Abstract
Nuclear receptors (NRs) are important pharmacological targets for a number of diseases, including cancer and metabolic disorders. To unmask the direct role of NR function it is fundamental to find the NR targets. During the last few years several NRs have been shown to affect microRNA expression, thereby modulating protein levels. The farnesoid X receptor (FXR), the main regulator of bile acid (BA) homeostasis, also regulates cholesterol, lipid and glucose metabolism. Here we used, for the first time, a proteomics approach on mice treated with a FXR ligand to find novel hepatic FXR targets. Nineteen spots with a more than two-fold difference in protein amounts were found by 2D-DIGE and 20 proteins were identified by MALDI-TOF MS as putative novel FXR targets. The most striking feature of the protein list was the great number of mitochondrial proteins, indicating a substantial impact of FXR activation on mitochondrial function in the liver. To examine if the differences found in the proteomics assay reflected differences at the mRNA level, a microarray assay was generated on hepatic samples from wild type and FXR−/− mice treated with a FXR ligand and compared to vehicle treatment. At least six proteins were shown to be regulated only at a post-transcriptional level. In conclusion, our study provides the impetus to include proteomic analysis for the identification of novel targets of transcription factors, such as NRs. This article is part of a Special Issue entitled: Translating nuclear receptors from health to disease.
Abbreviations: NR, nuclear receptor; FXR, farnesoid X receptor; BA, bile acid; FXR−/−, FXR loss-of-function
Keywords: Nuclear receptor, FXR, Proteomic analysis, Expression profiling, Post-transcriptional modification, Mitochondrial
Research highlights
► Twenty proteins were identified as being potential novel hepatic FXR targets. ► Many of the potential FXR targets have a role in regulating mitochondrial function. ► Microarray analysis was made for a comparison of protein amounts with mRNA levels. ► Six of the proteins found by 2D-DIGE seem to be regulated only at the protein level.
1. Introduction
Nuclear receptors (NRs) are important pharmacological targets for treatment of a number of diseases, including cancer and metabolic disorders [1]. Great efforts are made to find substances that are specific to a NR subtype or that affect only certain aspects of the NR function [2]. To understand all the effects and possible side effects of NR activation or repression it is fundamental to find the NR targets. Since NRs are transcription factors, it is natural to look for tissue specific expression patterns and their target genes by gene expression profiling [3,4]. A lot of information has been gained from such studies. However, bearing in mind the diverse ways of regulation of RNAs and proteins, including the effects of microRNAs on mRNA translation and degradation, maybe a proteomics approach should be applied as well. Indeed, during the last few years several NRs have been shown to affect microRNA expression, thereby modulating protein levels [5–10].
The farnesoid X receptor (FXR), the main regulator of bile acid (BA) homeostasis [11–13], also regulates cholesterol, lipid and glucose metabolism [14]. The importance of FXR for the metabolic homeostasis in the gut-liver axis has been revealed in whole body and tissue specific FXR loss-of-function (FXR−/−) mouse models [15–17]. Furthermore, FXR has been shown to play a role in processes such as liver regeneration [18], carcinogenesis [19–21], inflammation and bacterial overgrowth in the intestine [22,23]. There are therapeutic potentials for selective FXR modulators in diseases such as the metabolic syndrome, diabetes, gallstone disease, hypertriglyceridemia, steato-hepatitis and colon cancer [14,21]. A recent study of genomic FXR binding in mouse liver and intestine suggests a greater number of FXR target genes than is known thus far [24]. Also a high degree of tissue-specific binding was revealed, where only 11% of the binding sites were shared between the tissues, indicating a high degree of tissue-specific effects of FXR activation. The hepatic FXR target genes found up till now encode proteins involved in BA, lipid and glucose metabolism as well as in the detoxification of xenobiotics [25]. In a recent study FXR has also been shown to inhibit the expression of a microRNA, miR-34a [8].
To find hepatic FXR targets we used, for the first time, a proteomics approach with mice treated with the potent FXR ligand 6-ethyl chenodeoxycholic acid (6-ECDCA or INT-747) [26]. In addition, a microarray assay was carried out to examine if the differences found in the proteomics assay reflected differences at the mRNA level.
2. Materials and methods
2.1. Animals and treatments
Ten weeks old wild type C57BL/6J male mice and FXR−/− C57BL/6J male mice were treated with 10 mg/kg/day INT-747 (Intercept Pharmaceuticals) or only the vehicle, 1% methylcellulose, by gavages for at least three days. The animals were fasted overnight and given their last gavages 3 h before the sacrifice. The liver samples were snap-frozen in liquid nitrogen and kept at −80 °C until used. The Ethical Committee of the Consorzio Mario Negri Sud approved this experimental set-up, which was also certified by the Italian Ministry of Health according with internationally accepted guidelines for the animal care.
2.2. Proteomic analysis
2.2.1. Liver protein extraction for proteomic analysis
Individual mouse liver samples were ground into powder under liquid nitrogen, dissolved in a buffer containing 5 M urea, 2 M thiourea, 2% (w/v) CHAPS, 2% (w/v) Zwittergent 3-10 detergent (Calbiochem, Merck Biosciences, Darmstadt, Germany) 50 mM DTT (Sigma Aldrich, Milano, Italy) and protease inhibitor cocktail set III (Calbiochem). After a centrifugation at 100,000g for 30 min at 12 °C the pellets were discarded and the supernatants taken as the cytosol fraction. The protein content was determined by ETTAN™ procedure, using a protein assay kit from GE Healthcare (Chalfont St. Giles, Bucks, UK).
2.2.2. Two-dimensional difference gel electrophoresis (2D-DIGE) and quantitative gel image analysis
Five vehicle and five INT-747 samples (each 50 μg of protein) were labeled separately with either 200 pmol Cy3 or Cy5, and the internal standard (25 μg of each of the ten samples) was labeled with Cy2. One vehicle, INT-747 and standard sample forming a set of Cy2, Cy3 and Cy5 labeled samples were combined for each of five gels and were diluted in the rehydration solution, composed of 5 M urea, 2 M thiourea, 2% (w/v) CHAPS, 2% (w/v) Zwittergent, 40 mM DTT and 0.5% IPG buffer for pH 3–10 linear gradient (GE Healthcare). Isoelectric focusing (IEF) was carried out on immobilized IPG strips with a broad pH 3–10 linear gradient, by using an IPGphor Isoelectric Focusing System (GE Healthcare). After a rehydration step at 30 V for 16 h, focusing started at 200 V. The voltage was increased step by step to 1000 V, then gradually up to 8000 V and kept constant for further 5 h for a total 46,000 Vh. Following IEF, individual protein strips were reduced by rocking for 15 min in a solution containing 6 M urea, 50 mM Tris–HCl, pH 8.8, 30% (v/v) glycerol, 2% (w/v) SDS, 1% DTT. Proteins were subsequently alkylated by replacing DTT with 100 mM iodoacetamide for 15 min. The strips were placed on the top of 12.5% SDS-PAGE (160 × 160 × 1 mm) and run at 10 mA, for molecular size electrophoresis. Protein size was determined by running standard protein markers (Rainbow, GE Healthcare), in the range of 14.3–220.0 kDa.
Images were visualized using the pharos-FX imager from Bio-Rad. The gels were scanned using a 488 nm laser and an emission filter of 530 nm BP (band Pass) 40, a 532 nm laser and an emission filter of 695 nm DF (discriminating filter) 50, a 635 nm laser and 695 nm DF 55 emission filter to acquire the Cy2, Cy3, and Cy5 image respectively. All gels were scanned at 200 μm resolution. Images were then processed using the PD-Quest software (Bio-Rad) protocol. Protein spots were matched and gels were normalized using the internal standard present in all gels.
An overall total of around 1500 protein spots were visualized in the present study and a p-value < 0.05 (Student's t-test) was considered statistically significant. Only the spots showing at least a two-fold difference were further analyzed.
2.2.3. Protein identification by MALDI-TOF MS analysis
An additional gel was made using 300 μg of total protein pooled from each of the five vehicle and INT-747 samples analyzed run under conditions identical to the analytical gels except that the proteins were unlabeled (non DIGE). Selected protein spots were in situ digested and analyzed by MALDI-TOF MS. Briefly, protein bands were excised from SDS-PAGE and after washing, cysteins were reduced with DTT and alkylated with iodoacetamide. Gels were digested in situ by incubation with sequencing-grade trypsin (Promega, Madison, WI, USA) in 40 mM ammonium bicarbonate under slight shaking on a thermomixer at 37 °C overnight [27]. The reaction was stopped with H2O/TFA 0.1% at 30 °C, for 15 min. Tryptic peptides were extracted, desalted with ZipTip C18 columns (Millipore Corp, Bedford, MA, USA), eluted and crystallized in 50% (v/v) ACN/H2O saturated solution of alfa-cyano-4-hydroxy-cinnamic acid. Peptide mass spectra were obtained by a time-of-flight mass spectrometer (Reflex IV®, Bruker Daltonics, Bremen, Germany), equipped with a nitrogen laser with an emission wavelength of 337 nm. Mass spectra were acquired in positive ion Reflectron-mode with delayed extraction and an accelerating voltage of 20 kV. An external calibration was performed for each measurement, using a mixture of seven standard peptides (average mass accuracy better than 20 ppm). All mass spectra were acquired using a minimum number of 250 laser shots. Spectra were internally calibrated with trypsin autolysis products. Peptide matching and protein searches were performed submitting peptide mass lists to database search on NCBInr and/or SWISS PROT, using the MASCOT and ProFound search engines. The main search parameters were as follows: no restriction on molecular weight and isoelectric point (MW and pI); taxonomy, mause; one missed cleavage allowed; carboxymethylation of cystein; oxidation of methionine; 50–100 ppm peptide mass tolerance. Proteins listed as significant matches in MASCOT were considered when a threshold score allowing a p < 0.05 was achieved.
2.2.4. Western blot analysis
Western blot analysis was performed on pooled mouse liver samples, five in each group, processed as described above. Total proteins were quantified by ETTAN™ procedure, using a protein assay kit from GE Healthcare (Chalfont St. Giles, Bucks, UK).
For one-dimensional (1-DE) gel electrophoresis, samples of cytosol fractions were dissolved in SDS sample buffer, composed of 12.5 mM Tris–HCl, pH 6.8, 1% SDS, 10% glycerol, 0.025% bromophenol blue, boiled for 10 min, and applied to 12.5% (w/v) SDS-PAGE. Following 1-DE separation, proteins were transferred to nitrocellulose and after transfer, the nitrocellulose blots were checked by Ponceau red staining to ensure an homogeneous transfer efficiency. The blots were then probed with a primary antibody anti-GSTM1 (kindly provided by Dr. B. Favaloro, Ce.S.I., University G. D'Annunzio, Chieti, Italy), ATP5A (C-15):sc-49162 antibody , Laminin-R antibody (G-7):sc-74531,(Santa Cruz Biotechnology Inc, Santa Cruz, CA, USA), Annexin V antibody (ab 14196) Abcam plc,330 Cambridge science park, Cambridge CB4 OFL, UK. Blots were visualized by ECL chemiluminescence detection system (GE Healthcare) according to the manufacturer. Protein abundance was quantified by densitometric analysis, with Quantity ONE software (Bio-Rad) and the results normalized against β-Actin.
2.3. Real-time quantitative PCR (RT-qPCR)
Total RNA was extracted using QIAzol (Qiagen) and the integrity of the RNA was assessed on a formaldehyde gel. cDNA was synthesized from total RNA by High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) after DNase I treatment using the DNA-free Kit from Ambion. Relative amounts of Shp and cyclophilin mRNA were obtained on a 7500 Fast Real-Time PCR System machine (Applied Biosystems) using Power CYBR Green (Applied Biosystems). Shp mRNA levels were normalized to the amounts of cyclophilin mRNA. The primers used are available upon request.
2.4. Microarray
Expression profiling was performed using the Illumina Mouse MG-6V2 BeadArray Expression with biological duplicates for each treatment and animal genotype. The RNA integrity was assessed using the BioRad Experion System. Amplification was made with 500 ng of total RNA using the Illumina TotalPrep RNA Amplification kit (Ambion). The quantity and quality of biotin-UTP incorporated cRNA was also assessed using the BioRad Experion System. 1.5 μg amplified cRNA from each sample was hybridized to the arrays according to the manufacturer's guidelines. The data were analyzed using the GenomeStudio software.
3. Results
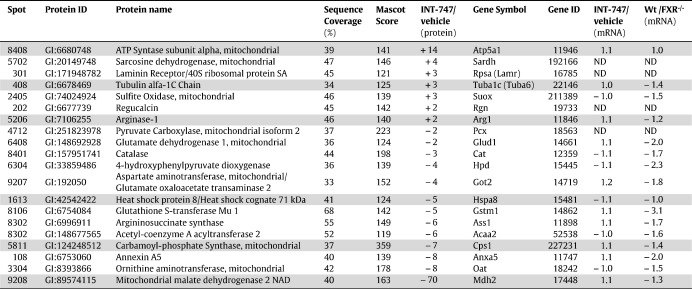
In a 2D-DIGE assay, comparing hepatic protein samples from INT-747 with vehicle treated mice, 19 spots were more than two-fold different in protein levels (Fig. 1A, Tables S1 and S2). For seven spots the amount of protein was higher in the INT-747 samples compared to vehicle, while in the rest it was reduced by the FXR ligand treatment. The proteins in the spots were identified by MALDI-TOF MS (Table 1). In spot 8302 two different proteins were found, argininosuccinate synthase and 3-ketoacylCoA thiolase. It is therefore unclear which protein, or if possibly both were down regulated in response to the FXR activation. In this study we also compared hepatic samples from FXR−/− mice, treated with the semi-synthetic bile acid INT-747 or vehicle, and no differences in protein amounts could be detected for any of the proteins in Table 1 (data not shown). This comparison was made as an objective control for non-FXR mediated effects that could relate to the weak agonistic effect of INT-747 on the membrane TGR5 receptor [28].
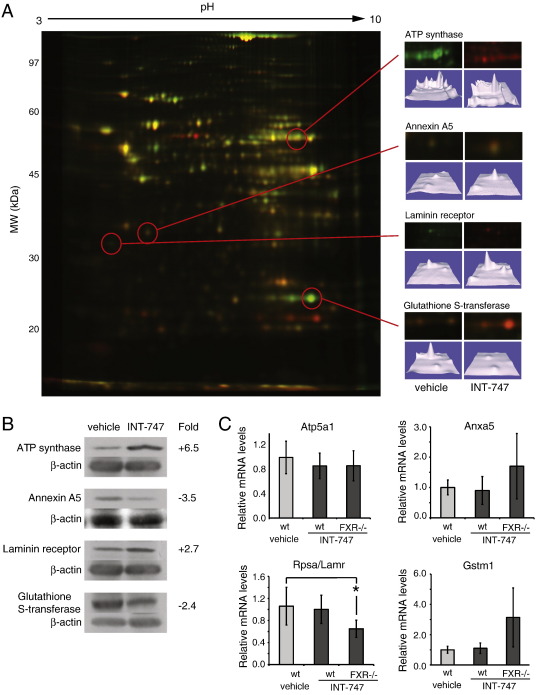
Fig. 1.
Identification of potential FXR targets. A) Hepatic cytosolic protein samples from wild type mice treated with INT-747 or vehicle were compared in a 2D-DIGE. Four examples of spots differentially expressed are shown and the proteins were identified by MALDI-TOF MS. The pH and molecular weights scales are indicated in the figure. B) Western blot on four of the proteins identified as potential FXR targets in the proteomics analysis. Pooled hepatic cytosolic protein samples from wild type mice treated with INT-747 or vehicle were compared (n = 5). The quantified results were normalized against β-actin. C) RT-qPCR on four genes corresponding to proteins identified as potential FXR targets in the proteomics analysis. Five individual samples were analyzed for each treatment and the error bars represent the standard deviation. *p < 0.05, Student's t-test.
Table 1.
Proteomics quotients for the identification of potential FXR targets along with corresponding mRNA quotients.
Gray shading, FXR targets showing a difference at the protein level but not at the mRNA level after FXR ligand treatment. ND, mRNA levels below background level in the microarray analysis, i.e. not detectable.
For four of the proteins, ATP synthase, annexin A5, the laminin receptor and glutathione S-transferase, the differences in protein levels were verified by the use of Western blot (Fig. 1B). All four proteins showed a similar regulation as were seen by DIGE, but with a lower fold difference. This indicated a successful outcome of the proteomics analysis but even so, the results for the other proteins in Table 1 should be confirmed by Western blot to be considered as true FXR targets. All in all, 20 different proteins were identified as being potential novel FXR targets in the proteomic analysis and four of these were verified as bona fide FXR targets by Western blot.
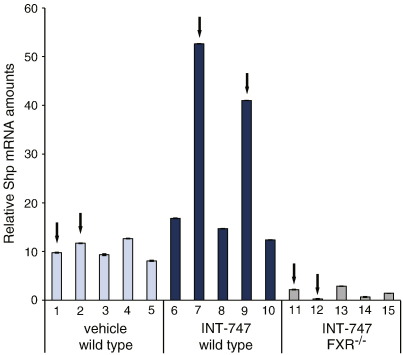
Subsequently, a microarray assay was carried out on hepatic samples from wild type and FXR−/− mice treated with INT-747 or vehicle to examine if the differences found in the proteomics assay reflected differences at the mRNA level. To ascertain FXR activation by the INT-747 treatment, the mRNA levels of the known FXR target gene Nr0b2, also called Shp, encoding the orphan NR small heterodimer partner (SHP), were assessed by RT-qPCR (Fig. S1). Samples with a high Shp mRNA expression after INT-747 treatment were chosen for the subsequent array experiment. Sequences corresponding to the pyruvate carboxylase (Pcx), regucalcin (Rgn), ribosomal protein SA (Rpsa) and sarcosine dehydrogenase (Sardh) genes (equivalent to the protein spots 4712, 202, 301 and 5702, respectively) yielded signals on the arrays that were below background levels, and were therefore considered as not detectable (Table 1). Comparing the array results of INT-747 treated with vehicle treated mice, no differences of at least 1.5-fold could be detected for the genes encoding the proteins identified as FXR targets in the proteomic analysis. In the comparison of wild type with FXR−/− mice, both INT-747 treated, four mRNAs turned out at least 2-fold different; annexin A5 (Anxa5), glutamate dehydrogenase 1 (Glud1), glutathione S-transferase, mu 1 (Gstm1) and 4-hydroxyphenylpyruvic acid dioxygenase (Hpd) (corresponding to protein spots 108, 6408, 8106 and 6304 respectively). Ten mRNAs differed at least 1.5-fold, adding acetyl-Coenzyme A acyltransferase 2 (Acaa2), argininosuccinate synthetase 1 (Ass1), catalase (Cat), glutamate oxaloacetate transaminase 2 (Got2), ornithine aminotransferase (Oat) and sulfite oxidase (Suox) (equivalent to protein spots 8302, 8302, 8401, 9207, 3304 and 2405 respectively) to the list. The less stringent 1.5-fold criteria still leaves six genes that were not considered differentially expressed, suggesting different amounts of protein after FXR ligand treatment due to other regulatory mechanisms beside effects on transcription. These sequences correspond to the genes arginase (Arg1), ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 (Atp5a1), carbamoyl-phosphate synthetase 1(Cps1), heat shock protein 8 (Hspa8), malate dehydrogenase 2 (Mdh2) and tubulin, alpha 1C (Tuba1c/Tuba6) (equivalent to protein spots 5206/5207, 8408, 5811, 1613, 9208 and 408, respectively).
Finally, the microarray results for four of the genes, corresponding to the FXR targets corroborated by Western blot (Atp5a1, Anxa5, Rpsa and Gstm1), were verified by RT-qPCR showing a very good correlation with the microarray data (Fig. 1C), with the exception of Rpsa/Lamr which was not detectable in the microarray analysis.
4. Summary and conclusions
Gene expression analysis has been the method of choice to identify NR targets. Very few studies of proteomic analysis of NR targets have been published thus far. However, considering regulatory mechanisms acting directly on the protein level, such as the effects of microRNAs, proteomics could be of great importance also for the NR field. In fact, in the present study, none of the proteins found to be potential FXR targets in the proteomics assay have previously been shown to be regulated by FXR. One of the most striking features of the protein list generated by our proteomics assay is the great number of mitochondrial proteins, indicating a novel and substantial impact of FXR activation on mitochondrial function in the liver. Another noticeable feature is the number of proteins involved in the urea cycle and metabolism of amino groups.
Two of the proteins verified as bona fide FXR targets have been implicated in hepatocellular carcinoma (HCC), the laminin receptor and glutathione-S transferase. The laminin receptor is an extracellular matrix glycoprotein involved in a wide variety of biological processes such as cell adhesion, differentiation, migration and metastasis. Increased levels of the laminin receptor have been correlated to HCC [29–31]. For glutathione S-transferase, an enzyme detoxifying electrophilic compounds by conjugating them with glutathione, null mutations have been linked with an increase in a number of cancers, including HCC when combined with alcohol intake [32,33]. ATP synthase subunit alpha is a part of the mitochondrial ATP synthase that catalyzes ATP synthesis and involvement of FXR activation in the oxidative phosphorylative process has not been shown before. Also ATP synthase has been shown to be connected with liver cancer. Yamada et al. found increased levels of ATP synthase in hepatoblastomas as compared to normal liver tissue [34]. Annexin A5 is a calcium-dependent phospholipid binding protein inhibiting phospholipase A2 and protein kinase C, and is widely used as a marker for apoptosis. Future studies should focus on dissecting the translational relevance of the present data in terms of in vivo metabolic pathways.
At least six of the proteins found in the proteomics assay seem to be regulated at a post-transcriptional level. It is possible, or perhaps even likely, that other proteins in Table 1 are post-transcriptionally regulated since the criteria we set to discover differences in mRNA amounts were at a low stringency. The comparison between the wild type mice treated or not with the FXR agonist did not reveal any differences at the mRNA level for the proteins in Table 1, even at a fold difference of 1.5. In fact, none of the mRNAs showed a difference greater than 1.2. Only the comparison between the wild type and FXR−/− mice showed differences in mRNA amounts. However, the comparison between wild type and FXR−/− animals is a more artificial set-up, relating to the presence and absence of FXR, rather than the actual activation of the receptor. Moreover, the INT-747 samples used for the microarray were selected for a high FXR activation by assessing the mRNA levels of the FXR target gene Shp, to ascertain the identification of FXR targets regulated on the mRNA level. Selecting samples with a high Shp induction created a bias toward finding transcriptionally regulated FXR targets, which could mean an incidence of false positives. Thus, the absence of differences in mRNA levels, in the INT-747 versus vehicle treated wild type mice, for the proteins identified as novel targets, in those selected samples, further increases the value of our finding. Recently a microRNA, miR-34a, was shown to be inhibited by FXR [8] suggesting one possible mechanism for the post-translational regulation of FXR targets. The FXR induced protein SHP was shown to interact with p53, thereby inactivating the transactivation of the miR-34a promoter, leading to an increase in Sirtuin 1.
In conclusion, we could find novel FXR targets by a proteomic approach. Since at least six of these were indicated to be post-transcriptionally regulated, these potential FXR targets could not have been found by gene expression techniques. Our laboratory is currently investigating the physiological relevance of the new FXR target pathways in different in vivo models. The knowledge of the FXR driven regulatory pathways is of great importance since soon a novel FXR ligand will probably enter the clinic. Perhaps more importantly, our study provides the impetus to include proteomic analysis for the identification of novel targets of transcription factors, such as NRs.
The following are the supplementary materials related to this article.
Fig. S1.
mRNA expression of the FXR target gene Shp. Hepatic RNA samples from wild type mice treated with INT-747 or vehicle and FXR−/− mice treated with INT-747 were assessed for Shp mRNA expression by RT-qPCR. Arrows indicate samples chosen for the subsequent microarray analysis.
Proteomics values from individual gels.
Statistical information of the DIGE
Acknowledgments
We thank S. Murzilli for her precious help during the study; Dr. D. J. Mangelsdorf (Southwestern Medical Center at Dallas, Texas) for providing us with FXR null mice; Dr. L. Adorini (Intercept Italia, Perugia, Italy) for providing us with INT747 compound. A. Moschetta is funded by the Italian Association for Cancer Research (AIRC, IG 10416), Italian Ministry of University (FIRB IDEAS RBID08C9N7), Italian Ministry of Health (Young Researchers Grant GR-2008-1143546), European Community's Seventh Framework Programme FP7/2007-2013 under grant agreement no. 202272 (LipidomicNet), Telethon (GPP08259), Cariplo (Milan) and Natural Pharma International (NPI Biotech) and University of Bari (Italy).
Footnotes
This article is part of a Special Issue entitled: Translating nuclear receptors from health to disease.
Contributor Information
Cissi Gardmo, Email: gardmo@negrisud.it.
Antonio Moschetta, Email: moschetta@negrisud.it.
References
- 1.Mangelsdorf D.J., Thummel C., Beato M., Herrlich P., Schutz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R.M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chawla A., Repa J.J., Evans R.M., Mangelsdorf D.J. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 3.Yang X., Downes M., Yu R.T., Bookout A.L., He W., Straume M., Mangelsdorf D.J., Evans R.M. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 4.Bookout A.L., Jeong Y., Downes M., Yu R.T., Evans R.M., Mangelsdorf D.J. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.G. Di Leva, P. Gasparini, C. Piovan, A. Ngankeu, M. Garofalo, C. Taccioli, M.V. Iorio, M. Li, S. Volinia, H. Alder, T. Nakamura, G. Nuovo, Y. Liu, K.P. Nephew, C.M. Croce, MicroRNA cluster 221–222 and estrogen receptor alpha interactions in breast cancer, J Natl Cancer Inst 102 706–721. [DOI] [PMC free article] [PubMed]
- 6.Yamagata K., Fujiyama S., Ito S., Ueda T., Murata T., Naitou M., Takeyama K., Minami Y., O'Malley B.W., Kato S. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol. Cell. 2009;36:340–347. doi: 10.1016/j.molcel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 7.Rainer J., Ploner C., Jesacher S., Ploner A., Eduardoff M., Mansha M., Wasim M., Panzer-Grumayer R., Trajanoski Z., Niederegger H., Kofler R. Glucocorticoid-regulated microRNAs and mirtrons in acute lymphoblastic leukemia. Leukemia. 2009;23:746–752. doi: 10.1038/leu.2008.370. [DOI] [PubMed] [Google Scholar]
- 8.J. Lee, A. Padhye, A. Sharma, G. Song, J. Miao, Y.Y. Mo, L. Wang, J.K. Kemper, A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition, J Biol Chem 285 12604–12611. [DOI] [PMC free article] [PubMed]
- 9.Song G., Wang L. Transcriptional mechanism for the paired miR-433 and miR-127 genes by nuclear receptors SHP and ERRgamma. Nucleic Acids Res. 2008;36:5727–5735. doi: 10.1093/nar/gkn567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X., Gocek E., Liu C.G., Studzinski G.P. MicroRNAs181 regulate the expression of p27Kip1 in human myeloid leukemia cells induced to differentiate by 1,25-dihydroxyvitamin D3. Cell Cycle. 2009;8:736–741. doi: 10.4161/cc.8.5.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parks D.J., Blanchard S.G., Bledsoe R.K., Chandra G., Consler T.G., Kliewer S.A., Stimmel J.B., Willson T.M., Zavacki A.M., Moore D.D., Lehmann J.M. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 12.Makishima M., Okamoto A.Y., Repa J.J., Tu H., Learned R.M., Luk A., Hull M.V., Lustig K.D., Mangelsdorf D.J., Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 13.Wang H., Chen J., Hollister K., Sowers L.C., Forman B.M. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 14.Modica S., Moschetta A. Nuclear bile acid receptor FXR as pharmacological target: are we there yet? FEBS Lett. 2006;580:5492–5499. doi: 10.1016/j.febslet.2006.07.082. [DOI] [PubMed] [Google Scholar]
- 15.Sinal C.J., Tohkin M., Miyata M., Ward J.M., Lambert G., Gonzalez F.J. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 16.Kok T., Hulzebos C.V., Wolters H., Havinga R., Agellon L.B., Stellaard F., Shan B., Schwarz M., Kuipers F. Enterohepatic circulation of bile salts in farnesoid X receptor-deficient mice: efficient intestinal bile salt absorption in the absence of ileal bile acid-binding protein. J. Biol. Chem. 2003;278:41930–41937. doi: 10.1074/jbc.M306309200. [DOI] [PubMed] [Google Scholar]
- 17.Kim I., Ahn S.H., Inagaki T., Choi M., Ito S., Guo G.L., Kliewer S.A., Gonzalez F.J. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res. 2007;48:2664–2672. doi: 10.1194/jlr.M700330-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Huang W., Ma K., Zhang J., Qatanani M., Cuvillier J., Liu J., Dong B., Huang X., Moore D.D. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312:233–236. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 19.Yang F., Huang X., Yi T., Yen Y., Moore D.D., Huang W. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007;67:863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- 20.Kim I., Morimura K., Shah Y., Yang Q., Ward J.M., Gonzalez F.J. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis. 2007;28:940–946. doi: 10.1093/carcin/bgl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Modica S., Murzilli S., Salvatore L., Schmidt D.R., Moschetta A. Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res. 2008;68:9589–9594. doi: 10.1158/0008-5472.CAN-08-1791. [DOI] [PubMed] [Google Scholar]
- 22.Inagaki T., Moschetta A., Lee Y.K., Peng L., Zhao G., Downes M., Yu R.T., Shelton J.M., Richardson J.A., Repa J.J., Mangelsdorf D.J., Kliewer S.A. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl Acad. Sci. U.S.A. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenzo-Zuniga V., Bartoli R., Planas R., Hofmann A.F., Vinado B., Hagey L.R., Hernandez J.M., Mane J., Alvarez M.A., Ausina V., Gassull M.A. Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology. 2003;37:551–557. doi: 10.1053/jhep.2003.50116. [DOI] [PubMed] [Google Scholar]
- 24.A.M. Thomas, S.N. Hart, B. Kong, J. Fang, X.B. Zhong, G.L. Guo, Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine, Hepatology 51 1410–1419. [DOI] [PMC free article] [PubMed]
- 25.Fiorucci S., Rizzo G., Donini A., Distrutti E., Santucci L. Targeting farnesoid X receptor for liver and metabolic disorders. Trends Mol. Med. 2007;13:298–309. doi: 10.1016/j.molmed.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Pellicciari R., Fiorucci S., Camaioni E., Clerici C., Costantino G., Maloney P.R., Morelli A., Parks D.J., Willson T.M. 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J. Med. Chem. 2002;45:3569–3572. doi: 10.1021/jm025529g. [DOI] [PubMed] [Google Scholar]
- 27.Shevchenko A. Evaluation of the efficiency of in-gel digestion of proteins by peptide isotopic labeling and MALDI mass spectrometry. Anal. Biochem. 2001;296:279–283. doi: 10.1006/abio.2001.5321. [DOI] [PubMed] [Google Scholar]
- 28.Pellicciari R., Sato H., Gioiello A., Costantino G., Macchiarulo A., Sadeghpour B.M., Giorgi G., Schoonjans K., Auwerx J. Nongenomic actions of bile acids. Synthesis and preliminary characterization of 23- and 6,23-alkyl-substituted bile acid derivatives as selective modulators for the G-protein coupled receptor TGR5. J. Med. Chem. 2007;50:4265–4268. doi: 10.1021/jm070633p. [DOI] [PubMed] [Google Scholar]
- 29.Ding S.J., Li Y., Shao X.X., Zhou H., Zeng R., Tang Z.Y., Xia Q.C. Proteome analysis of hepatocellular carcinoma cell strains, MHCC97-H and MHCC97-L, with different metastasis potentials. Proteomics. 2004;4:982–994. doi: 10.1002/pmic.200300653. [DOI] [PubMed] [Google Scholar]
- 30.Zheng S., Ruan Y., Wu Z., Tang J. The relationship between 67KD laminin receptor expression and metastasis of hepatocellular carcinoma. J. Tongji Med. Univ. 1997;17:200–202. doi: 10.1007/BF02895618. 224. [DOI] [PubMed] [Google Scholar]
- 31.Xu L., Hui L., Wang S., Gong J., Jin Y., Wang Y., Ji Y., Wu X., Han Z., Hu G. Expression profiling suggested a regulatory role of liver-enriched transcription factors in human hepatocellular carcinoma. Cancer Res. 2001;61:3176–3181. [PubMed] [Google Scholar]
- 32.White D.L., Li D., Nurgalieva Z., El-Serag H.B. Genetic variants of glutathione S-transferase as possible risk factors for hepatocellular carcinoma: a HuGE systematic review and meta-analysis. Am. J. Epidemiol. 2008;167:377–389. doi: 10.1093/aje/kwm315. [DOI] [PubMed] [Google Scholar]
- 33.Covolo L., Gelatti U., Talamini R., Garte S., Trevisi P., Franceschi S., Franceschini M., Barbone F., Tagger A., Ribero M.L., Parrinello G., Donadon V., Nardi G., Donato F. Alcohol dehydrogenase 3, glutathione S-transferase M1 and T1 polymorphisms, alcohol consumption and hepatocellular carcinoma. ItalyCancer Causes Control. 2005;16:831–838. doi: 10.1007/s10552-005-2302-2. [DOI] [PubMed] [Google Scholar]
- 34.Yamada S., Ohira M., Horie H., Ando K., Takayasu H., Suzuki Y., Sugano S., Hirata T., Goto T., Matsunaga T., Hiyama E., Hayashi Y., Ando H., Suita S., Kaneko M., Sasaki F., Hashizume K., Ohnuma N., Nakagawara A. Expression profiling and differential screening between hepatoblastomas and the corresponding normal livers: identification of high expression of the PLK1 oncogene as a poor-prognostic indicator of hepatoblastomas. Oncogene. 2004;23:5901–5911. doi: 10.1038/sj.onc.1207782. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteomics values from individual gels.
Statistical information of the DIGE