented an opportunity to investigate the impact on transcription switching to an alternative telomere and these cells displayed a substantial delay in this process [24•].
Evidence indicating a role for the telomere itself in VSG ES silencing comes from studies on repressor/activator protein 1 (RAP1). This telomere-associated protein is also essential for growth, and RNA interference-based knockdown produced cells expressing multiple VSGs on the surface [11]. RAP1 may recruit SIR2rp1 and additional factors, thereby mediating short-range telomeric silencing, as in yeast [25] and long-range VSG ES silencing (see Figure 2). Intriguingly, sustained VSG ES silencing at a chromosome end lacking RAP1-binding sites, the terminal telomeric repeats [26], may indicate the presence of silent compartments containing subtelomere clusters.
Telomeres and chromatin are central to tightly regulating interaction between the nuclear pool of RNA polymerase and VSG genes. It seems likely that the silencing mechanism targets all telomeres in the context of a dominant, and currently mysterious, anti-silencing machine or factor that compels VSG ESs to obey the rules of allelic exclusion. This latter activity apparently acts in a telomere-specific manner. A better understanding of the exclusion process might reveal targets that can be exploited for therapy. Indeed, at least one of the factors linked to silencing, the DAC3 deacetylase, represents a potentially druggable target [20•].
Control of VSG expression site recombination
Subtelomeres tend to be ‘fragile sites’ that are prone to rapid gene turnover and increased rates or sequence exchange. As such, these hotbeds of innovation are ideal sites for contingency genes such as VSGs [3]. VSG switching by recombination most commonly occurs by gene conversion reactions that copy a silent VSG into the active ES, replacing the VSG that was previously transcribed (Figure 1). This mitotic process requires considerable mechanistic flexibility, since gene conversion reactions have been documented using donor VSGs from three distinct genomic locations: the silent ESs, the telomeres of African trypanosome-specific minichromosomes, and from the subtelomeric VSG arrays [3]. Gene conversion of array VSGs can recombine complete genes into the ES, or can generate novel VSGs (‘mosaics’) by segmental gene conversion reactions using multiple VSG pseudogenes [27]. A role for homologous recombination (HR) [28] in these gene conversion processes was first revealed by mutating RAD51, the key enzyme of homology-directed DNA strand exchange, resulting in impaired VSG switching [29]. Subsequent analysis confirmed the importance of RAD51-directed strand exchange. Mutation of at least one of four T. brucei RAD51 paralogues also impairs switching [30] (R Dobson et al., unpublished), as does mutation of the T. brucei orthologue of BRCA2 [31•], a breast cancer oncogene that co-ordinates loading of RAD51 onto processed DNA double strand breaks (DSBs) [28]. In part, this is mediated through conserved BRC repeats, which are dramatically expanded in T. brucei BRCA2 [31•]; possibly an example of adaptations imposed on the HR machinery by VSG switching. An important recent development is the use of a yeast meganuclease (I-SceI) for the controlled generation of a chromosomal DSB [32•]; at a chromosome internal locus this results in a temporal cascade of cell cycle stalling, accumulation of RAD51 in subnuclear foci and predominant allelic HR. The system has also now been used for genetic dissection of chromatin control of DSB repair (DSBR) in T. brucei (Glover et al., unpublished); a histone acetyltransferase and a histone deacetylase have been shown to impact DNA resection and RAD51 filament disassembly respectively.
Beyond the HR strand exchange step, we have much to learn about the upstream and downstream processes in VSG switching, and alternative recombination pathways. 70 bp repeats are a key element of VSG switching as they flank >90% of VSGs [27], meaning they can provide upstream sequence homology for recombination as well as ES-specific initiation. Recent work suggested an initiating role for DSBs at 70 bp repeats upstream of the active VSG [33•]: naturally occurring breaks were detected in this region, and the generation of an I-SceI-induced DSB adjacent to the 70 bp repeats increased the rate of switching. A key factor in the detection of DSBs and in nucleolytic processing to allow RAD51 filament formation is the Mre11-Rad50-Xrs2/Nbs1 (MRX) complex [34]. Mutation of MRE11 in T. brucei reveals a role in HR repair [35,36], but not in VSG switching [35], meaning VSG switch-initiating breaks may not be DSBs, at least initially, or that other factors assume an MRX-like function during VSG switching.
A number of pathways may contribute to VSG switching (Table 1, reviewed in [6]) but the recombination steps that operate downstream of RAD51-mediated strand exchange have been explored to only a limited extent. Break-induced replication (BIR) has gained prominence recently [37], at least in part because of roles in telomere maintenance in yeast and mammals [38]. BIR, involving telomere-proximal VSGs and associated 70 bp repeats, might be an adaptation of backup telomere maintenance pathways to satisfy the demands of VSG switching [33•,39]. However, a mechanistic demonstration of BIR, rather than gene conversion, in VSG switching is still needed [40]. For instance, the DNA polymerases (Pols) that catalyse DNA synthesis during VSG switching remain unknown. Though HR in eukaryotes relies on Pols α, δ and ɛ, B family replication enzymes [41], other work has suggested roles for Y family Pols in recombination [42].
Table 1.
DSBR pathways and their possible contribution to antigenic variation in T. brucei.
| Pathway | Sub-pathway | Features | Proposed role in VSG switching | Ref. |
|---|---|---|---|---|
| Homologous recombination (HR) | Gene conversion (GC) | Copying and replacement of a segment of DNA using flanking homologies | Any VSG (fragment) with sufficient homology could be copied into the active ES by this typically RAD51-dependent mechanisma | [29] |
| Single-strand annealing (SSA) | Deletion of a segment of DNA using flanking homologies | None | ||
| Break-induced replication (BIR) | Copying and replacement of a segment of DNA to the chromosome end using a single region of homology | The subset of telomeric VSGs could be copied into the active ES by this typically RAD51-dependent mechanisma | [33] | |
| End-joining (EJ) | Non-homologous EJ (NHEJ) | Re-ligation of broken strands typically with small deletions | None — not seen in T. brucei | |
| Microhomology-mediated EJ (MMEJ, aka micro-SSA) | Deletion of a segment of DNA using flanking microhomologies of 5–20 bp. Gene conversion (see above) can be mediated by one-sided MMEJ | MMEJ-based equivalents of (one-sided) GC and BIR would be predicted to be RAD51-dependentand independent respectivelya (see above) | [32] | |
Recombination-based VSG switching operates via RAD51-dependent and independent mechanisms.
RAD51-mediated HR is clearly important in antigenic variation, but RAD51-independent pathways also operate [29,30,31•]. Deletions based on microhomology-mediated end-joining (MMEJ, aka micro-single-strand annealing) are readily detectable in T. brucei following an I-SceI-induced DSB [32•] and one-sided, MMEJ-based gene conversion also operates (Glover et al., unpublished). MMEJ has also been observed in T. brucei cell extracts [43] and following DNA transformation [44] and occurs in mutants lacking RAD51 and KU, a key component of non-homologous end-joining (NHEJ). The relationship between MMEJ in T. brucei and alternative end-joining (A-EJ) pathways in other eukaryotes [45] is currently unclear. Nevertheless, though MMEJ/A-EJ is considered subservient to NHEJ in other eukaryotes, significant roles in immunoglobulin gene rearrangements have been described [46]. In addition, evidence is emerging that MMEJ/A-EJ can be a significant route for chromosome rearrangements [47], a process that can shape genomes and may be suppressed by NHEJ [48]. Indeed, MMEJ may be coupled to BIR in such rearrangements [49]. Though KU70-80 is conserved, the Ligase IV-XRCC4 complex of NHEJ has not been found in the trypanosomatids, so these parasites may have evolved to enhance MMEJ.
Conclusions
Mono-telomeric VSG expression and recombination are central to the process of antigenic variation in African trypanosomes and it will be important to understand the machinery underlying both of these processes. The recent work highlighted above has begun to illuminate both areas with chromatin and epigenetics emerging as prominent features. Additional regulators will emerge and further studies on interactions, mapping of epigenetic marks and their functional analysis, nuclear location and cell-cycle control will provide further insights while forward genetic approaches may reveal further novelty and could shed some light on the machinery required for mutually exclusive escape from silencing. Further characterization of DSB processing, HR and MMEJ, their contribution to the various VSG recombination reactions and their genetic dissection, are also needed. Finally, now the importance of the terminal telomeric repeats is established for gene silencing, other subtelomeric sequences may be found to serve cis-regulatory functions in spreading heterochromatin, serving as transcription boundaries or enhancers or promoting instability and recombination.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
Acknowledgements
DH's research is supported by grants from the Wellcome Trust (Project grants 083648 and 089172). RMcC's research is supported by grants from the Wellcome Trust and Medical Research Council. We also thank our LSHTM colleague, Martin Taylor, for comments on the draft manuscript.
References
- 1.Horn D. The molecular control of antigenic variation in Trypanosoma brucei. Curr Mol Med. 2004;4:563–576. doi: 10.2174/1566524043360078. [DOI] [PubMed] [Google Scholar]
- 2.Hertz-Fowler C., Figueiredo L.M., Quail M.A., Becker M., Jackson A., Bason N., Brooks K., Churcher C., Fahkro S., Goodhead I. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One. 2008;3:e3527. doi: 10.1371/journal.pone.0003527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horn D., Barry J.D. The central roles of telomeres and subtelomeres in antigenic variation in African trypanosomes. Chromosome Res. 2005;13:525–533. doi: 10.1007/s10577-005-0991-8. [DOI] [PubMed] [Google Scholar]
- 4.Alsford S., Horn D., Glover L. DNA breaks as triggers for antigenic variation in African trypanosomes. Genome Biol. 2009;10:223. doi: 10.1186/gb-2009-10-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCulloch R., Horn D. What has DNA sequencing revealed about the VSG expression sites of African trypanosomes? Trends Parasitol. 2009;25:359–363. doi: 10.1016/j.pt.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Morrison L.J., Marcello L., McCulloch R. Antigenic variation in the African trypanosome: molecular mechanisms and phenotypic complexity. Cell Microbiol. 2009;11:1724–1734. doi: 10.1111/j.1462-5822.2009.01383.x. [DOI] [PubMed] [Google Scholar]
- 7.Field M.C., Carrington M. The trypanosome flagellar pocket. Nat Rev Microbiol. 2009;7:775–786. doi: 10.1038/nrmicro2221. [DOI] [PubMed] [Google Scholar]
- 8.Schwede A., Carrington M. Bloodstream form trypanosome plasma membrane proteins: antigenic variation and invariant antigens. Parasitology. 2010:1–11. doi: 10.1017/S0031182009992034. [DOI] [PubMed] [Google Scholar]
- 9.Gottschling D.E., Aparicio O.M., Billington B.L., Zakian V.A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 10.Glover L., Horn D. Repression of polymerase I-mediated gene expression at Trypanosoma brucei telomeres. EMBO Rep. 2006;7:93–99. doi: 10.1038/sj.embor.7400575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Yang X., Figueiredo L.M., Espinal A., Okubo E., Li B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell. 2009;137:99–109. doi: 10.1016/j.cell.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration that transcription was derepressed at silent ESs when the telomere-associated protein, RAP1, was depleted.
- 12.Vanhamme L., Poelvoorde P., Pays A., Tebabi P., Van Xong H., Pays E. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol Microbiol. 2000;36:328–340. doi: 10.1046/j.1365-2958.2000.01844.x. [DOI] [PubMed] [Google Scholar]
- 13.Navarro M., Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414:759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- 14.Sutherland H., Bickmore W.A. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 15•.Figueiredo L.M., Cross G.A. Nucleosomes are depleted at the VSG expression site transcribed by RNA polymerase I in African trypanosomes. Eukaryot Cell. 2010;9:148–154. doi: 10.1128/EC.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of the two reports showing that chromatin displays a disordered structure at the active ES.
- 16•.Stanne T.M., Rudenko G. Active VSG expression sites in Trypanosoma brucei are depleted of nucleosomes. Eukaryot Cell. 2010;9:136–147. doi: 10.1128/EC.00281-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of the two reports showing that chromatin displays a disordered structure at the active ES.
- 17•.Landeira D., Bart J.M., Van Tyne D., Navarro M. Cohesin regulates VSG monoallelic expression in trypanosomes. J Cell Biol. 2009;186:243–254. doi: 10.1083/jcb.200902119. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration of premature separation of active ES sister chromatids and increased transcriptional switching when cohesin components were depleted.
- 18.Alsford S., Kawahara T., Isamah C., Horn D. A sirtuin in the African trypanosome is involved in both DNA repair and telomeric gene silencing but is not required for antigenic variation. Mol Microbiol. 2007;63:724–736. doi: 10.1111/j.1365-2958.2006.05553.x. [DOI] [PubMed] [Google Scholar]
- 19.Kawahara T., Siegel T.N., Ingram A.K., Alsford S., Cross G.A., Horn D. Two essential MYST-family proteins display distinct roles in histone H4K10 acetylation and telomeric silencing in trypanosomes. Mol Microbiol. 2008;69:1054–1068. doi: 10.1111/j.1365-2958.2008.06346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Wang Q.P., Kawahara T., Horn D. Histone deacetylases play distinct roles in telomeric VSG expression site silencing in African trypanosomes. Mol Microbiol. 2010;77:1237–1245. doi: 10.1111/j.1365-2958.2010.07284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration that transcription was derepressed at silent ES promoters when the histone acetyltransferase, DAC3, was depleted. DAC1 was also shown to antagonize SIR2rp1-dependent telomeric silencing.
- 21.Kristjuhan A., Wittschieben B.O., Walker J., Roberts D., Cairns B.R., Svejstrup J.Q. Spreading of Sir3 protein in cells with severe histone H3 hypoacetylation. Proc Natl Acad Sci USA. 2003;100:7551–7556. doi: 10.1073/pnas.1332299100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J., Zhou B.O., Lenzmeier B.A., Zhou J.Q. Histone deacetylase Rpd3 antagonizes Sir2-dependent silent chromatin propagation. Nucleic Acids Res. 2009;37:3699–3713. doi: 10.1093/nar/gkp233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes K., Wand M., Foulston L., Young R., Harley K., Terry S., Ersfeld K., Rudenko G. A novel ISWI is involved in VSG expression site downregulation in African trypanosomes. EMBO J. 2007;26:2400–2410. doi: 10.1038/sj.emboj.7601678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.Figueiredo L.M., Janzen C.J., Cross G.A. A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 2008;6:e161. doi: 10.1371/journal.pbio.0060161. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration that ESs were partially derepressed and that the kinetics of a transcriptional ES switch were slow in cells lacking the histone H3K76me3 methyltransferase, DOT1B.
- 25.Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- 26.Glover L., Alsford S., Beattie C., Horn D. Deletion of a trypanosome telomere leads to loss of silencing and progressive loss of terminal DNA in the absence of cell cycle arrest. Nucleic Acids Res. 2007;35:872–880. doi: 10.1093/nar/gkl1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcello L., Barry J.D. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res. 2007;17:1344–1352. doi: 10.1101/gr.6421207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 29.McCulloch R., Barry J.D. A role for RAD51 and homologous recombination in Trypanosoma brucei antigenic variation. Genes Dev. 1999;13:2875–2888. doi: 10.1101/gad.13.21.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Proudfoot C., McCulloch R. Distinct roles for two RAD51-related genes in Trypanosoma brucei antigenic variation. Nucleic Acids Res. 2005;33:6906–6919. doi: 10.1093/nar/gki996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31•.Hartley C.L., McCulloch R. Trypanosoma brucei BRCA2 acts in antigenic variation and has undergone a recent expansion in BRC repeat number that is important during homologous recombination. Mol Microbiol. 2008;68:1237–1251. doi: 10.1111/j.1365-2958.2008.06230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration that antigenic variation was impaired in cells lacking the RAD51-binding protein, BRCA2, and detailing a recent evolutionary adaptation of the protein.
- 32•.Glover L., McCulloch R., Horn D. Sequence homology and microhomology dominate chromosomal double-strand break repair in African trypanosomes. Nucleic Acids Res. 2008;36:2608–2618. doi: 10.1093/nar/gkn104. [DOI] [PMC free article] [PubMed] [Google Scholar]; Establishment of inducible meganuclease cleavage system and assays for the study of chromosomal DSBR in T. brucei.
- 33•.Boothroyd C.E., Dreesen O., Leonova T., Ly K.I., Figueiredo L.M., Cross G.A., Papavasiliou F.N. A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature. 2009;459:278–281. doi: 10.1038/nature07982. [DOI] [PMC free article] [PubMed] [Google Scholar]; Demonstration that meganuclease cleavage triggered replacement of the active VSG with a VSG from another ES, possibly via a break-induced replication mechanism. The detection of natural DSBs at the active ES is consistent with the idea that DSBR naturally brings about antigenic variation.
- 34.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson N.P., McCulloch R., Conway C., Browitt A., Barry J.D. Inactivation of Mre11 does not affect VSG gene duplication mediated by homologous recombination in Trypanosoma brucei. J Biol Chem. 2002;277:26185–26193. doi: 10.1074/jbc.M203205200. [DOI] [PubMed] [Google Scholar]
- 36.Tan K.S., Leal S.T., Cross G.A. Trypanosoma brucei MRE11 is non-essential but influences growth, homologous recombination and DNA double-strand break repair. Mol Biochem Parasitol. 2002;125:11–21. doi: 10.1016/s0166-6851(02)00165-2. [DOI] [PubMed] [Google Scholar]
- 37.McEachern M.J., Haber J.E. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 38.Cesare A.J., Reddel R.R. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 39.Dreesen O., Li B., Cross G.A. Telomere structure and function in trypanosomes: a proposal. Nat Rev Microbiol. 2007;5:70–75. doi: 10.1038/nrmicro1577. [DOI] [PubMed] [Google Scholar]
- 40.Barry D., McCulloch R. Molecular microbiology: a key event in survival. Nature. 2009;459:172–173. doi: 10.1038/459172a. [DOI] [PubMed] [Google Scholar]
- 41.Lydeard J.R., Jain S., Yamaguchi M., Haber J.E. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 42.Lehmann A.R. New functions for Y family polymerases. Mol Cell. 2006;24:493–495. doi: 10.1016/j.molcel.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 43.Burton P., McBride D.J., Wilkes J.M., Barry J.D., McCulloch R. Ku heterodimer-independent end joining in Trypanosoma brucei cell extracts relies upon sequence microhomology. Eukaryot Cell. 2007;6:1773–1781. doi: 10.1128/EC.00212-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conway C., Proudfoot C., Burton P., Barry J.D., McCulloch R. Two pathways of homologous recombination in Trypanosoma brucei. Mol Microbiol. 2002;45:1687–1700. doi: 10.1046/j.1365-2958.2002.03122.x. [DOI] [PubMed] [Google Scholar]
- 45.McVey M., Lee S.E. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boboila C., Jankovic M., Yan C.T., Wang J.H., Wesemann D.R., Zhang T., Fazeli A., Feldman L., Nussenzweig A., Nussenzweig M. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci USA. 2010;107:3034–3039. doi: 10.1073/pnas.0915067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang F., Carvalho C.M., Lupski J.R. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simsek D., Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hastings P.J., Ira G., Lupski J.R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ekwall K. Genome-wide analysis of HDAC function. Trends Genet. 2005;21:608–615. doi: 10.1016/j.tig.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 51.Halme A., Bumgarner S., Styles C., Fink G.R. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116:405–415. doi: 10.1016/s0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]