Abstract
Gene expression is a fundamentally stochastic process, with randomness in transcription and translation leading to significant cell-to-cell variations in mRNA and protein levels. This variation appears in organisms ranging from microbes to metazoans and its characteristics depend both on the biophysical parameters governing gene expression and on gene network structure. Stochastic gene expression can have important consequences for cellular function, being beneficial in some contexts and harmful in others. These situations include stress response, pathogenesis, metabolism, development, cell cycle, circadian rhythms and aging.
Introduction
Life is a study in contrasts between randomness and determinism: from the chaos of biomolecular interactions to the precise coordination of development, living organisms are able to resolve these two seemingly contradictory aspects of their internal workings. Scientists often reconcile the stochastic and the deterministic by appealing to the statistics of large numbers, thus diminishing the importance of any one molecule in particular. However, cellular function often involves small numbers of molecules, of which perhaps the most important example is DNA. It is this molecule, usually present in just one or few copies per cell, that gives organisms their unique genetic identity. But what about genetically identical organisms grown in homogenous environments? To what degree are they unique? Increasingly, researchers have found that even genetically identical individuals can be very different, and that some of the most striking sources of this variability are random fluctuations in the expression of individual genes. Fundamentally, this is because the expression of a gene involves the discrete and inherently random biochemical reactions involved in the production of mRNAs and proteins. The fact that DNA (and hence the genes encoded therein) is present in very low numbers means that these fluctuations do not just average away but can instead lead to easily detectable differences between otherwise identical cells; in other words, gene expression must be thought of as a stochastic process.
The experimental observation that the levels of gene expression vary from cell-to-cell is certainly not new. In 1957, Novick and Weiner showed that the production of beta-galactosidase in individual cells was highly variable and random, with induction increasing the proportion of cells expressing the enzyme rather than increasing every cell's expression level equally (Novick and Weiner, 1957). Such early studies were hindered, however, by the lack of reliable single-cell assays of gene expression. One of the first studies to use an expression reporter in single-cells to examine the stochastic underpinnings of expression variability is the pioneering work of Ko et al., 1990. They examined the effect of different doses of glucocorticoid on the expression of a glucocorticoid-responsive transgene encoding beta-galactosidase and found that the cell-to-cell variability in the expression of the transgene was surprising large. Moreover, increasing the dose led to an increased frequency of cells displaying a high level of expression rather than a uniform increase in expression in every cell; that is, dose dependence was a consequence of changing the probability that an individual cell would express the gene at a high level.
Yet despite the potential biological consequences of random cellular variability (Spudich and Koshland, 1976), several years would pass before theoretical work ignited much of the present interest in stochastic gene expression (McAdams and Arkin, 1997; Arkin et al. 1998) They modeled gene expression using a stochastic formulation of chemical kinetics derived by Gillespie (1977), predicting that in some biologically realistic parameter ranges, protein numbers could fluctuate markedly within individual cells. Theythen extended their analysis to model the circuit underlying the decision between lysis and lysogeny of the phage-lambda, showing that stochastic effects in the expression of key regulators could explain why some cells activated the lytic pathway whereas others followed the lysogenic pathway. The notion that stochastic effects in gene expression could have important biological implications has motivated much research in the field and has only recently been explored experimentally.
Since this early research, the study of stochastic gene expression has blossomed into a rich field, with researchers from a diverse set of backgrounds working on a wide range of problems. The field is also notable for its strong interplay between theory and experiment, with many scientists making significant contributions to both. In this review, we will describe these researchers' efforts to characterize the underlying phenomenon through a host of organisms using a variety of experimental and theoretical methods. We will then highlight some recent endeavors trying to tie stochastic gene expression to biological phenomena.
Noisy bugs
The first attempts to characterize stochastic gene expression were born from experiments in synthetic biology in which experimenters found that noisy behavior in gene expression was interfering with the operation of engineered genetic circuits. One example is the “repressilator”, a synthetic network of repressors that was capable of producing oscillations in gene expression (Elowitz and Leibler, 2000). The authors found that the oscillations were subject to marked fluctuations in their period and magnitude, and conjectured that stochastic effects in gene expression were causing these effects. In another study explicitly aimed at controlling fluctuations, Becskei and Serrano (2000) showed that engineering a circuit with negative feedback could reduce cell-to-cell variability in expression. Although these experiments showed that noise in gene expression was important and could even be controlled, the molecular basis for the observed variability remained unclear.
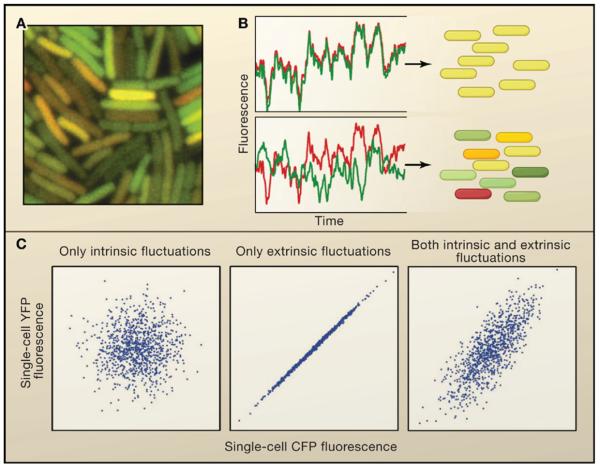
The first experiments to explore the causes of stochastic gene expression were the landmark studies of Elowitz et al. (2002) and Ozbudak et al. (2002). Elowitz et al. introduced the concepts of extrinsic and intrinsic noise in gene expression (analyzed mathematically by Swain et al., 2002). These ideas are perhaps most easily explained through example. In their experiments, Elowitz et al. quantified the variability in the expression from a promoter in E. coli by introducing two copies of the same promoter into the genome of E. coli, one driving the expression of cyan fluorescent protein (CFP) and the other driving the expression of yellow fluorescent protein (YFP) (Figures 1A and 1B). In this setup, extrinsic fluctuations are those that affect the expression of both copies of the gene equally in a given cell, such as variations in the numbers of RNA polymerases or ribosomes. Intrinsic fluctuations are those due to the randomness inherent to transcription and translation; being random, they should affect each copy of the gene independently, adding uncorrelated variations in levels of CFP and YFP levels (Figure 1C). They found that both sources of noise can be significant depending on the promoter. Later time-lapse measurements showed that in bacteria, the time scale for intrinsic fluctuations is less than 9 minutes, whereas extrinsic fluctuations exert their effects on time scales of about 40 minutes, or roughly the length of the cell cycle (Rosenfeld et al., 2005).
Figure 1. Intrinsic and extrinsic contributions to noise in gene expression.
A) A fluorescence image of individual E. coli displaying marked cell-to-cell variability in the expression of two identically regulated fluorescent proteins. B) Schematic depiction of the temporal behaviors of extrinsic noise (upper) and intrinsic noise (lower). C) Expected cell-to-cell variations when fluctuations are intrinsic, extrinsic or both. (A and B adapted from Elowitz et al., 2002).
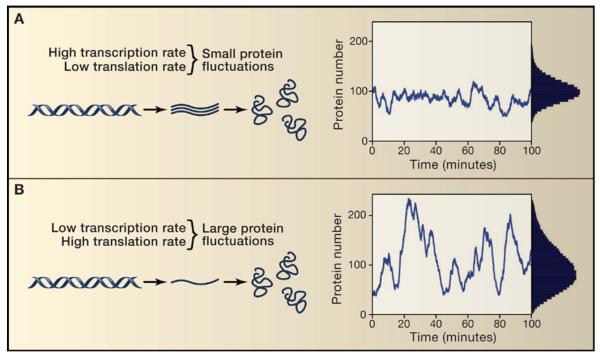
Ozbudak et al. (2002) obsereved that variability in the expression of a gene expressing GFP driven by an inducible promoter in B. subtilis depended on the underlying biochemical rates of transcription and translation. In these experiments transcription rates were controlled by varying the level of induction and the translation rate was altered by introducing mutations into the ribosomal binding site. This verified a stochastic theory of intrinsic noise they had developed predicting how noise in gene expression would change as these parameters were altered (Thattai and van Oudenaarden, 2001) (Figures 2A and 2B). In particular, the theory predicted that noise (measured by the standard deviation in protein expression level divided by the mean) would depend inversely on the rate of transcription but would not depend on the rate of translation. This is because proteins are produced in translational “bursts” from individual transcripts; the concept of bursts in gene expression continues to play an important role in current research, especially in higher eukaryotes.
Figure 2. Noise in prokaryotic gene expression depends on the rates of transcription and translation.
A) When the transcription rate is high, variability in protein levels is low, but B) when the transcription rate is lowered and the translation rate is raised, gene expression is far noisier, even at the same mean, as shown in Ozbudak et al. (2002).
Recently, a set of exciting single-molecule experiments have observed translational bursts in individual living bacteria. To count the number of proteins per cell, Cai et al., 2006 used used two methods: one involving microfluidics, in which they quantified the number of beta-galactosidase enzymes in a cell by monitoring its enzymatic activity, and one involving direct visualization of single YFP molecules tethered to the cellular membrane (Yu et al., 2006). Both studies showed that proteins were synthesized in rapid, burst-like fashion.
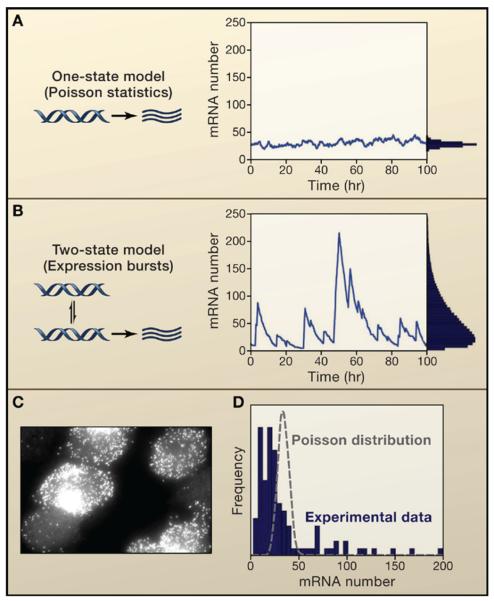
Another study (Golding et al., 2005) used the MS2-GFP method (Bertrand et al., 1998; Beach et al., 1999), which allows one to monitor the transcription of individual mRNA molecules in real time. This is accomplished by introducing a repeated sequence motif into the 3′ untranslated region of the mRNA to which a fusion of the MS2 coat protein and GFP binds, thus rendering the mRNA molecule fluorescent. According to the model presented in Figure 3A, one would expect that mRNA molecules are produced at a steady rate according to the statistics of a Poisson process. The authors found, however, that the mRNA molecules were themselves produced in transcriptional bursts, as if the gene itself was randomly switching back and forth between transcriptionally active and inactive states. This finding mirrors results obtained for eukaryotes described below. It would be interesting to combine these different measurements of the dynamics of individual mRNAs and proteins, given the role that competition between translation and mRNA degradation may play in stochastic gene expression (Yarchuk et al., 1998).
Figure 3. The Contribution of Transcriptional Bursts to Cell-to-Cell Variability.
A) Transcription without bursts with a relatively small amount of noise. B) Bursts in transcription can cause significantly higher variability, even when producing the same mean number of transcripts. C) In situ detection of individual mRNA molecules reveals large cell-to-cell variability in mammalian cells. D) Experimental histogram of mRNA numbers. The grey dashed line depicts the theoretical distribution one would expect in the absence of transcriptional bursts. (C and D adapted from Raj et al., 2006)
Eukaryotes and the burst hypothesis
After these experiments in bacteria, researchers began to investigate stochastic gene expression in eukaryotes, initially focusing on yeast. Almost immediately, several reports seemed to indicate that the sources of variability in gene expression in yeast are different from those in bacteria in a number of important ways (Becskei et al., 2005; Blake et al., 2006; Blake et al., 2003; Raser and O'Shea, 2004). These studies all examined the relationship between the mean level of expression and the variation about that mean, a relationship that is in theory qualitatively different depending on the sources of noise. In all these studies, the relationship predicted by the simple model in Figure 2 was insufficient to explain the experimental observations. These observations were, however, compatible with models of transcriptional bursts in which the gene itself randomly transitioned between states of transcriptional activity and inactivity (Figure 3B). Such models of transcriptional bursting add another important source of stochasticity beyond random events in transcription and translation, which have now been analyzed theoretically in some detail (Friedman et al., 2006; Karmakar and Bose, 2004; Kepler and Elston, 2001; Pedraza and Paulsson, 2008).
That such models are required to explain eukaryotic data but not most prokaryotic data (Cai et al., 2006; Maamar et al., 2007; Yu et al., 2006), with an important exception (Golding et al., 2005), strongly suggests that some regulator of gene expression specific to eukaryotes is responsible. The most likely candidate for this is remodeling of chromatin: when the surrounding chromatin is in an open, acetylated state, the gene is able to transcribe relatively freely, whereas when chromatin is in a condensed state, transcription is repressed. Although there is still no direct evidence that chromatin remodeling is responsible for stochastic changes in gene activity, several studies have tried to link chromatin-related events to stochastic gene expression by indirect means. These include positional effects like measuring correlations between proximally located genes (Becskei et al., 2005; Raj et al., 2006) or altering the behavior of chromatin remodeling agents (Raser and O'Shea, 2004; Xu et al., 2006). However, global studies of noise in yeast (Bar-Even et al., 2006; Newman et al., 2006) have shown that the presence of chromatin remodeling complexes is neither necessary nor sufficient for the expression of a gene to be noisy; also, factors such as the location and number of transcription factor binding sites can control noise (Murphy et al., 2007).
In yeast noise in gene expression is primarily extrinsic in origin (Becskei et al., 2005; Colman-Lerner et al., 2005; Raser and O'Shea, 2004; Volfson et al., 2006), resulting in correlated fluctuations between different genes. Sources identified thus far for this extrinsic noise are cell size (Raser and O'Shea, 2004; Newman et al., 2006; Volfson et al., 2006), variations in common upstream factors (Becskei et al., 2005; Volfson et al., 2006) and chromosomal location (Becskei et al., 2005); by contrast, extrinsic variability in prokaryotic gene expression is thought to stem mostly from variations in upstream factors (Elowitz et al., 2002). There is some debate as to the role of differences in cell-cycle and cell size, with some data (Raser and O'Shea, 2004) showing that extrinsic variability remains even after controlling for these variables, whereas other data indicates that a stringent analysis of size and shape by flow cytometry can account for most of the extrinsic noise (Newman et al., 2006). Generally, one of the difficulties in studying extrinsic variability is its catchall nature: the lack of any specific mechanism makes its analysis rather phenomenological. Although there is some knowledge of the time scales over which extrinsic noise operates (Rosenfeld et al., 2005) and theoretical analyses of the effects that it might have (Shahrezaei et al., 2008) (Paulsson, 2004), understanding extrinsic noise remains an unresolved problem in the field.
Higher eukaryotes: noisier than expected
Meanwhile, work has begun on systematically examining cell-to-cell variability in gene expression in higher eukaryotes. A priori, one might expect that higher eukaryotes, with their larger size and numbers of molecules, might exhibit less variability than prokaryotes and yeast. On the other hand, the prevalence of transcriptionally-silenced heterochromatin would argue that slow, random events of gene activation and inactivation would lead to much larger fluctuations than in unicellular organisms. As it happens, the latter is the case, with a growing body of evidence that fluctuations in higher eukaryotes can be remarkably large.
Interestingly, the study of expression variability in higher eukaryotes began well before the recent heightened interest in stochastic gene expression. Beginning with the aforementioned work of Ko et al. (1990), several other reports indicated that gene expression in mammalian cells was variable, stemming from short, rare events of active transcription (Ross et al., 1994; Newlands et al., 1998; Takasuka et al., 1998; White et al., 1995).
Many of these early experiments were limited by the difficulties inherent to measuring gene expression in single-cells in higher eukaryotes. One problem is sensitivity: owing to their large cellular volumes, even moderately expressed fluorescent proteins can be difficult to detect. Another problem is the lack of tools available to manipulate these organisms genetically. To circumvent these problems, researchers have come up with many new ways of assaying gene expression at the single-cell level to measure cell-to-cell variability.
One approach is to measure mRNAs rather than proteins. For instance, utilizing the MS2-GFP method of mRNA detection (Beach et al., 1999; Bertrand et al., 1998), Chubb et al. (2006) showed that a developmental gene in Dictyostelium discoideum is transcribed in a pulsatile fashion, directly demonstrating the burst hypothesis by watching mRNAs accumulate and dissipate from active and inactive sites of transcription in real time. In comparison with the less intense bursts observed using a similar approach in bacteria (Golding et al., 2005), the authors found that the bursts were less frequent but longer lasting. In contrast with earlier bacterial models, this shows that bursts in gene expression are the primary intrinsic cause of cell-to-cell variability.
One can also measure mRNA numbers in single cells across a population using variants of fluorescence in situ hybridization (FISH) capable of detecting individual mRNA molecules (Femino et al., 1998; Raj et al., 2006; Raj et al., 2008). Raj et al. (2006) combined single molecule FISH with statistical analysis to show that individual mammalian cells transcribed a stably integrated transgene in infrequent but potent bursts, resulting in large cell-to-cell variations in mRNA number (Figures 3C and 3D) that correlated with the presence or absence of active sites of transcription [seen also by (Voss et al., 2006)]. These bursts were correlated between genes that were located proximally to each other but not between genes that were distally located, providing another clue that chromatin remodeling may be responsible for genes transitioning between an active and inactive state: “opening” of the chromatin surrounding one gene is likely to open chromatin for neighboring genes, leading to correlations in their expression, whereas distant genes are not affected in this coordinated manner, resulting in uncorrelated expression. This behavior is also seen in globin expression (de Krom et al., 2002) and shows that genomic position can be important in interpreting the concepts of intrinsic and extrinsic noise.
Quantitative single-cell RT-PCR methods have been used to obtain cell-by-cell counts of endogenous mRNAs, thus circumventing issues associated with generating transgenic cell lines and organisms. By simultaneously measuring the numbers of five transcripts in individual pancreatic islet cells, Bengtsson et al. (2005) showed that the distributions of these mRNAs across the population were heavily skewed as in Figure 3D. Moreover, they measured correlations in the fluctuations in the expression of these genes, finding that two functionally related genes were highly correlated whereas the rest were uncorrelated, perhaps pointing to the existence of common regulators for the two genes. Such findings highlight the potential use of stochastic gene expression in uncovering the mechanisms of transcriptional regulation. One difficulty with this approach is the rigorous set of controls required to calibrate RT-PCR results in molecular units, a problem that can be obviated through the use of so-called “digital” RT-PCR. This method, in which cDNA reverse transcribed from an individual cell is fractionated into enough individual PCR reactions that each reaction will contain either 0 or 1 cDNAs, has been used to examine the expression of the PU.1 transcription factor in both hematopoetic stem cells and in myeloid progenitor cells (Warren et al., 2006), in which the authors observed marked heterogeneity in transcript levels.
Atlhough the evidence for transcriptional bursting continues to accumulate, little is known about the source of these bursts. As mentioned earlier, one possibility often posited is that stochastic events of chromatin remodeling could underlie the bursts by causing the gene to switch between transcriptionally active and inactive states (Becskei et al., 2005; Raj et al., 2006; Raser and O'Shea, 2004; Warren et al., 2006). In support of this view, direct visualization of chromatin remodeling has shown it to be a slow process that can act over a long range on a timescale of hours (Tumbar et al., 1999). However, there are other plausible mechanisms that might underlie transcriptional bursts. One possibility is the existence of pre-initiation complexes that form on the promoter region of the DNA and facilitate multiple rounds of RNA polymerase II transcription events (Blake et al., 2006; Blake et al., 2003). If such complexes exist only for short periods of time, they could also result in pulsatile transcription. Another point to consider is that transcription doesn't take place in a uniform fashion throughout the genome but is concentrated in transcriptional “factories” (Jackson et al., 1993; Wansink et al., 1993) to which active genes are recruited (Osborne et al., 2004). Remarkably, it appears that a limited number of these factories (on the order of hundreds) are responsible for most mRNA transcription in the cell; thus, competition for these factories could result in the stochastic expression of any given gene. Ultimately, understanding the biochemical origins of bursting may require the application of new (or perhaps combinations of old) techniques for imaging gene expression and genome organization in real-time, as cell-to-cell variability in population “snapshots” may not be sufficient to resolve the dynamics of the bursting mechanisms (Pedraza and Paulsson, 2008). Although difficult, the prize for such a technical feat would be a much deeper understanding of the transcriptional process.
The above studies examining mRNA copy number variation provide insights into the origins of noise, although they mostly fail to show how those mRNA fluctuations propagate to noise in protein levels. To examine noise in protein levels in human cells, Sigal et al. (2006) used a clever strategy to fluorescently tag endogenous proteins. They transfected a cell line with DNA containing artificial YFP exons that occasionally insert themselves into an intron, YFP is included in the protein encoded by the encapsulating gene. Using time-lapse microscopy, the authors were able to show that gene expression in individual cells was variable, but that the fluctuations were slowly varying in time; that is, it took multiple cell divisions before a highly expressing cell would become a lowly expressing cell and vice versa. Interestingly, they also found correlations between genes in the same pathway, but not between unrelated genes, echoing the results of Bengtsson et al. (2005).
Yet, the variability observed at the protein level by Sigal et al. (2006) seems generally much smaller than that observed at the mRNA level in the aforementioned studies, with the distribution of mRNAs being much more heavily skewed (Figure 3B). How might such a discrepancy be resolved? One answer may be methodological: by screening for cells expressing a detectable amount of YFP, the proteins with YFP insertions obtained by Sigal et al. may be biased towards heavily or constitutively expressing genes with less variability, an interpretation supported by the fact that variability in the number of GAPDH mRNAs is lower than other genes (Warren et al., 2006). It is also possible that protein stability plays a role in the relationship between mRNA and protein variability (Raj et al., 2006). Short-lived proteins will track mRNA levels very closely, leading to protein distributions that resemble (and correlate strongly with) mRNA distributions. However, if the proteins degrade slowly (as is the case for YFP), then the large pool of older proteins will buffer the rapid fluctuations in mRNA; that is, mRNA bursts may serve only to “top up” protein levels. In this case, mRNA and protein levels do not strongly correlate.
Networked noise
Building on these studies elucidating the sources and characteristics of noise, researchers went on to study the effects of noise in simple synthetic genetic networks. One example is transcriptional cascades, which are a common regulatory motif, particularly in development. First, researchers investigated the effect of noise in an upstream gene on noise in a downstream gene. This was done using multiple fluorescent reporters to quantify the relative contributions of variability in the upstream gene, global noise due to effects such as cell size, and also noise intrinsic to the expression of the downstream gene (Pedraza and van Oudenaarden, 2005; Rosenfeld et al., 2005). They found that variability can be transmitted from the upstream gene to the downstream gene, adding substantially to the noise inherent in downstream gene's expression. Further study of cascades showed that longer genetic cascades can filter out rapid fluctuations at the expense of amplifying noise in the timing of the propagated signal (Hooshangi et al., 2005). Mathematical analysis has also shown that stochastic behavior can have the counterintuitive effect of actually lowering transmitted variability (Paulsson and Ehrenberg, 2000; Thattai and van Oudenaarden, 2002).
Negative or positive feedback are other very common types of regulation in genetic networks. In these types of feedback loops the protein encoded by a gene negatively or positively influences its own transcription. Negative feedback can reduce the effects of noise because fluctuations above and below the mean are pushed back towards the mean, as has been predicted theoretically (Savageau, 1974; Thattai and van Oudenaarden, 2001) and demonstrated experimentally (Austin et al., 2006; Becskei and Serrano, 2000; Dublanche et al., 2006).
In the presence of positive feedback, noise can result in much more dramatic behavior. Positive feedback can act as a switch, in which a small amount of expression from a given gene can serve to further activate expression of the gene itself, eventually flipping the gene from an “off” state to an “on” state. In the presence of cooperativity, though, a cell can remain in the “off” state indefinitely, as cooperativity creates a threshold that the protein level must surpass in order to trigger the feedback. In that case, occasional large fluctuations in gene expression can serve to randomly activate the switch and push the cell into the on state (Hasty et al., 2000). This bistable expression pattern has been observed in synthetic systems with positive feedback switches (Becskei et al., 2001; Gardner et al., 2000; Isaacs et al., 2003; Kramer and Fussenegger, 2005) and also in several naturally occurring genetic positive feedback loops (Acar et al., 2005; Maamar and Dubnau, 2005; Maamar et al., 2007; Suel et al., 2006; Suel et al., 2007; Smits et al., 2005). The existence of multiple phenotypic profiles also appears in more complex biological networks, as we shall see in the next section.
Noise in its natural context
Researchers have only recently begun to explore the role fluctuations play in biological situations. One can imagine two roles for noise in cellular function: one is as a nuisance that serves as an impediment to reliable behavior, and one is as a source of variability that cells may exploit. In the remainder of the review, we first focus on cases where noise is beneficial and then discuss the potential negative effects of noise, drawing on examples in organisms ranging from microbes to metazoans.
Useful unicellular variability
In unicellular organisms, one can make the argument that variability could be very useful in that it would allow heterogenous phenotypes even in clonal populations, enabling a population of organisms to “commit” certain subpopulations to different behaviors. Variability in a population is enhanced by networks that can produce multiple, mutually exclusive profiles of gene expression profiles (such as ON and OFF expression of a particular gene) within single organisms. These states are “bistable” (or multistable) in the sense that small variations in expression are insufficient to cause the organism to flip from one state to another and are often heritable, providing a mechanism for epigenetic inheritance (Ptashne, 2007). Occasionally, however, a large stochastic fluctuation in gene expression can induce a transition from one state to another, an idea that underlies many of the following studies.
Metabolism
Metabolic networks are an important and perhaps surprising class of genetic networks exhibiting multistability with stochastic transitions. Despite being some of the most extensively studied gene networks in existence, only recently have researchers begun to examine the behavior of metabolic genes at the single-cell level, yielding unanticipated results. For instance, following up on the pioneering studies of Novick and Weiner, it was found that the lactose utilization network in E. coli displayed an “all or none” type of behavior and that single cells stochastically transition between these two states (Mettetal et al., 2006; Ozbudak et al., 2004). Such behavior has also been seen in cells that were all initially in an uninduced state, arguing that some stochastic mechanism must have caused the network to switch from the off to the on expression state. In another example the galactose utilization network in yeast also displays strikingly bimodal patterns in the expression of the GAL family of genes responsible for galactose metabolism (Acar et al., 2005). The authors explained this using a model in which fluctuations in the GAL3 gene were responsible for transitions between the ON and OFF states. Then they altered the expression of a key feedback component of the network, thereby changing the degree to which the fluctuations were buffered and thus modulating the frequency of the stochastic transitions. The dynamics of these switching events has also been analyzed using time-lapse microscopy, yielding fascinating results (Kaufmann et al., 2007). There, the authors found that not only were the states themselves heritable, but the transition itself was heritable in that related yeast cells appeared to switch in a correlated fashion. Again, the authors were able to explain their results using a stochastic model, with the key feature being stochastic bursts in GAL80 expression. In all of these studies, however, the link between stochastic switching and stochastic gene expression has been implicit rather than explicit, with more experiments being required to validate the models.
Of course, these results raise the inevitable question of why genetically identical populations would display such marked phenotypic variability in their metabolic pathways. One idea is that having individual cells stochastically switch between activating or inactivating a metabolic pathway could confer a fitness advantage to the overall population in fluctuating environments (Kussell and Leibler, 2005; Thattai and van Oudenaarden, 2004; Wolf et al., 2005). Intuitively, this benefit arises from a tradeoff between anticipation and sensing of food sources. Cells can either directly sense food in the environment before activating their metabolic networks, or they can choose to stochastically commit some fraction of the population to having those metabolic networks active in anticipation of the arrival of a new food source. The cost of the former strategy is slow response time and implementation of the sensing apparatus, whereas the latter strategy essentially sacrifices some fraction of the population to suboptimal growth. These studies have shown that stochastic switching is a viable alternative to sensing and that it is most effective when the switching rate is closely tuned to the rate at which the environment fluctuates. Experimentally, Acar et al. (2008) tested these theories by monitoring the growth rate of a yeast strain with a controllable rate of switching in a periodically fluctuating environment and show that fast switchers do indeed grow faster in rapidly fluctuating environments whereas slow switchers do better when environmental changes come more slowly. Furthermore, Blake et al. (2006) showed that expression variability, even in the absence of discrete fit and unfit expression states, can be beneficial in times of stress. It is likely, however, that in real biological systems, cells rely on some combination of variability in gene expression and sensing in their stress responses; elucidating this interplay in biological contexts could have broad implications for microbial growth strategies.
Microbial stress responses
Another case of bet-hedging in microbial populations is in the response to cellular stress, such as lack of food or exposure to antibiotics. A particularly nice example of the former that has garnered considerable recent attention is the phenomenon of competence in B. subtilis. B. subtilis has the remarkable ability to take up DNA from the environment (called competentence), which exhibits itself upon the entry to stationary phase through the activation of a quorum sensing mechanism. Interestingly, only a small fraction (roughly 10-20%) of the cells become competent, while the rest remain in a vegetative state. This phenotypic variability was first observed over 40 years ago (Nester and Stocker, 1963) and is the result of a positive feedback loop in which the transcription factor ComK promotes its own expression: when the feedback loop is activated, high levels of ComK are produced, activating a host of downstream genes involved in DNA uptake, whereas non-competent cells produce only low basal amounts of ComK (Maamar and Dubnau, 2005; Smits et al., 2005; Suel et al., 2006). The resulting bimodal expression pattern is easily visualized using fluorescent proteins.
A natural hypothesis is that spontaneous fluctuations in comK expression of sufficient magnitude can cause a non-competent cell to transition to competence. To test this notion, Maamar et al. (2007) quantified comK expression in individual non-competent cells by using single-molecule FISH to count the number of comK transcripts. They showed that increasing the mean level of comK transcription resulted in an increase in the percentage of competent cells, presumably because the fraction of cells with fluctuations in ComK above a certain threshold also increased. To test that possibility directly, they increased the comK transcription rate while lowering the translation rate, which reduces noise in gene expression while leaving the mean expression level unchanged (Ozbudak et al., 2002; Thattai and van Oudenaarden, 2001). Lowering the noise should reduce the number of cells whose fluctuations cross the threshold for competence, and indeed the authors found that the number of competent cells was dramatically reduced, demonstrating the importance and utility of noise theory in biological situations. Another recent study (Suel et al., 2007) showed that reducing total cellular noise also resulted in a lower percentage of competent cells. To achieve this overall noise reduction, they used a special mutant that is unable to septate, resulting in very large cells with multiple genomes. In these large cells, the impact of all fluctuations is reduced, given that the cell is in some ways the “average” of many smaller cells, with ever larger cells consequently having ever lower overall fluctuations. The authors found that these larger cells did in fact display commensurately fewer transitions to the competent state. Overall, though, it is important to note that the low number of comK transcripts measured (Maamar et al., 2007) and the non-uniformity of the duration of competence episodes (Suel et al., 2007) imply that this system has evolved to be purposefully imprecise, a feature that cells may exploit in other situations.
Stochastic effects coupled with positive feedback can also lead to variability in the timing of particular molecular events such as the onset of meiosis in yeast (Nachman et al., 2007). In this work the timing between introduction of environmental stress and the onset of meiosis in individual cells was highly variable. This variability seemed not to depend on position in the cell cycle or other external factors, but rather was heavily dependent on noise in the expression of the meiotic regulator Ime1 (although cell-size did appear to have a strong effect). Together, these studies paint a picture in which noise in gene expression can lead to random fates at random times when stressed, a surprising finding that may ultimately prove remarkably prevalent.
Pathogens
Heterogeneous phenotypes in clonal populations can also be medically relevant. One example is bacterial persistence in the face of antibiotic exposure. Persistent cells grow at a much slower rate than non-persistent cells, but are able to survive antibiotic treatment. The existence of persistent subpopulations of Mycobacterium tuberculosis, Staphylococcus aureus, and Pseudomonas aeruginosa among other is thought to be a major obstacle to effective treatment (Stewart et al., 2003). Interestingly, the work of Balaban et al. (2004) showed that a small persistent subpopulation exists even in untreated cultures of E. coli and that these persistent cells are generated continuously during growth. Although not much is known about how the underlying network can result in such disparate phenotypes, it is entirely possible that stochastic gene expression could play a significant role in establishing non-genetic heterogeneity in these populations.
Another example of heterogeneity in a pathogen is that of the latent phase of HIV infection. Upon infection with the HIV virus, a small pool of latent CD4 T lymphocytes forms containing stably integrated but non-expressing virus. The low level of expression of the virus in this population of cells renders them difficult to target pharmacologically, making latency a serious impediment to effective treatment. Weinberger et al. (2005) showed that one explanation for the latent and active expression patterns is a positive feedback loop mediated by the Tat protein. They showed that stochastic fluctuations in Tat expression can interact with the feedback loop to create populations of cells with high and low levels of viral expression Interestingly, though, later work (Weinberger et al., 2007) showed that Tat positive feedback did not serve to maintain the “ON” state (as in the competence network in B. subtilis) but rather that the heterogeneity was caused by large transient bursts of expression that positive feedback served to amplify rather than stabilize (Weinberger et al., 2008).
Random Developments
As the above examples demonstrate, there is a clear rationale for using stochastic gene expression to create a diversity of phenotypes, namely that isogenic populations of viruses, bacteria and yeast cannot display heterogeneity in any other way. However, in many higher eukaryotes, population diversity largely arises from genetic and environmental diversity, making the argument for utilizing stochastic gene expression less plausible. In development, for example, one would imagine that a deterministic execution of the developmental program would be critical to producing functional tissues, with organism-to-organism variations reflecting genetic rather than stochastic differences. Yet even in development, researchers are finding many interesting examples of stochastic cell-fate decisions linked to stochastic gene expression.
One celebrated example of stochastic gene expression having an important role in development is the expression of different odorant receptors in different sensory neurons in mice. Olfaction presents an interesting regulatory challenge, as there are over a thousand different odorant receptors, each of which must be expressed differentially in individual neurons to confer distinctive sensitivity. Developing a regulatory network capable of such complex decision making is prohibitively complex, so the mouse adopts a much simpler “Monte-Carlo” strategy in which each neuron randomly expresses a particular odorant receptor (Vassar et al., 1993) in a mutually exclusive fashion (Tsuboi et al., 1999). A fascinating line of further inquiry would be to determine the stochastic mechanisms responsible for these choices during the development of the olfactory epithelium and elucidation of the network responsible for “locking in” a particular decision once made.
Another particularly nice instance in which stochastic gene expression has been explicitly linked to a cell fate decision is photoreceptor expression in Drosophila eyes. The Drosophila eye consists of a large number of optical units called ommatidia, each of which contains two cells that in turn express one of a specific pair of photoreceptors, either Rh3 and Rh5 (for blue sensitive ommatidia) or Rh4 and Rh6 (for yellow sensitive ommatidia). Wernet et al. (2006) showed that this decision is almost exclusively due to the stochastic expression of the spineless gene during mid-pupation, with stochastically large levels of spineless expression resulting in the adoption of the yellow fate in roughly 70% of the ommatidia.
The process of hematopoiesis, in which progenitor stem cells differentiate into the various types of blood cells, is another example in which cellular differentiation may be stochastic (Enver et al., 1998; Hume, 2000). To link this stochastic differentiation to variations in gene expression, Chang et al. (2008) showed that variability in the expression of the stem cell marker Sca-1 in individual cells correlated strongly with the probability of that cell to choose an erythroid or myeloid lineage. Moreover, microarray analysis on the populations of cells expressing high and low levels of Sca-1 showed transcriptome-wide variability, indicating that the fluctuations were not limited only to a small set of genes. It would be interesting to see how widespread these massively correlated fluctuations are in other examples of stochastic differentiation and if these correlations stem from an unknown master regulator or arise from noise in many parts of a large interlocking genetic network.
Shutting out the noise
Despite these examples of organisms exploiting noise, it is possible, if not probable, that noise in gene expression is more generally an obstacle that organisms must overcome to achieve robust function. Less is currently known about the mechanisms by which the effects of noise are minimized, likely due to the difficulty in studying a phenomenon that by definition is invariant to perturbations. In fact, much of the focus on the benefits of noise reflects the fact that studying the consequences of stochastic gene expression is much easier when the phenomenon in question is itself stochastic. Nevertheless, progress has been made in understanding how organisms tolerate noise, from the basics of cellular function through development.
Genomic approaches
One way to find evidence for the deleterious effects of noise is to make comprehensive measurements of noise over a large number of genes and look for evidence that noise has been selected against in certain sets of genes. This was the approach taken by Newman et al. (2006) and Bar-Even et al. (2006), with the former measuring the noise in expression in over 2500 genes in yeast and the latter examining fewer genes (43) but in a variety of environmental conditions. Both studies reached strikingly similar conclusions, finding that noise stemmed mostly from randomness in mRNA synthesis and destruction and that genes with higher levels of expression generally exhibited less variability from cell to cell. This latter point highlights a potential tradeoff between the level of noise in gene expression and the metabolic cost of maintaining a large number of proteins. They also found that stress response genes, which are typically non-essential, tended to be noisy, reflecting the potential benefits of noise in this class of gene (Blake et al., 2006). In contrast, genes involved in protein synthesis and degradation were much less variable, implying that genes essential for cellular function require more precise expression levels. The regularity of these essential genes may be achieved in a number of ways such as genomic positioning of essential genes in areas of open chromatin that are presumably less noisy (Batada and Hurst, 2007). This correlative data does not prove the case, however, and an explicit test that noise in essential genes is deleterious would be fruitful in this regard.
Noise minimization and compensation in gene networks
Given that genes often interact in networks, it is also important to understand how the effects of noise are minimized in particular genetic networks. To study this more complex problem, Kollman et al.(2005) in their study of the chemotaxis network of E. coli, began with several plausible biochemical models of chemotaxis. Each model possessed the fundamental property of precise adaptation of pathway activity to local food signals, but varied in their ability to tolerate noise. Upon measuring the noise and correlations in the expression of several key components of the pathway, they found that the model that most successfully tolerated such noise described a network similar to the one found in E. coli, suggesting that the endogenous network may have evolved to tolerate noise while avoiding the costs associated with high levels of protein expression.
Another example of noise-resistance in a signaling pathway is the mating pheromone response pathway in yeast studied by Colman-Lerner et al. (2005). Through the use of dual reporters inspired by Elowitz et al. (2002), they quantified all the different sources of cell-to-cell variability in their system, with the primary distinction being between random biochemical events in the propagation of the signal itself and preexisting differences in cells' capacity to respond to the signal. They found that most of the variability observed was due to preexisting cellular differences, corroborating other claims that variability in yeast is largely extrinsic (Raser and O'Shea, 2004; Volfson et al., 2006). Interestingly, though, they found a surprising negative correlation between the signaling capacity of the pathway in individual cells and the capacity to express the pathway's target gene in those same cells. The implication is that variability in the signaling pathway is compensated for at the level of gene expression, thus allowing the cell to produce a robust gene expression profile despite large differences in signaling capacities.
Noise resistance has also driven much research into the networks underlying circadian rhythms, biochemical oscillations present in organisms ranging from cyanobacteria to humans that are entrained by periodic exposure to sunlight but are capable of “free-running” without any external signals. These oscillations display a remarkable fidelity in their duration from cycle to cycle, but the source of this reliability in still unclear and may depend on properties of the network used to implement the oscillator. For instance cyanobacteria, despite possessing perhaps the simplest known clock, produce very regular oscillations. Notably, the proteins involved can oscillate in vitro in the absence of any transcriptional regulation at all (Nakajima et al., 2005), but presumably variability in the numbers of these proteins in individual cells can cause cells to lose synchrony. Indeed, gene expression variability during the clock cycle has many interesting properties (Chabot et al., 2007). Another possibility is that cell-cell communication might allow cells to compensate for the fluctuations in the oscillations of individual cells. This is not the case in cyanobacteria, however, given that when one places two cells at different phases of the circadian cycle next to each other, their progeny robustly maintain the two different cycles inherited from the parents (Mihalcescu et al., 2004).
In higher organisms, transcriptional regulation plays a key role in the generation of circadian rhythms, and single cell experiments have shown the performance of the clock in individual mammalian cells can be rather poor, with strikingly variable periods observed both in culture (Nagoshi et al., 2004) and in whole organisms (Liu et al., 1997). There is some evidence behind the general consensus that cell-cell communication allow all the cells in an organism's pacemaker to maintain its phase (Liu et al., 1997), but it would be interesting to explore how noise in gene expression contributes to dephasing individual cells, especially given recent theories claiming that even these networks seem to have some noise-resistant properties (Barkai and Leibler, 2000; Forger and Peskin, 2005). More generally, such results could apply to other kinds of genetic oscillators like the cell-cycle, where recent work has shown that noise is a key factor in cell-cycle timing variability (Di Talia et al., 2007).
Noise minimization in development
So far, little work has been done on the role of noise in gene expression in development, probably due to difficulties in obtaining quantitative measurements. However, one excellent example of a developmental buffer against noise is the activity of Hsp90 in Arabidopsis (Queitsch et al., 2002), the inhibition of which reveals the effects of genetic and environmental variability. Surprisingly, this same inhibition results in marked developmental variability even in relatively isogenic populations, most likely stemming from stochastic effects. An exciting avenue for further research would be to try and link stochastic gene expression to phenotypic diversity (familiar to geneticists as the common phenomena of partial penetrance or variable expressivity of phenotypes).
Stochastic gene expression and aging
Another line of evidence that noise is undesirable comes from research showing that aging is correlated with increased noise in gene expression. In one case, researchers showed that the expression of a variety of housekeeping and cell-type specific genes in individual murine cardiac myocytes become increasingly stochastic as the organism aged (Bahar et al., 2006). They further found that treating cells isolated from young animals with hydrogen peroxide also produces an increase in expression variability, perhaps indicating that oxidative damage may be a factor. Similar stochastic effects have been seen in aging murine muscle tissues (Newlands et al., 1998).
Conversely, the stochastic expression of a gene may actually be responsible for determining lifespan in C. elegans (Rea et al., 2005). The authors found that the level of expression of a reporter expressed from a heat shock promoter in response to environmental stress on the first day of adulthood was remarkably stochastic and moreover predicted the lifespan of the organism. Although the mechanisms underlying these stochastic phenomena are still unclear, it is possible that aging may be surprisingly dependent on the effects of stochastic gene expression.
Conclusion
We would like to emphasize that despite the flurry of activity in the area of stochastic gene expression over the last several years, the field is still remarkably young, with many significant discoveries likely to come in the future. Basic measurements of cell-to-cell variability in higher eukaryotes are still in their infancy, and single-molecule techniques have shown that surprises still lurk even in supposedly well-characterized systems such as E. coli. Moving forward, researchers have also started to examine biological consequences of noise—already, there are more and more examples of noise being beneficial in isogenic populations, a trend we expect to continue. We anticipate more studies highlighting how cells control and tolerate noise to produce reliable behavior. Of course, the most exciting discoveries are those that are completely unexpected, and given the fundamental nature of stochastic gene expression, it may prove important in unpredictable ways in experimental systems both new and old.
Acknowledgements
We would like to thank Michael Laub, Ido Golding, Jim Collins and Jeff Gore for many helpful comments on the manuscript. We also apologize to any authors whose work we were unable to mention due to space constraints. A.v.O was supported by NSF grant PHY-0548484 and NIH grants R01-GM068957 and R01-GM077183. A.R. is supported by NSF Fellowship DMS-0603392 and a Burroughs Wellcome Fund Career Award at the Scientific Interface.
References
- Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- Acar M, Mettetal JT, van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nat Genet. 2008;40:471–475. doi: 10.1038/ng.110. [DOI] [PubMed] [Google Scholar]
- Arkin A, Ross J, McAdams HH. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics. 1998;149:1633–1648. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin DW, Allen MS, McCollum JM, Dar RD, Wilgus JR, Sayler GS, Samatova NF, Cox CD, Simpson ML. Gene network shaping of inherent noise spectra. Nature. 2006;439:608–611. doi: 10.1038/nature04194. [DOI] [PubMed] [Google Scholar]
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, Vijg J. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Bar-Even A, Paulsson J, Maheshri N, Carmi M, O'Shea E, Pilpel Y, Barkai N. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- Barkai N, Leibler S. Circadian clocks limited by noise. Nature. 2000;403:267–268. doi: 10.1038/35002258. [DOI] [PubMed] [Google Scholar]
- Batada NN, Hurst LD. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat Genet. 2007;39:945–949. doi: 10.1038/ng2071. [DOI] [PubMed] [Google Scholar]
- Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol. 1999;9:569–578. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- Becskei A, Kaufmann BB, van Oudenaarden A. Contributions of low molecule number and chromosomal positioning to stochastic gene expression. Nat Genet. 2005;37:937–944. doi: 10.1038/ng1616. [DOI] [PubMed] [Google Scholar]
- Becskei A, Seraphin B, Serrano L. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. Embo J. 2001;20:2528–2535. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- Bengtsson M, Stahlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15:1388–1392. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Blake WJ, M KA, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- Chabot JR, Pedraza JM, Luitel P, van Oudenaarden A. Stochastic gene expression out-of-steady-state in the cyanobacterial circadian clock. Nature. 2007;450:1249–1252. doi: 10.1038/nature06395. [DOI] [PubMed] [Google Scholar]
- Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner A, Gordon A, Serra E, Chin T, Resnekov O, Endy D, Pesce CG, Brent R. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437:699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- de Krom M, van de Corput M, von Lindern M, Grosveld F, Strouboulis J. Stochastic patterns in globin gene expression are established prior to transcriptional activation and are clonally inherited. Mol Cell. 2002;9:1319–1326. doi: 10.1016/s1097-2765(02)00558-0. [DOI] [PubMed] [Google Scholar]
- Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature. 2007;448:947–951. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- Dublanche Y, Michalodimitrakis K, Kummerer N, Foglierini M, Serrano L. Noise in transcription negative feedback loops: simulation and experimental analysis. Mol Syst Biol. 2006;2:41. doi: 10.1038/msb4100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Enver T, Heyworth CM, Dexter TM. Do stem cells play dice? Blood. 1998;92:348–351. discussion 352. [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Forger DB, Peskin CS. Stochastic simulation of the mammalian circadian clock. Proc Natl Acad Sci U S A. 2005;102:321–324. doi: 10.1073/pnas.0408465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N, Cai L, Xie XS. Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Phys Rev Lett. 2006;97:168302. doi: 10.1103/PhysRevLett.97.168302. [DOI] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Gillespie DT. Exact Stochastic Simulation of Coupled Chemical-Reactions. Journal of Physical Chemistry. 1977;81:2340–2361. [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Hasty J, Pradines J, Dolnik M, Collins JJ. Noise-based switches and amplifiers for gene expression. Proc Natl Acad Sci U S A. 2000;97:2075–2080. doi: 10.1073/pnas.040411297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooshangi S, Thiberge S, Weiss R. Ultrasensitivity and noise propagation in a synthetic transcriptional cascade. Proc Natl Acad Sci U S A. 2005;102:3581–3586. doi: 10.1073/pnas.0408507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume DA. Probability in transcriptional regulation and its implications for leukocyte differentiation and inducible gene expression. Blood. 2000;96:2323–2328. [PubMed] [Google Scholar]
- Isaacs FJ, Hasty J, Cantor CR, Collins JJ. Prediction and measurement of an autoregulatory genetic module. Proc Natl Acad Sci U S A. 2003;100:7714–7719. doi: 10.1073/pnas.1332628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. Embo J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmakar R, Bose I. Graded and binary responses in stochastic gene expression. Phys Biol. 2004;1:197–204. doi: 10.1088/1478-3967/1/4/001. [DOI] [PubMed] [Google Scholar]
- Kaufmann BB, Yang Q, Mettetal JT, van Oudenaarden A. Heritable stochastic switching revealed by single-cell genealogy. PLoS Biol. 2007;5:e239. doi: 10.1371/journal.pbio.0050239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler TB, Elston TC. Stochasticity in transcriptional regulation: origins, consequences, and mathematical representations. Biophys J. 2001;81:3116–3136. doi: 10.1016/S0006-3495(01)75949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko MS. A stochastic model for gene induction. J Theor Biol. 1991;153:181–194. doi: 10.1016/s0022-5193(05)80421-7. [DOI] [PubMed] [Google Scholar]
- Ko MS, Nakauchi H, Takahashi N. The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. Embo J. 1990;9:2835–2842. doi: 10.1002/j.1460-2075.1990.tb07472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmann M, Lovdok L, Bartholome K, Timmer J, Sourjik V. Design principles of a bacterial signalling network. Nature. 2005;438:504–507. doi: 10.1038/nature04228. [DOI] [PubMed] [Google Scholar]
- Kramer BP, Fussenegger M. Hysteresis in a synthetic mammalian gene network. Proc Natl Acad Sci U S A. 2005;102:9517–9522. doi: 10.1073/pnas.0500345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- Liu C, Weaver DR, Strogatz SH, Reppert SM. Cellular construction of a circadian clock: period determination in the suprachiasmatic nuclei. Cell. 1997;91:855–860. doi: 10.1016/s0092-8674(00)80473-0. [DOI] [PubMed] [Google Scholar]
- Maamar H, Dubnau D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol. 2005;56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc Natl Acad Sci U S A. 1997;94:814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettetal JT, Muzzey D, Pedraza JM, Ozbudak EM, van Oudenaarden A. Predicting stochastic gene expression dynamics in single cells. Proc Natl Acad Sci U S A. 2006;103:7304–7309. doi: 10.1073/pnas.0509874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalcescu I, Hsing W, Leibler S. Resilient circadian oscillator revealed in individual cyanobacteria. Nature. 2004;430:81–85. doi: 10.1038/nature02533. [DOI] [PubMed] [Google Scholar]
- Murphy KF, Balazsi G, Collins JJ. Combinatorial promoter design for engineering noisy gene expression. Proc Natl Acad Sci U S A. 2007;104:12726–12731. doi: 10.1073/pnas.0608451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman I, Regev A, Ramanathan S. Dissecting timing variability in yeast meiosis. Cell. 2007;131:544–556. doi: 10.1016/j.cell.2007.09.044. [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Nester EW, Stocker BA. Biosynthetic Latency In Early Stages Of Deoxyribonucleic Acidtransformation In Bacillus Subtilis. J Bacteriol. 1963;86:785–796. doi: 10.1128/jb.86.4.785-796.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlands S, Levitt LK, Robinson CS, Karpf AB, Hodgson VR, Wade RP, Hardeman EC. Transcription occurs in pulses in muscle fibers. Genes Dev. 1998;12:2748–2758. doi: 10.1101/gad.12.17.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- Novick A, Weiner M. Enzyme Induction as an All-or-None Phenomenon. Proc Natl Acad Sci U S A. 1957;43:553–566. doi: 10.1073/pnas.43.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Lim HN, Shraiman BI, Van Oudenaarden A. Multistability in the lactose utilization network of Escherichia coli. Nature. 2004;427:737–740. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]
- Paulsson J, Ehrenberg M. Random signal fluctuations can reduce random fluctuations in regulated components of chemical regulatory networks. Phys Rev Lett. 2000;84:5447–5450. doi: 10.1103/PhysRevLett.84.5447. [DOI] [PubMed] [Google Scholar]
- Paulsson J. Summing up the noise in gene networks. Nature. 2004;427:415–418. doi: 10.1038/nature02257. [DOI] [PubMed] [Google Scholar]
- Pedraza JM, Paulsson J. Effects of molecular memory and bursting on fluctuations in gene expression. Science. 2008;319:339–343. doi: 10.1126/science.1144331. [DOI] [PubMed] [Google Scholar]
- Pedraza JM, van Oudenaarden A. Noise propagation in gene networks. Science. 2005;307:1965–1969. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- Ptashne M. On the use of the word ‘epigenetic’. Curr Biol. 2007;17:R233–236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using sets of singly labeled probes. Nat Meth. 2008 September 21; doi: 10.1038/nmeth.1253. published online. doi ##################. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea SL, Wu D, Cypser JR, Vaupel JW, Johnson TE. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat Genet. 2005;37:894–898. doi: 10.1038/ng1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- Ross IL, Browne CM, Hume DA. Transcription of individual genes in eukaryotic cells occurs randomly and infrequently. Immunol Cell Biol. 1994;72:177–185. doi: 10.1038/icb.1994.26. [DOI] [PubMed] [Google Scholar]
- Savageau MA. Comparison of classical and autogenous systems of regulation in inducible operons. Nature. 1974;252:546–549. doi: 10.1038/252546a0. [DOI] [PubMed] [Google Scholar]
- Shahrezaei V, Ollivier JF, Swain PS. Colored extrinsic fluctuations and stochastic gene expression. Mol Syst Biol. 2008;4:196. doi: 10.1038/msb.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal A, Milo R, Cohen A, Geva-Zatorsky N, Klein Y, Liron Y, Rosenfeld N, Danon T, Perzov N, Alon U. Variability and memory of protein levels in human cells. Nature. 2006;444:643–646. doi: 10.1038/nature05316. [DOI] [PubMed] [Google Scholar]
- Smits WK, Eschevins CC, Susanna KA, Bron S, Kuipers OP, Hamoen LW. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol Microbiol. 2005;56:604–614. doi: 10.1111/j.1365-2958.2005.04488.x. [DOI] [PubMed] [Google Scholar]
- Spudich JL, Koshland DE., Jr. Non-genetic individuality: chance in the single cell. Nature. 1976;262:467–471. doi: 10.1038/262467a0. [DOI] [PubMed] [Google Scholar]
- Stewart GR, Robertson BD, Young DB. Tuberculosis: a problem with persistence. Nat Rev Microbiol. 2003;1:97–105. doi: 10.1038/nrmicro749. [DOI] [PubMed] [Google Scholar]
- Suel GM, Garvia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- Suel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Tunability and noise dependence in differentiation dynamics. Science. 2007;315:1716–1719. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci U S A. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasuka N, White MR, Wood CD, Robertson WR, Davis JR. Dynamic changes in prolactin promoter activation in individual living lactotrophic cells. Endocrinology. 1998;139:1361–1368. doi: 10.1210/endo.139.3.5826. [DOI] [PubMed] [Google Scholar]
- Thattai M, van Oudenaarden A. Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci U S A. 2001;98:8614–8619. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattai M, van Oudenaarden A. Attenuation of noise in ultrasensitive signaling cascades. Biophys J. 2002;82:2943–2950. doi: 10.1016/S0006-3495(02)75635-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattai M, van Oudenaarden A. Stochastic gene expression in fluctuating environments. Genetics. 2004;167:523–530. doi: 10.1534/genetics.167.1.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi A, Yoshihara S, Yamazaki N, Kasai H, Asai-Tsuboi H, Komatsu M, Serizawa S, Ishii T, Matsuda Y, Nagawa F, Sakano H. Olfactory neurons expressing closely linked and homologous odorant receptor genes tend to project their axons to neighboring glomeruli on the olfactory bulb. J Neurosci. 1999;19:8409–8418. doi: 10.1523/JNEUROSCI.19-19-08409.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Sudlow G, Belmont AS. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol. 1999;145:1341–1354. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- Volfson D, Marciniak J, Blake WJ, Ostroff N, Tsimring LS, Hasty J. Origins of extrinsic variability in eukaryotic gene expression. Nature. 2006;439:861–864. doi: 10.1038/nature04281. [DOI] [PubMed] [Google Scholar]
- Voss TC, John S, Hager GL. Single-cell analysis of glucocorticoid receptor action reveals that stochastic post-chromatin association mechanisms regulate ligand-specific transcription. Mol Endocrinol. 2006;20:2641–2655. doi: 10.1210/me.2006-0091. [DOI] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Bryder D, Weissman IL, Quake SR. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci U S A. 2006;103:17807–17812. doi: 10.1073/pnas.0608512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell. 2005;122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Weinberger LS, Schenk T. An HIV feedback resistor: auto-regulatory circuit deactivator and noise buffer. PLoS Biology. 2007;5(1):e9. doi: 10.1371/journal.pbio.0050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger LS, Dar RD, Simpson ML. Transient-mediated fate determination in a transcriptional circuit of HIV. Nat Genet. 2008;40:466–470. doi: 10.1038/ng.116. [DOI] [PubMed] [Google Scholar]
- Wernet MF, Mazzoni EO, Celik A, Duncan DM, Duncan I, Desplan C. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 2006;440:174–180. doi: 10.1038/nature04615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MR, Masuko M, Amet L, Elliott G, Braddock M, Kingsman AJ, Kingsman SM. Real-time analysis of the transcriptional regulation of HIV and hCMV promoters in single mammalian cells. J Cell Sci. 1995;108(Pt 2):441–455. doi: 10.1242/jcs.108.2.441. [DOI] [PubMed] [Google Scholar]
- Wolf DM, Vazirani VV, Arkin AP. Diversity in times of adversity: probabilistic strategies in microbial survival games. J Theor Biol. 2005;234:227–253. doi: 10.1016/j.jtbi.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Xu EY, Zawadzki KA, Broach JR. Single-cell observations reveal intermediate transcriptional silencing states. Mol Cell. 2006;23:219–229. doi: 10.1016/j.molcel.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Yarchuk O, Jacques N, Guillerez J, Dreyfus M. Interdependence of translation, transcription and mRNA degradation in the lacZ gene. J Mol Biol. 1998;226:581–596. doi: 10.1016/0022-2836(92)90617-s. [DOI] [PubMed] [Google Scholar]
- Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]