Abstract
Purpose
Treatment of malignant pleural mesothelioma (MPM) with Ranpirnase (Onconase) results in disruption of protein translation and cell apoptosis. We hypothesize that Onconase acts via down regulation of nuclear factor kappa B (NFKβ) by specific microRNAs (miRNA) and that interference of this pathway could have implications for MPM resistance to chemotherapy.
Experimental Design
Three immortalized MPM cell lines (H2959, H2373, and H2591) were exposed to Onconase at 0–20 µg/mL. Cell counts were measured at 48 and 72 hours. Gene expression in miRNA-enriched RNA was validated by RT-PCR. The functional implications of miRNA expression were evaluated by transfecting miRNA mimics or inhibitors into MPM cell lines, and performing Matrigel™ invasion, cell proliferation, soft agar colony formation, and scratch closure assays. Effects on NFKβ expression and downstream targets including ABC transporters, BCL-xl, and IAP were assessed by RT-PCR and Western Blotting.
Results
Treatment with 20µg/mL of Onconase significantly decreased cell count and invasion. Hsa-miR-17* was significantly upregulated and hsa-miR-30c significantly down-regulated by Onconase treatment in all cell lines. Forced expression of hsa-miR-17* mimic and hsa-miR-30c inhibitor each significantly decreased functional activity of Onconase in all assays. NFKB1(p50) expression and downstream targets were also decreased with Onconase treatment as well as with forced expression miRNA mimic and inhibitors.
Conclusions
Onconase treatment caused a significant decrease in cell proliferation, invasion, and in expression of certain miRNAs. Recapitulation of the resultant miRNA expression pattern with hsa-miR-17* mimic and hsa-miR-30c inhibitor resulted in downregulation of NFKB1 and reduced malignant behavior in functional assays. Thus, Onconase likely exerts its anti-tumor effect through these miRNAs.
Keywords: Mesothelioma, microRNA, multidrug resistance
Introduction
Malignant pleural mesothelioma (MPM) is a rare disease with approximately 2000–3000 new cases in the United States diagnosed annually.(Kaufman and Pass 2008) Over the last 20 years, the incidence of MPM has steadily increased, with 70–80% of cases implicating asbestos exposure as the dominate risk factor.(Kaufman and Pass 2008; Robinson and Lake 2005) Management of MPM is predominately with chemotherapy and radiation due its diffuse and invasive nature, which often precludes surgical resection. Consideration for surgery either as an extrapleural pneumonectomy or pleurectomy/decortication is dependent on the disease stage, pulmonary function, and surgeon experience.(Beck et al. 2008) Current first line therapy for unresectable disease includes combination pemextred and cisplatin, a treatment plan with a high rate of failure due to chemoresistance or the presence of advanced disease (Pass et al. 2004; Pavlakis and Vogelzang 2006; Carbone et al. 2007). Onconase (Ranpirnase), a member of the pancreatic RNAase A superfamily of ribonucleases, is a chemotherapeutic agent which has demonstrated selective antitumor activity in a variety of human malignancies including MPM (Wu et al. 1993). Onconase exerts its antiproliferative and cytotoxic effects by interfering with cell cycle regulation and induction of programmed cell death and intermediates in ranpirnase mechanism of action have been described in many publications (Porta et al. 2008; Lee and Raines 2008). Activation of MAPK and an increase in JNK activity was first described in HeLa cells(Iordanov et al. 2000). Ranpirnase has been shown to activate caspase 3 and decrease Bcl2 concomitant with inducing apoptosis (Ardelt et al. 2007). Ranpirnase decreases nuclear factor kappa-light-chain-enhancer of activated B cells (NFkappaB, p50), a transcription factor that has also been strongly implicated in cancer cell chemo-resistance, anti-apoptosis and induction of cell proliferation(Ardelt et al. 2003). Growth suppression induced by Onconase is closely coordinated with a down-regulation in the steady state and subcellular distribution of NF-kappaB (Tsai et al. 2004), In fact, the reduction of NF-kappaB expression by onconase appears to coincide or even precede the growth suppression, which suggests a causal relationship. Onconase also enhances tumor cell sensitivity to a number of cytotoxic agents with diverse mechanisms of action, making it ideal for use in combination therapy (Zhao et al. 2008; Mikulski et al. 1992; Rybak et al. 1996). In a recent Phase IIIb clinical trial reported at ASCO 2009, Onconase added to Doxorubicin therapy demonstrated significant efficacy in patients with MPM that failed prior chemotherapy. Median survival was prolonged in this Onconase-Doxorubicin treated subgroup of previously treated mesothelioma patients compared to patients who received Doxorubicin alone (p=0.016) (Reck et al. 2009).
Although it is believed that tRNA is chiefly affected by Ranpirnase, other down-stream RNA species that mediate the cytotoxic effects of Onconase have not been previously described (Wu et al. 1993; Zhao et al. 2008; Mikulski et al. 1992; Deptala et al. 1998). MicroRNAs (miRNAs) are small noncoding RNAs that negatively regulate protein-coding genes, and provide insight into gene regulation and cell behavior. The relationship between Onconase treatment and miRNA and how it intermediates and controls cell proliferation, invasion, migration, and apoptosis has not been investigated. Moreover, further validation of the Onconase regulation of NFKβ expression and whether microRNAs influenced by Onconase are involved in this regulation remains speculative. We hypothesize that (1) Onconase acts via specific miRNAs (2) these miRNAs will have similar effect on MM cells as Onconase itself and (3) these effects are involved in the regulation of NFKβ and its downstream targets. An elucidation of these pathways could give insight into the ability of Onconase to act as a chemosensitizing adjunctive therapy.
Results
Onconase Treatment – Cell Count and Cell Functionality
Cell lines were subjected to treatment with Onconase at 0–20 µg/mL. A significant decrease in cell proliferation was observed in all three cell lines with 20 µg/mL Onconase treatment at 48 and 72 hours as shown (Supplemental Fig. 1A) and 20 µg/mL Onconase significantly reduced the invasive capacities of H2595, H2373 and H2591 cells (Supplemental Fig. 1B). By contrast, control cells demonstrated a 2-fold increase in cell number at 48 hours, and a 3-fold increase at 72 hours. Similar effects were seen with 5–10 µg/mL Onconase at 48 and 72 hours, but to a lesser extent than seen with treatment at 20 µg/mL (Supplemental Figure 2). . RT-PCR validation of ABCB1 and NFKB(p50 subunit) in all three cell lines showed decreased levels of their expression by Onconase treatment at 48 and 72 hours (Supplemental Fig. 3). These findings were validated at protein levels by Western Blotting (Supplemental Fig. 4)
miRNAExpression
Mirs that were influenced by Onconase treatment are seen in Table 1, and the associated Heatmap (Figure 1). Hsa-miR-17* and hsa-miR-30c demonstrated the most significant changes in miRNA expression following Onconase treatment across cell lines and were selected for further study based on these data. Treatment with Onconase at 20 µg/mL for 72 hours resulted in upregulated expression of hsa-miR-17* by 30 fold compared to controls (p ≤ 10−8), and downregulated expression of hsa-miR-30c by 41% in cell lines compared to controls (p ≤ 10−8) based on results obtained from the miRNA arrays. These microRNA array results from Onconase treatment were validated by RT-PCR. A dose dependent effect of Onconase on the expression of these miRs was seen (Figure 2). hsa-miR-17* had minimal native expression in the mesothelioma cell lines studied and was upregulated 2.46 to 3.23 fold by treatment with 20 ug/ml Onconase. Conversely, hsa-miR-30c has high endogenous expression in the absence of Onconase and was down regulated 61–63% following treatment with Onconase.
Table 1.
Significantly Changed Micrornas With Mesothelioma Cell Line Onconase Treatment
| Parametric p- value |
FDR | Mean of intensities Onconase |
Mean of intensities Control |
Fold- change |
MicroRNA | |
|---|---|---|---|---|---|---|
| 1 | < 1e-07 | < 1e-07 | 1518.4072631 | 50.2959051 | 30.1894808 | hsa-miR-17* |
| 2 | < 1e-07 | < 1e-07 | 407.3114758 | 133.4896675 | 3.0512584 | hsa-miR-769-3p |
| 3 | < 1e-07 | < 1e-07 | 3247.1378431 | 5515.6325766 | 0.5887154 | hsa-miR-30c |
| 4 | < 1e-07 | < 1e-07 | 9152.7590494 | 25788.858063 | 0.3549114 | hsa-miR-23a |
| 5 | < 1e-07 | < 1e-07 | 1891.0935228 | 3244.7084036 | 0.5828239 | hsa-miR-125b |
| 6 | < 1e-07 | < 1e-07 | 16597.7904741 | 6016.4929054 | 2.7587152 | hsa-miR-565 |
| 7 | < 1e-07 | < 1e-07 | 335.217613 | 792.1658504 | 0.4231659 | hsa-miR-125a-5p |
| 8 | < 1e-07 | < 1e-07 | 2724.4955954 | 5161.3129688 | 0.5278687 | hsa-miR-30b |
| 9 | < 1e-07 | < 1e-07 | 391.554316 | 96.9952605 | 4.0368397 | hsa-miR-124* |
| 10 | < 1e-07 | < 1e-07 | 1128.473283 | 2152.3264268 | 0.524304 | hsa-miR-99a |
| 11 | < 1e-07 | < 1e-07 | 1232.615019 | 2445.7869319 | 0.5039748 | hsa-let-7a |
| 12 | < 1e-07 | < 1e-07 | 707.5745087 | 1325.044174 | 0.5340007 | hsa-miR-30e |
| 13 | < 1e-07 | < 1e-07 | 12289.9606867 | 25663.4000117 | 0.4788906 | hsa-miR-222 |
| 14 | < 1e-07 | < 1e-07 | 877.0648344 | 1413.1624322 | 0.6206398 | hsa-miR-99b |
| 15 | < 1e-07 | < 1e-07 | 1408.6432393 | 2445.7256307 | 0.5759613 | hsa-miR-103 |
| 16 | 1e-07 | 1.4e-06 | 541.0343315 | 155.3695451 | 3.4822418 | hsa-miR-640 |
| 17 | 1e-07 | 1.4e-06 | 9956.774553 | 18131.8047399 | 0.5491331 | hsa-miR-23b |
| 18 | 2e-07 | 2.7e-06 | 595.5519873 | 1187.8930299 | 0.5013515 | hsa-let-7d |
| 19 | 3e-07 | 3.5e-06 | 21.9060305 | 253.9964753 | 0.0862454 | hsa-miR-32 |
| 20 | 3e-07 | 3.5e-06 | 3906.9784929 | 6220.6028033 | 0.6280707 | hsa-miR-202* |
Figure 1.
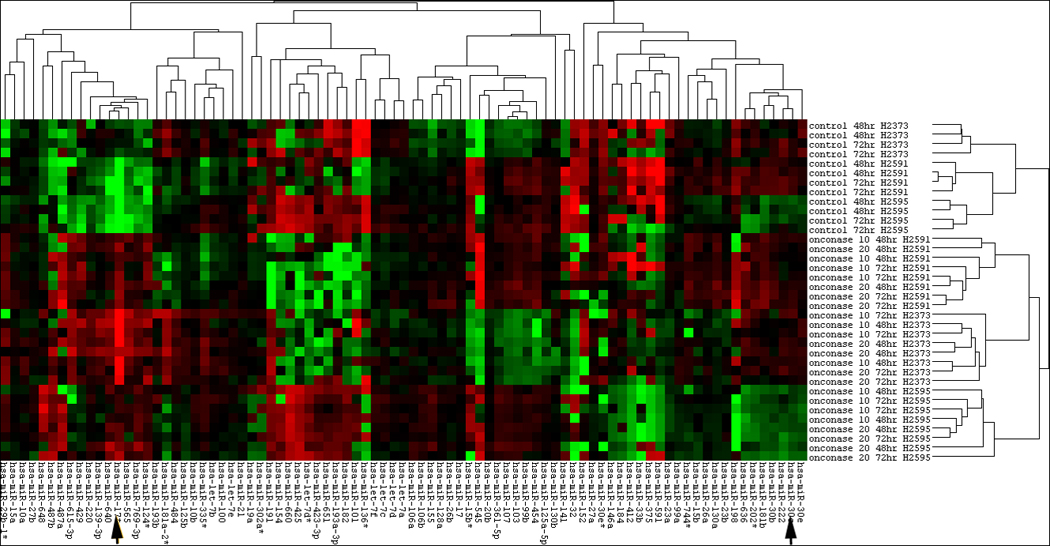
Heat map of microRNA expression with Onconase treatment of mesothelioma cell lines. Control conditions are seen in the upper panels and dose and time changes are seen in the lower panels. Hsa-mir-17* and hsa-mir-30c are seen at the arrows.
Figure 2.
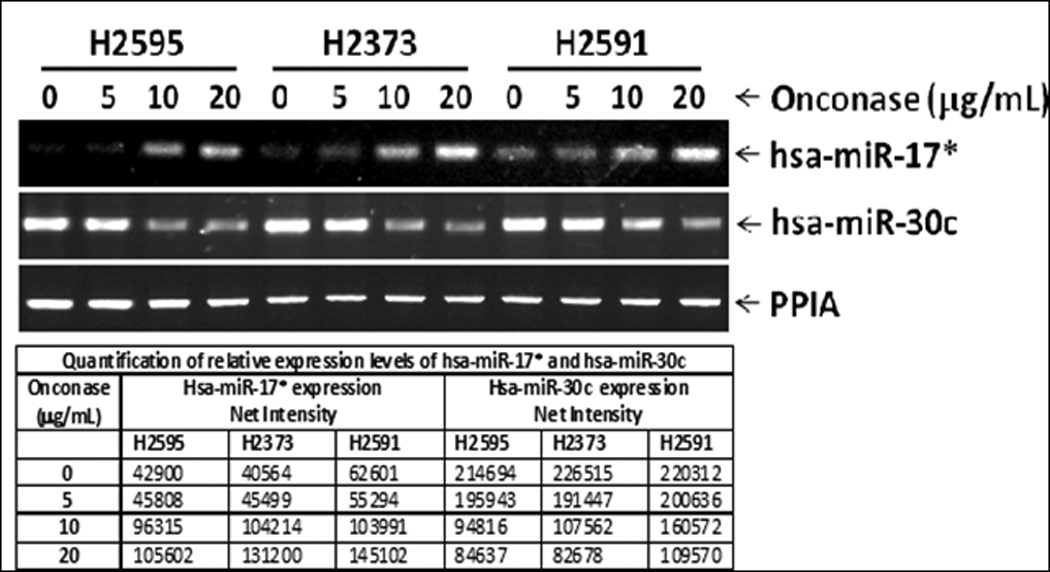
Dose effect of Onconase on expression of hsa-mir-17*, which increased in a dose dependent fashion, and has-mir-30c which decreased similarly in a dose-dependent fashion. Relative expression levels relative to PPIA for each cell line and each microRNA are seen below.
Transfection and Functional Assays
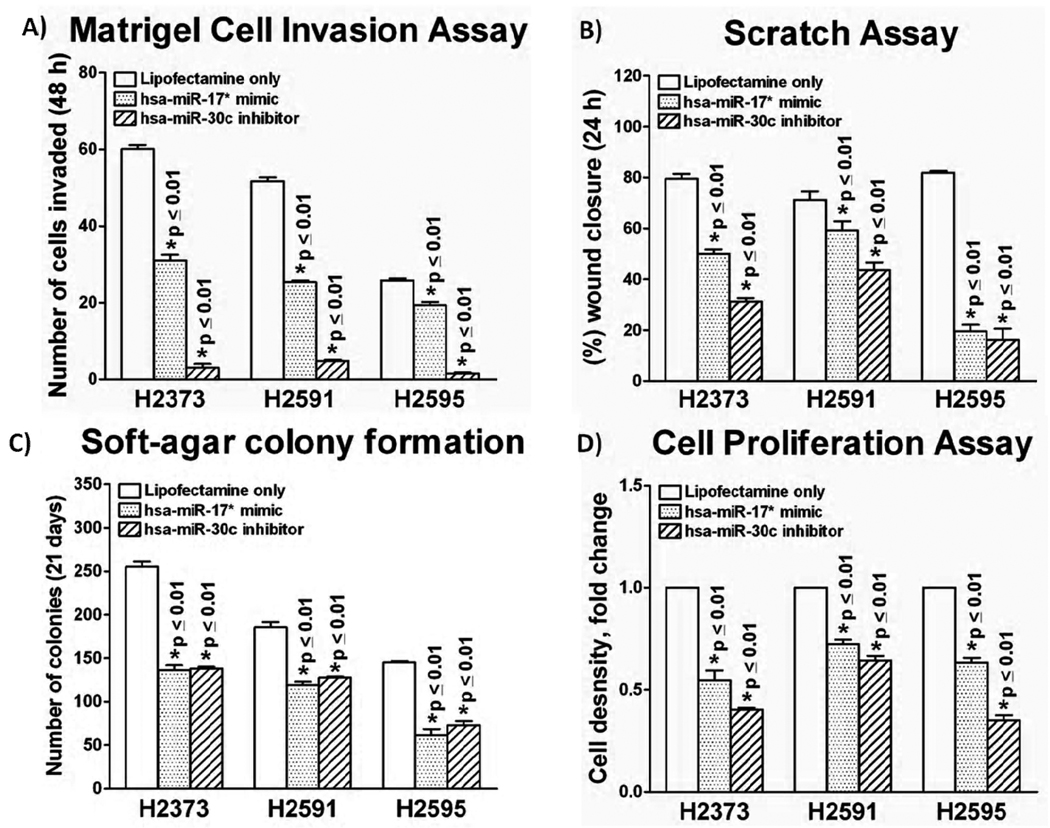
We were successful in transfecting hsa-miR 17* mimic and hsa-30c inhibitor in all three cell lines (Fig. 3A and 3B). Hsa-miR-17* mimic transfection decreased cell invasion by 31–48% in all cell lines (p ≤ 0.004) compared to controls, while hsa-30c inhibitor expression decreased cell invasion by 81 – 96% in all cell lines (p ≤ 0.001) compared to controls (Fig. 4A). Scratch closure assay demonstrated a similar trend, with significant reduction in closure with expression of the hsa-miR-17* mimic (p ≤ 0.005) and hsa-miR-30c inhibitor (p ≤ 0.001) (Fig. 4B). Anchorage independent growth was reduced 31 – 47% by hsa-miR-17* mimic expression compared to controls (p ≤ 0.004), and 32 to 46% by hsa-miR-30c inhibitor expression compared to controls (p ≤ 0.001) (Fig. 4C). Cell proliferation assay also demonstrated a similar effect, with 25 – 33% reduction in cell number following forced expression of hsa-miR-17* mimic (p ≤ 0.004), and a 35 – 80% reduction with forced expression of hsa-miR-30c inhibitor (p ≤ 0.001). (Fig. 4d) Overall, forced expression of both the hsa-miR-17* mimic and hsa-miR-30c inhibitor resulted in a significant decrease in functional activity in all four assays with each of the three MPM cell lines. Conversely, cell invasion results were reversed when the cells were forcibly expressed with hsa-miR-17* inhibitor and hsa-miR-30c mimic (Figure 5). These data with the two microRNAs are consistent with the effects that Onconase itself had on proliferation and invasion, and suggest that miR-17* has tumor suppressor properties while miR- 30c may, in fact, stimulate oncogenic properties of mesothelioma cell lines(Saxena et al. 2002).
Figure 3.
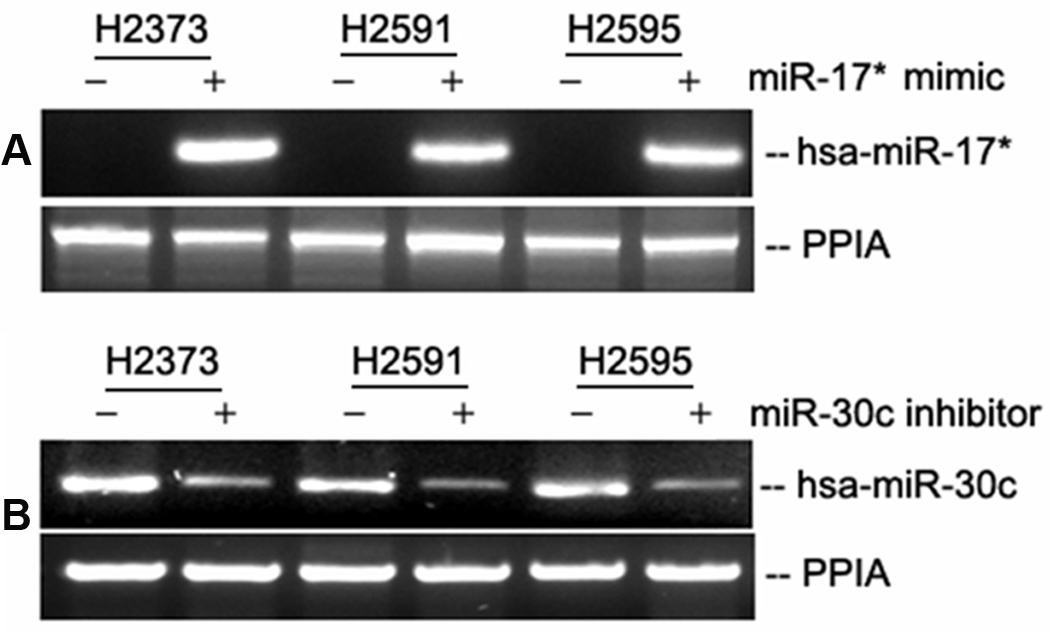
Verification of hsa-miR-17* mimic and hsa-miR-30c inhibitor expression effects following transfection of plasmids into untreated MPM cell lines. Native cells have low levels of hsa-miR-17* expression, while transfected cells showed a strong signal. All native cell lines express hsa-miR-30c, which is decreased following transfection of hsa-miR-30c inhibitor. Expression of the mimics and inhibitors parallels the effect of Onconase treatment on the MPM cell lines.
Figure 4.
Bar graph depiction of results from functional assays with forced expression of hsa-miR-17* mimic and hsa-miR-30c inhibitor. Forced expression of both hsa-miR-17* mimic and hsa-miR-30c inhibitor had a significant effect on functional activity in all cell lines in each of the four assays. (A) In Matrigel™ invasion, hsa-miR-17* mimic decreased invasion by 31 to 48% in all cell lines (p ≤ 0.01) compared to controls, while hsa-30c inhibitor decreased cell invasion by 81 to 96% in all cell lines (p ≤ 0.01) compared to controls. (B) Scratch closure assay demonstrated a similar trend, with 23 to 75% reduced closure with forced expression of hsa-miR-17* mimic (p ≤ 0.01), and 42 to 77% reduced closure with expression of hsa-miR-30c inhibitor (p ≤ 0.01). (C) Anchorage-independent growth was reduced 31 to 47% by hsa-miR-17* mimic (p ≤ 0.01), and 32 to 46% by hsa-miR-30c inhibitor (p ≤ 0.01). (D) Cell proliferation showed similar trends, with 25 to 33% reduction in proliferation with forced expression of hsa-miR-17* mimic (p ≤ 0.01), and a 35 to 80% reduction with forced expression of hsa-miR-30c inhibitor (p ≤ 0.01).
Figure 5.
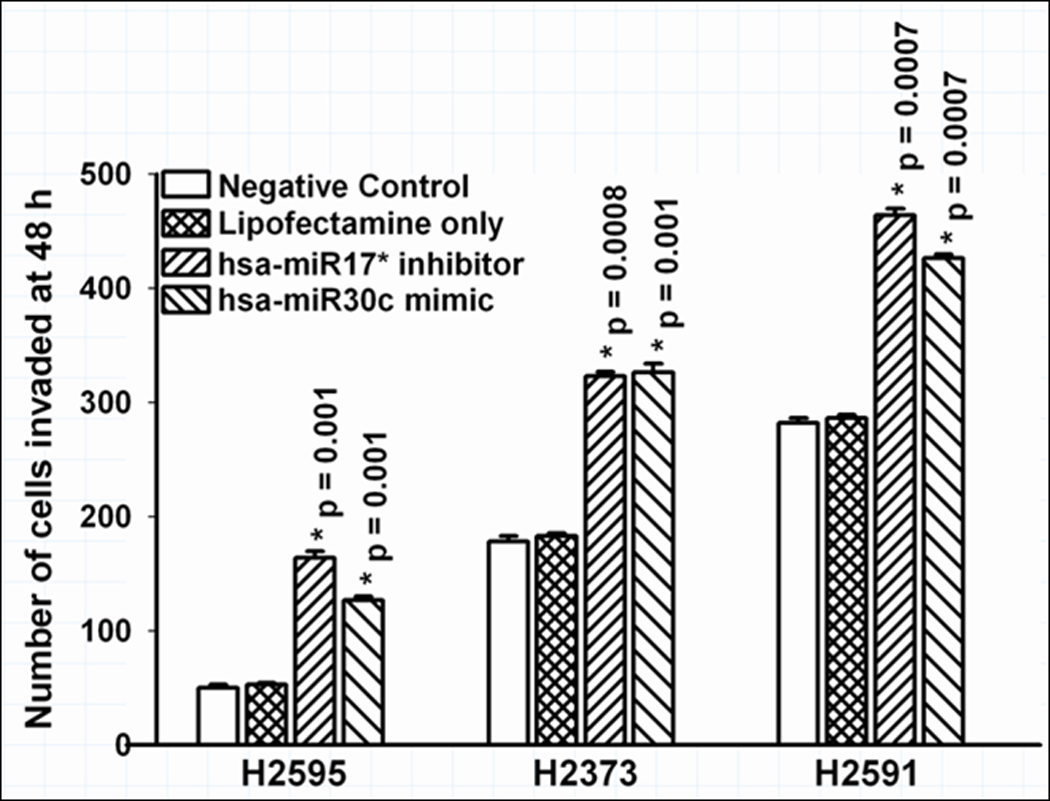
Effect of knockdown of miR-17* and forced expression of miR-30c on invasion of mesothelioma cell lines, i.e. the converse of the experiments seen in Figure 4. Invasion was promoted by interfering with the tumor suppressive effects of miR-17* and forced expression of mir-30c similarly cause increased invasion.
ABCB1, ABCC1 NFKβ, and downstream targets of NFKβ expression
RT-PCR demonstrated that the forced expression of hsa-miR-17* mimic and hsa-miR-30c inhibitor reduced the expression of ABCB1 and NFKB1 compared to controls(Figure 6A) and we validated this finding by Western blotting (Fig. 6B). Overall, a recapitulation of decreased expression of these two genes with Onconase incubation was seen with the forced expression of a hsa-miR-17* mimic and hsa-miR-30c inhibitor (Supplementary Figure 3).
Figure 6.
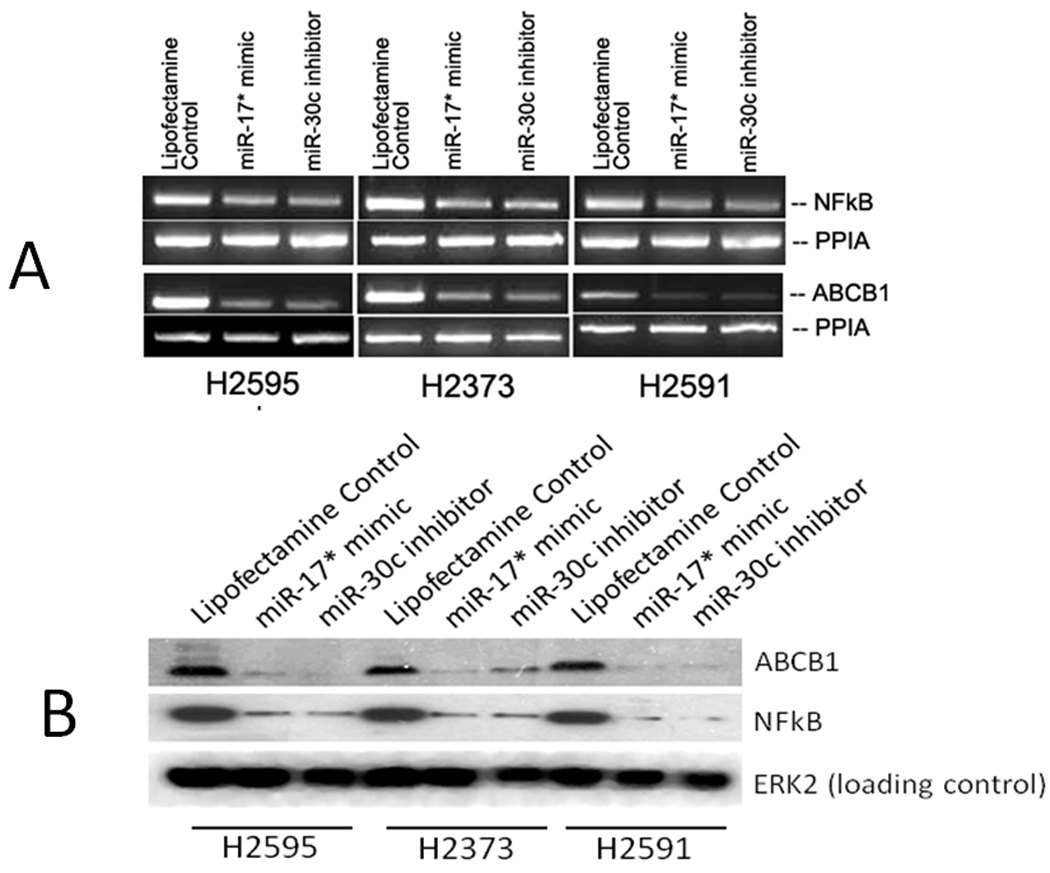
Forced expression of hsa-miR-17* mimic and hsa-miR-30c inhibitor on expression of NFKB(p50) and ABCB1. As seen with Onconase (Supplementary Figure 3), gene expression (A) decreased in all three cell lines. Decrease in protein expression was confirmed by Western Blot (B).
Reporter Assay microRNA Binding Assays
In RT-PCR and Western blot analysis, forced expression of miR-17* mimic and miR-30c inhibitor decreased the NFKβ and ABCB1 levels at both mRNA and protein. To further demonstrate whether these miRNAs truly target endogenous NFKβ (p50) and ABCB1, we co-transfected HEK 293T cells with miRNAs and reporter plasmids. Luciferase assays indicated that the introduction of hsa-miR-17* mimic significantly (p = 0.004) decreased the luciferase activity of NFKβ to 58% compared to negative control (Fig. 7). This reduced luciferase activity after hsa-miR-17* mimic transfection provides evidence that the miRNA under study is directly involved in the regulation of NFKβ gene. Introduction of hsa-miR-30c mimic showed no effect on NFKβ luciferase reporter. On the other hand, neither transfection of hsa-miR-17* mimic and hsa-miR-30c inhibitor had an effect on ABCB1 indicating that these miR’s have an indirect effect on ABCB1 gene regulation possibly through other targets (Figure 8).
Figure 7.
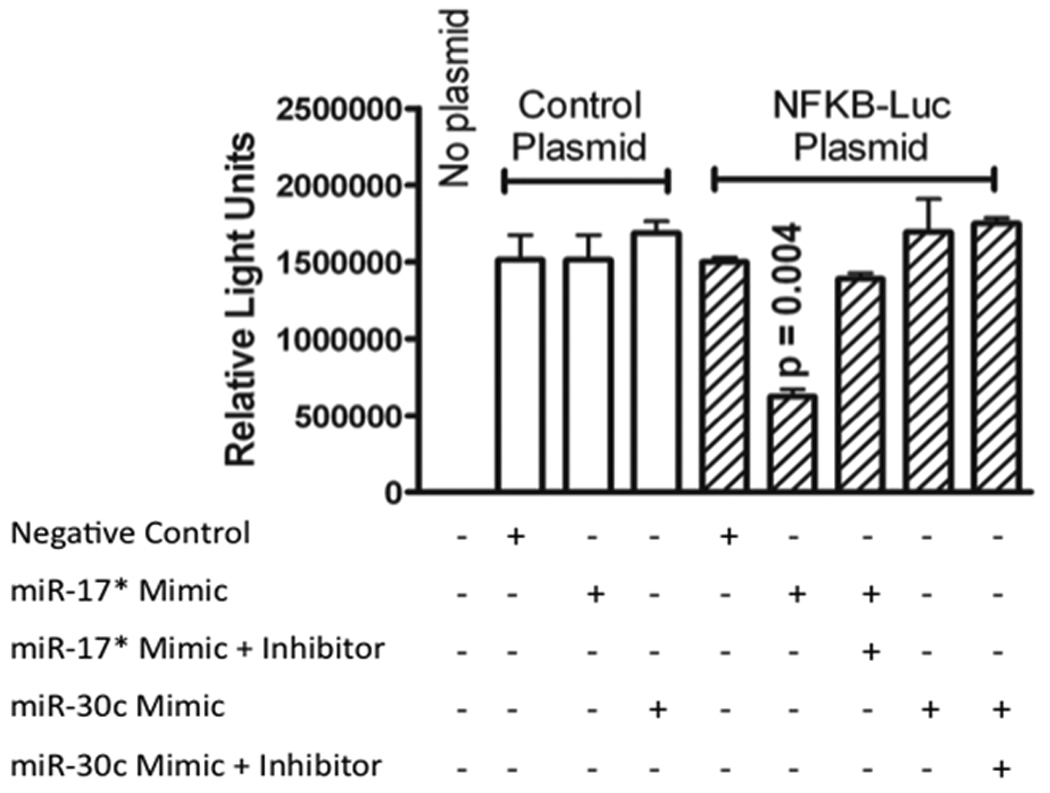
Reporter assay for effects of hsa-mir-17* and has-mir-30c on NFKB. Hsa-miR-17* mimic significantly (p = 0.004) decreased the luciferase activity of NFKβ compared to negative control (and the effect was completely reversed with the addition of the microRNA inhibitor. Introduction of hsa-miR-30c mimic showed no effect on NFKβ luciferase reporter.
Figure 8.
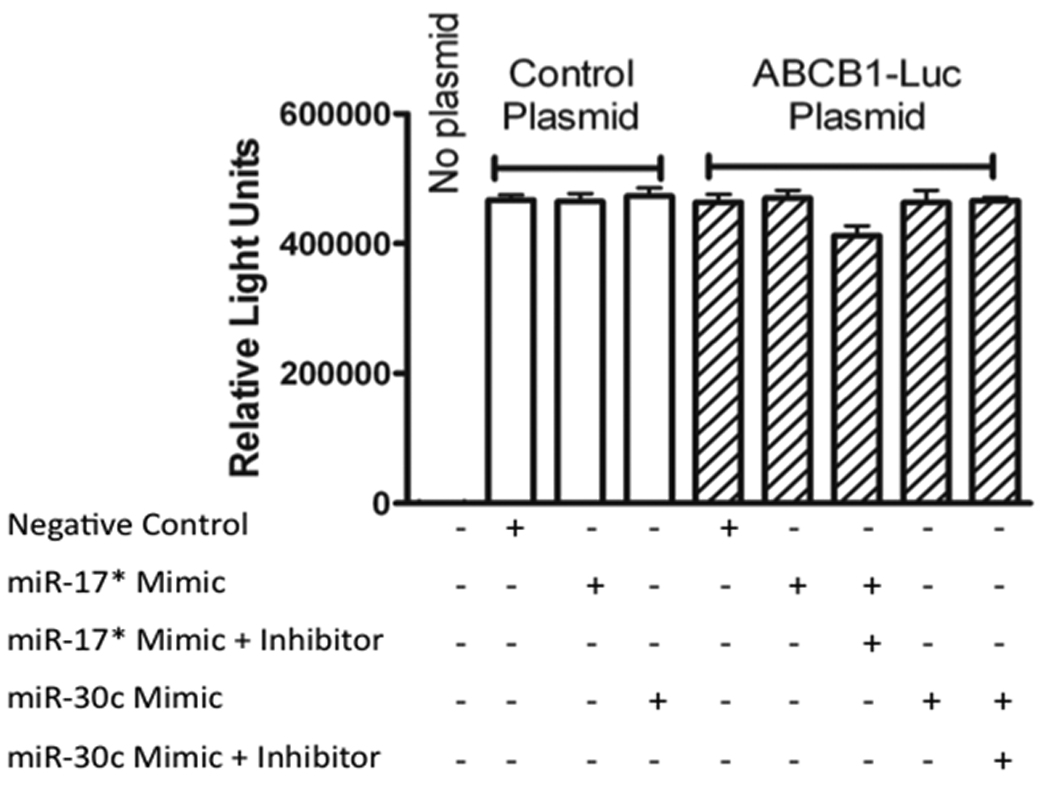
Reporter assay for effects of hsa-mir-17* and has-mir-30c on ABCB1. No evidence for direct control of the gene by either microRNA was seen.
RT-PCR evaluation of survival genes downstream of NFKβ including BCL-xL, cIAP1, as well as another known target influencing cellular potential for invasion (MMP9, gelatinase B) revealed decreased expression of these genes with hsa-miR- 17* mimic and hsa-miR-30c inhibitor transfection (Fig 9, Supplemental Figure 5). From these data we deduce that Onconase treatment affects these specific miRNAs, which are involved in downstream regulation of pathways responsible for the malignant behavior of these cell lines.
Figure 9.
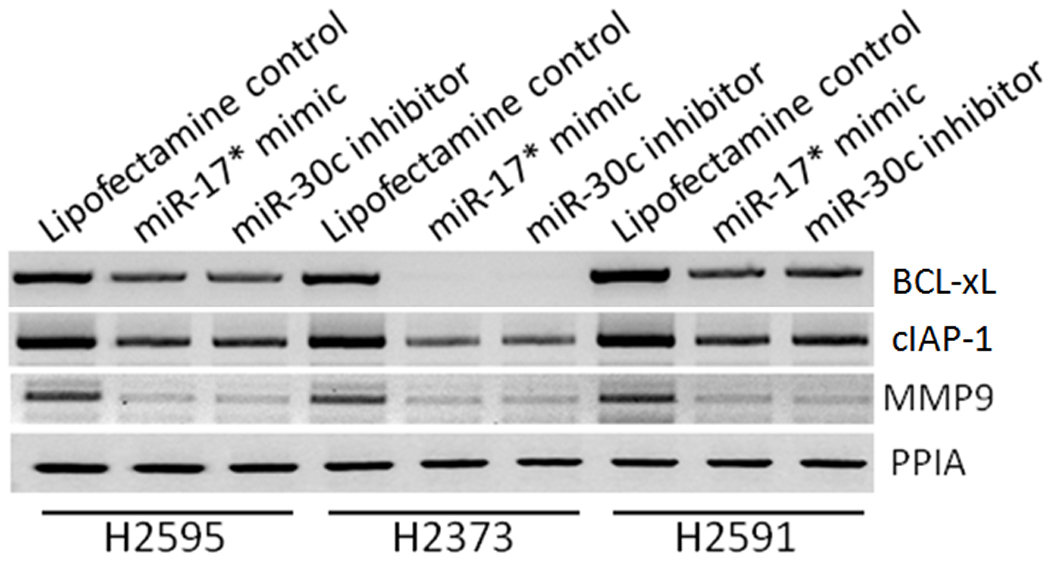
Effect of Onconase-associated microRNAs on downstream NFKβ targets and NFKβ-associated invasive genes. Both survival genes BCL-xL and xIAP1 had reduced expression with forced expression of the microRNAs as well as MMP9. All three of these genes have been associated with MPM.
Discussion
Onconase, a homolog of bovine pancreatic ribonuclease (RNase A), exerts its biological effect through selective cleavage of intracellular tRNA phosphodiester bonds and inhibition of protein synthesis (Singh et al. 2007; Saxena et al. 2002; Halicka et al. 2002). In addition, Onconase selectively alters expression of genes involved in cell cycle regulation and programmed cell death; with decreased expression of BCL-2 (anti-apoptotic), and upregulation of BAX (pro-apoptotic) expression in vitro(Ardelt et al. 2007). Onconase’s unique biochemical structure and unusually high conformational stability lends itself to enhanced intracellular survival and restricted inactivation by native cytoplasmic ribonuclease inhibitors (Dickson et al. 2005; Kobe and Deisenhofer 1996). These properties result in significant cytostatic and cytotoxic activity against various human tumors in both in vitro and in vivo models (Beck et al. 2008). In vitro studies have demonstrated Onconase’s efficacy as a single agent in lung cancer models with selective tumor cell death and preservation of surrounding normal cells (Lee 2008). These data also correlate with studies that demonstrate Onconase-specific inhibition of cell growth in human malignancies such as lung, colorectal, breast, and esophageal carcinoma (Beck et al. 2008; Lee et al. 2007; Kim et al. 2007). Onconase also exhibits synergistic properties with various chemotherapeutic agents in vitro (Beck et al. 2008) but a mechanism for its chemosensitizing abilities has never been elucidated.
miRNA’s are important functional RNA units, 21 to 23 nucleotides in length, which have a selective capacity to regulate gene expression, enzymatic activity, and regulatory cell functions by interfering with mRNA transcription and translation (Ambros 2004). miRNAs exist in short stem-loop structures called pre-miRNA, and are processed into mature functional miRNA species within the cell cytoplasm (Ambros et al. 2003). Mature miRNA molecules are complementary to one or more mRNA molecules, and can interfere with translation by restricting ribosomal progression (Ambros 2004; Wang et al. 2004). Selective miRNA expression is thought to be important to cancer cell transformation and survival by down-regulating of specific gene products that are important to cell cycle regulation and programmed cell death (Bartel 2004). Expression profiles associated with a variety of human malignancies have characterized over 200–300 different regulatory miRNAs, however less than 10% of those characterized have been specifically correlated with a particular pathway or mechanism of action (Ambros 2004).
In our study, Onconase caused dose-specific inhibition of proliferative and invasive properties of mesothelioma cells. Hsa-miR-17* and hsa-miR-30c, the two miRNAs most significantly affected by Onconase treatment, have not previously been reported as important regulators in MPM (Kasashima et al. 2004; Lagos-Quintana et al. 2003). These miRNAs were initially isolated from murine tumor cells, and then have been identified in a variety of human malignancies. However, to date no functional data has been reported to suggest specific behaviors or mechanisms by which these miRNAs regulate gene expression. Our demonstration of Onconase-mediated changes in miRNA expression profiles with significant up regulation of hsa-miR-17* and down regulation of hsa-miR-30c paired with decreased malignant activity in MPM cell lines forced to over-express hsa-miR-17* mimic and hsa-miR-30c inhibitor, is the first to implicate these miRNAs in MPM. Moreover, the downstream miRNA’s targets that regulate gene expression and alter cellular behavior following Onconase treatment have not been previously described.
NFKβ plays a key role in asbestos induced malignant transformation. Asbestos induced malignant transformation of mesothelial cells is due to induction of a profound inflammatory reaction, phagocytosis of fibers with subsequent macrophage differentiation, and the release of numerous cytokines and reactive oxygen species that are mutagenic(Robledo and Mossman 1999). In vitro models evaluating the impact of asbestos on cell cycle regulation and survival have discovered a significant relationship between activation of NFKβ pathways and cellular protection from asbestos induced cell death (Yang et al. 2008; Yang et al. 2006). This effect has been verified by selective inhibition of NFKβ, which subjects the exposed cells to programmed cell death (Yang et al. 2006). NFKβ is a regulatory transcription protein complex that mediates a variety of cellular and inflammatory processes involved in cellular stress, apoptosis, and malignant transformation (Gilmore 1999). Down regulation of NFKβ reduces tumorigenic properties of malignant epithelial cells, and has been demonstrated in both in vivo and in vivo models to be an important component in molecular pathways that involve tumor cell invasion (Gilmore 2006; Gilmore 2003; Micheau and Tschopp 2003; Barnhart and Peter 2003). To date, specific miRNAs that mediate NFKβ expression and reduce the invasive potential of malignant cells have not been described. We have demonstrated a direct relationship between Onconase mediated hsa-miR-17* up regulation and the expression of NFKB1 that encodes an NFKβ subunit p50 in three MPM cell lines. We have also demonstrated that forced overexpression of these miRNA’s result in decreased malignant behavior observed in cell lines evaluated by in vitro functional assays due to the selective downregulation of cell survival genes and those associated with increased invasion. Therefore, we believe Onconase has a specific effect on NFKβ, and that hsa-miR-17* and hsa-miR-30c are specifically relevant intermediates in this pathway. Proof for this hypothesis is demonstrated with our data in mesothelioma cell lines, which investigates important cell survival genes downstream from NFKβ. Soini (Soini et al. 1999) has reported that all mesothelioma cell lines are positive for bcl-X, and other investigators have demonstrated that introduction of antisense oligonucleotides specific to Bcl-xL mRNA, can sensitize MPM cells to chemotherapeutic agents (Cao et al. 2007). Additionally, the growth of established mouse flank human tumor xenografts is reduced with intra-tumor administration of antisense oligoncleotides to BCL-x (Littlejohn et al. 2008). Another NFKβ target, IAP1, is over-expressed in the disease (Gordon et al. 2007; Symanowski et al. 2009) and is responsible for a large degree of the resistance of cultured MPM cells to cisplatin. As detailed above, miR-17c* and inhibitor of miR-30 suppressed expression of these genes in our MPM cell lines, possibly explaining enhanced sensitivity of chemotherapy drugs when used in combination with Onconase.
Another challenge in the development of novel chemotherapeutic treatments is the phenomenon of multidrug resistance by tumor cells, a process which most commonly occurs by up regulation of ATP-binding cassette (ABC) transporters. ABC transporters facilitate translocation of a variety of substrates across the cell membrane, including metabolic products and exogenous substrates such as chemotherapy drugs, providing a protective mechanism for malignant cells (Davidson et al. 2008). Structural domains specific to ABC transmembrane proteins which facilitate movement of hydrophobic solutes common to many chemotherapeutic agents have been a focus of study (Jones and George 2004; Jones and George 2004). Inhibition of ABC transporters has been proven to be an efficacious technique for improvements in cancer therapy, and currently tyrosine kinase inhibitors are being investigated due to their preferential inhibition of these multidrug resistant transmembrane proteins (Shukla et al. 2009; Shi et al. 2007; Shi et al. 2009). ABCB1 and ABCC1, two structurally similar ABC transporters, have been previously implicated in chemotherapy resistance in human malignancies (Gillet et al. 2007; O'Donnell et al. 2005). Our work demonstrates high native expression of multidrug resistance genes in MPM, and agrees with previous data regarding ABC transporters and MPM (Soini et al. 2001; Khokhar et al. 2001). Onconase treatment significantly decreased expression of ABCB1 in cell lines which natively expressed those ABC transporters, and the effect of Onconase associated microRNAs is not a direct effect on ABCB1. It is curious that the only subgroup exhibiting benefit of Onconase in the recent randomized trial were those Onconase-treated patients who had received previous cytotoxic chemotherapy. These results require validation in a larger clinical study but the mechanism for these findings could be related to microRNA-induced effects on NFKβ.
Aggressive protocols of combination chemotherapy with or without targeted therapies are likely the direction of future cancer therapy. Our data suggest that Onconase may significantly enhance the biologic activity of additional chemotherapeutic treatments through diverse mechanisms, making it an ideal agent for multi-drug therapies. The full therapeutic potential of Onconase may also be realized by exploiting its distinctive structural and functional features to produce more powerful variants or novel antitumor drugs.
Materials and Methods
Cell lines
Mesothelioma cell lines H2373, H2591, and H2595 were maintained in 1x Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA). Mesothelioma cell lines in this study were established in our laboratory from the surgical specimens of Dr. Harvey I. Pass as described (Pass et al. 1995).
Onconase treatment
Cells were plated at a density of 2 × 106 cells / plate in 10-cm plates in duplicates. Cells in culture were treated with 0, 1.25, 2.5, 5, 10 or 20 µg/mL Onconase (Alpha Cell Corporation, Paramus, NJ) for 48 and 72h.
RNA isolation and miRNA expression profiles
miRNA-enriched RNA was isolated from treated and untreated cultured cells using mirVANA miRNA isolation kit (Ambion, Foster City, CA). miRNA expression profiles generated on OSUCCC microarrays were quantiles normalized, analyzed as previously reported (19) at Ohio State University and sorted for significance.
RT-PCR
From the microRNA expression miR’s with the greatest fold change and highest significance, hsa-miR-17* and hsa-miR-30c were selected for further study. RT-PCR validation of onconase effect on the expression of hsa-miR-17* and hsa-miR-30c was performed using stem-loop PCR technology(Chen et al. 2005; Varkonyi-Gasic et al. 2007) (56, 57). miRNA sequences are obtained from miRbase (http://www.mirbase.org). The stem-loop technique involves a reverse transcription step to synthesize artificial cDNA of the miR followed by PCR amplification. Reverse transcription was accomplished with initial incubation at 16°C for 30 minutes followed by gradual annealing and extension for 60 cycles at 30°C for 30 sec, 42°C for 30 sec and 50°C for 30 sec, and a final extension at 85°C for 5 minutes using SuperScript™ II Reverse Transcriptase Kit (Invitrogen, Carlsbad, CA) using miR specific stem-loop RT primers. PCR amplification was carried out in a total volume of 20 µL using the Clontech Advantage™ 2 PCR Kit (Clontech, Mountain View, CA) and optimized concentrations of MgCl2. The reactions were performed with denaturing for 15 seconds at 94°C, annealing for 30 seconds at 57°C, and elongation for 30 seconds at 70°C for a total of 35 cycles; the final extension at 72°C for 5 minutes. RT-PCR products were analyzed on 4% MetaPhor™ agarose (Lonza, Rockland ME) gels.
For hsa-miR-17*, 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACCTACAAGTGCC-3’ stem-loop RT primer was used for reverse transcription, and 5’-ACTGCAGTGAAGGCAC-3’(sense) and 5’-GTGCAGGGTCCGAGGT-3’ (antisense) primers were used for PCR amplification. For hsa-miR-30c, 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACGCTGAGAGTGT-3’ stem-loop RT primer was used for reverse transcription, and 5’-GCGTGTAAACATCCTACAC-3’ (sense) and 5’-GTGCAGGGTCCGAGGT-3’ (antisense) primers were used for PCR amplification. The amplification of peptidylprolyl isomerase A (PPIA) with primers 5’- TCTGAGCACTGGAGAGAAAGG-3’ (sense) and 5’-GGAAAACATGGAACCC-3’ (antisense) served as controls for equal loading and integrity of all PCR experiments.
Quantification of RT-PCR products
The RT-PCR products were analyzed on 4% MetaPhor ™agaose (Lonza, Rockland, ME) gels and the images were captured using Kodak Imaging System 4000MM (Molecular Imaging, Woodbridge, CT). Relative signal intensities were quantified using Kodak Molecular Imaging Software V4.04. The relative expression levels were normalized to the loading control, PPIA.
miR Oligonucleotides and Transfections
Oligonucleotides miRIDIAN hsa-miR-17* mimic (C-300486-05) and hsa-miR-30c inhibitor (IH-300542-07) were purchased from Dharmacon RNAi Technologies (Lafayette, CO). Standard Lipofectamine2000 (Invitrogen, Carlsbad, CA) protocol was followed as recommended to transfect miRNA. Briefly, cells plated at 80–90% confluency were incubated in antibiotic free DMEM media for 1 h and then 40 nM of miRIDIAN miR-17* mimic or miR-30c inhibitor was complexed with lipofectimine and introduced into H2373, H2591 and H2595 cells. Transfected cells were incubated for 48 h. For negative control, cells were transfected with scrambled oligo (CN-001000-01-05, Dharmacon RNAi Technologies, Lafayette, CO). Pooled populations of transfected cells were assayed for proliferation, scratch assay, matrigel cell invasion and soft agar colony formation. Hsa-miR-17* mimic and hsa-miR-30 inhibitor expression in transfected cells was verified using stem-loop RT-PCR technology as described above.
Cell proliferation assay
Three thousand cells from each experimental cell line were plated in flat bottom 96-well plates in 100 mL DMEM and 10% FBS in six replicates and incubated at 37°C for 48 hours. 20 µL of Cell Titer-Blue reagent (Promega, Madison, Wisconsin) was added to each well and incubated for three hours at 37°C and 5% CO2. Proliferation was assessed by optical density (OD) values measured at 560/590λ using a universal plate reader (Universal Reader Victor; PerkinElmer Life and Analytical Sciences, Waltham, Massachusetts).
Scratch assay (wound closure assay)
A day prior to the experiment, 2 × 105 cells/well was seeded in 6-well plates. Using a 1 mL pipette tip, three vertical linear scratches were made per well. Cells were washed with 1× PBS and medium was changed. Plates were marked with a fine pen. Scratch widths (6 measurements per well) were measured using a light microscope at zero and 24 hours. The percent of the scratch closure was calculated.
Matrigel™ cell invasion assay
Cell invasion assay was performed using the BD Biocoat Matrigel™ Invasion Chambers (BD Biosciences, Franklin Lakes, NJ) according the manufacturer’s protocol. One million cells were seeded onto the inserts (8µM pore sized polycarbonate membrane) coated with a thin layer of Matrigel™ Basement Membrane Matrix (BD Biosciences, Franklin Lakes, NJ) diluted at 1:100 in PBS. The plates were incubated for 48 hours at 37°C and 5% CO2. Cells that invaded the Matrigel™ Matrix to the lower surface of the membrane were fixed and stained with Giemsa solution, and counted under a light microscope. Eight fields in 4 separate quadrants of each membrane were counted and averaged. The assay was performed in triplicates.
Soft agar colony formation assay
Base agar were prepared 30 minutes prior to cell plating by mixing equal volumes of 1% agarose at 40°C and pre-warmed 2× DMEM with 10% FBS, poured onto a 6-well plate (1.5 mL per well), and allowed to polymerize. The top agar was made by mixing equal volumes of warm 0.7% agarose and 2× DMEM with 10% FBS (total volume 2 mL per well), combined with five thousand cells from each experimental cell line. Top agar was poured onto the base agar and plates were incubated at 37°C and 5% CO2 for 21 days. Colonies were stained with 0.5 mL of 0.005% Crystal Violet for 1 hour and counted using a dissecting microscope.
NFKB1(p50), ABCB1, and BCL2L1 expression assessment
NFKB1, ATP-binding cassette transporters B1 (ABCB1), and Pro-survival Bcl-2 homologue (BCL2L1) were assessed in all native cell lines, Onconase treated cell lines, and cell lines with forced expression of the hsa-miR-17* mimic and hsa-miR-30c inhibitor utilizing primers designed from Dharmacon RNAi Technologies (Lafayette, CO). For NFKB1, the resulting primers, 5’-CCATATTTGGGAAGGCCTGAA-3’ (sense) and 5’-TGGTCCCACATAGTTGCAGA-3’ (antisense), amplify a 264 bp product. For ABCB1 RT-PCR, the resulting primers, 5’-CCATATTTGGGAAGGCCTGAA-3’ (sense) and 5’-TGGTCCCACATAGTTGCAGA-3’ (antisense), amplify a 157 bp product. For BCL2L1, 5’-GGAGCCACTGGCCACAGCAG-3’ (sense) and 5’-ATCCCAGCCGCCGTTCTCCT-3’ (antisense), amplify a 368 bp product. For cIAP-1, 5’-GCCGGGGTGCCTGTCTCAGAA-3’ (sense) and 5’-ACCACAGGCAAAGCAGGCTACC-3’ (antisense), amplify a 500 bp product. For MMP9, 5’- AAGCTGGACTCGGTCTTTGA -3’(sense) and 5’- TCAACTCACTCCGGGAACTC-3’ (antisense), amplify a 370 bp product. The reactions were performed as previously described. RT-PCR products were subjected to electrophoresis on 2% Tris-acetate EDTA agarose gel.
Western Blot Analysis
Total cell proteins were prepared by lysing the cells in 10 mM/L Tris (pH 7.5), 1 mM/L EDTA, 150 mM /L NaCl, 1% Triton X-100, 1 mM/L DTT, 10% glycerol for half-hour on ice. The lysates were centrifuged and cleared at 13K and total protein was estimated by BCA™ Protein Assay Kit (Thermoscientific, Rockford, IL). 30ug of samples were separated on 8–16% Tris-Glycine gel (Invitrogen,Carlsbad, CA) and transferred to PVDF membrane (Millipore,Bedford, MA). Membranes were blocked in 5% non-fat dry milk for 1 hour at room temperature followed by incubation in primary antibody for overnight at 4°C. The membrane was then incubated in secondary antibody conjugated with horseradish peroxidase for one hour at room temperature and were developed using the SuperSignal West Pico Chemiluminescent Substrate (Pierce,Rockford, IL). To ensure equal loading and transfer of proteins, membranes were stripped and reprobed with ERK2 antibody. Mouse monoclonal Mdr-1(ABCB1; sc-5551C), rabbit polyclonal NFkB (sc-372) and ERK2 (sc-154) were purchased from Santa Cruz Biotechnologies,Inc, Santa Cruz, CA.
Luciferase Assays for MicroRNA target sequences
Luciferase assays were carried out in HEK 293T cells to determine the effect of miRNAs, hsa-miR-17* and hsa-miR-30c mimic on the activity of NFKβ and ABCB1. The luciferase reporters, pEZX-Luc- NFKβ-3’-UTR, pEZX-Luc- ABCB1-3’-UTR and Luc-control plasmids were purchased from Genocoepia, USA. HEK 293T cells were transfected using lipofectamine 2000 (Invitrogen, USA) following manufacturer’s protocol. In brief, cells were seeded at 70% confluence in a 24-well plate and then transfected with 0.8 µg of DNA constructs along with 40 nM of miRNA Oligo’s. The cells were transferred to a 96-well plate 18 h after transfection and cultured for another 24 h. Luciferase activity was measured by luciferase assay kit (Genocoepia, USA) with Lumistar Galaxy (BMG Lab Technologies Incorporation, USA) Renilla luciferase was used for normalization. All the experiments were done in triplicates.
Statistical analysis
Bivariate correlations-Pearson coefficient and students two-sided T-tests were used to determine significance, which was reported as significant for p<0.05. All assays were performed in triplicates and statistical evaluations were performed using the SPSS software package (v.12). All values in the text and figures represent the means ± standard deviation.
Supplementary Material
Supplemental Figure 1: Cell lines subjected to Onconase treatment at 20 µg/mL (A) Control proliferation has been normalized to a value of 1 for each time point. Significantly decreased proliferation compared to control is seen at 48 and 72 hours. (B) Significantly decreased invasion of MPM cell lines at both time points.
Supplemental Figure 2: Dose effects of Onconase on the proliferation of three mesothelioma cell lines. Although effects were seen with as little as 5 ug/ml, maximum effects were seen with 20ug/ml.
Supplemental Figure 3: Cell lines subjected to Onconase treatment at 20 µg/mL for 72 hours resulted in decreased expression of multidrug resistance gene ABCB. NFKB(p50) expression was also reduced at both time points.
Supplemental Figure 4: Western Blot demonstrates that cell lines subjected to Onconase treatment at 20 µg/mL for 72 hours have decreased protein for multidrug resistance genes ABCB1 and NFKB. ERK2, unaffected by Onconase, was used as a loading control.
Supplemental Figure 5: Quantification of effects of Onconase-associated microRNAs on genes downstream of NFKB. The gene expression was uniformly decreased by hsa-mir-17* and has-mir-30c inhibitor and the degree of decrease was cell line dependent.
Acknowledgments
Ranpirnase was provided by AlphaCell Corporation, Paramus, NJ. We also wish to acknowledge the generous support of Belluck and Fox, LPA for this project.
Supported by NCI/NIH EDRN Biomarker Discovery Laboratory Grant UO1CA1111295 to Dr. Pass and1PO1 CA114047-D1A1 to Dr. Carbone.
Footnotes
Conflict of Interest: None of the authors have competing financial interests in relation to this work
Reference List
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Ardelt B, Ardelt W, Darzynkiewicz Z. Cytotoxic ribonucleases and RNA interference (RNAi) Cell Cycle. 2003;2:22–24. doi: 10.4161/cc.2.1.232. [DOI] [PubMed] [Google Scholar]
- Ardelt B, Juan G, Burfeind P, Salomon T, Wu JM, Hsieh TC, Li X, Sperry R, Pozarowski P, Shogen K, Ardelt W, Darzynkiewicz Z. Onconase, an anti-tumor ribonuclease suppresses intracellular oxidative stress. Int J Oncol. 2007;31:663–669. doi: 10.3892/ijo.31.3.663. [DOI] [PubMed] [Google Scholar]
- Barnhart BC, Peter ME. The TNF receptor 1: a split personality complex. Cell. 2003;114:148–150. doi: 10.1016/s0092-8674(03)00561-0. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Beck AK, Pass HI, Carbone M, Yang H. Ranpirnase as a potential antitumor ribonuclease treatment for mesothelioma and other malignancies. Future Oncol. 2008;4:341–349. doi: 10.2217/14796694.4.3.341. [DOI] [PubMed] [Google Scholar]
- Cao X, Rodarte C, Zhang L, Morgan CD, Littlejohn J, Smythe WR. Bcl2/bcl-x(L) Inhibitor Engenders Apoptosis and Increases Chemosensitivity in Mesothelioma. Cancer Biol Ther. 2007;6 doi: 10.4161/cbt.6.2.3626. [DOI] [PubMed] [Google Scholar]
- Carbone M, Albelda SM, Broaddus VC, Flores RM, Hillerdal G, Jaurand MC, Kjaerheim K, Pass HI, Robinson B, Tsao A. Eighth international mesothelioma interest group. Oncogene. 2007;26:6959–6967. doi: 10.1038/sj.onc.1210515. [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–364. doi: 10.1128/MMBR.00031-07. table. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deptala A, Halicka HD, Ardelt B, Ardelt W, Mikulski SM, Shogen K, Darzynkiewicz Z. Potentiation of tumor necrosis factor induced apoptosis by onconase. Int J Oncol. 1998;13:11–16. doi: 10.3892/ijo.13.1.11. [DOI] [PubMed] [Google Scholar]
- Dickson KA, Haigis MC, Raines RT. Ribonuclease inhibitor: structure and function. Prog Nucleic Acid Res Mol Biol. 2005;80:349–374. doi: 10.1016/S0079-6603(05)80009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet JP, Efferth T, Remacle J. Chemotherapy-induced resistance by ATP-binding cassette transporter genes. Biochim Biophys Acta. 2007;1775:237–262. doi: 10.1016/j.bbcan.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Gilmore TD. The Rel/NF-kappaB signal transduction pathway: introduction. Oncogene. 1999;18:6842–6844. doi: 10.1038/sj.onc.1203237. [DOI] [PubMed] [Google Scholar]
- Gilmore TD. The Re1/NF-kappa B/I kappa B signal transduction pathway and cancer. Cancer Treat Res. 2003;115:241–265. [PubMed] [Google Scholar]
- Gilmore TD. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- Gordon GJ, Mani M, Mukhopadhyay L, Dong L, Yeap BY, Sugarbaker DJ, Bueno R. Inhibitor of apoptosis proteins are regulated by tumour necrosis factor-alpha in malignant pleural mesothelioma. J Pathol. 2007;211:439–446. doi: 10.1002/path.2120. [DOI] [PubMed] [Google Scholar]
- Halicka DH, Pozarowski P, Ita M, Ardelt WJ, Mikulski SM, Shogen K, Darzynkiewicz Z. Enhancement of activation-induced apoptosis of lymphocytes by the cytotoxic ribonuclease onconase (Ranpirnase) Int J Oncol. 2002;21:1245–1250. doi: 10.3892/ijo.21.6.1245. [DOI] [PubMed] [Google Scholar]
- Iordanov MS, Ryabinina OP, Wong J, Dinh TH, Newton DL, Rybak SM, Magun BE. Molecular determinants of apoptosis induced by the cytotoxic ribonuclease onconase: evidence for cytotoxic mechanisms different from inhibition of protein synthesis. Cancer Res. 2000;60:1983–1994. [PubMed] [Google Scholar]
- Jones PM, George AM. The ABC transporter structure and mechanism: perspectives on recent research. Cell Mol Life Sci. 2004;61:682–699. doi: 10.1007/s00018-003-3336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasashima K, Nakamura Y, Kozu T. Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem Biophys Res Commun. 2004;322:403–410. doi: 10.1016/j.bbrc.2004.07.130. [DOI] [PubMed] [Google Scholar]
- Kaufman AJ, Pass HI. Current concepts in malignant pleural mesothelioma. Expert Rev Anticancer Ther. 2008;8:293–303. doi: 10.1586/14737140.8.2.293. [DOI] [PubMed] [Google Scholar]
- Khokhar NZ, She Y, Rusch VW, Sirotnak FM. Experimental therapeutics with a new 10-deazaaminopterin in human mesothelioma: further improving efficacy through structural design, pharmacologic modulation at the level of MRP ATPases, and combined therapy with platinums. Clin Cancer Res. 2001;7:3199–3205. [PubMed] [Google Scholar]
- Kim DH, Kim EJ, Kalota A, Gewirtz AM, Glickson J, Shogen K, Lee I. Possible mechanisms of improved radiation response by cytotoxic RNase, Onconase, on A549 human lung cancer xenografts of nude mice. Adv Exp Med Biol. 2007;599:53–59. doi: 10.1007/978-0-387-71764-7_8. [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease. A J Mol Biol. 1996;264:1028–1043. doi: 10.1006/jmbi.1996.0694. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I. Ranpirnase (Onconase), a cytotoxic amphibian ribonuclease, manipulates tumour physiological parameters as a selective killer and a potential enhancer for chemotherapy and radiation in cancer therapy. Expert Opin Biol Ther. 2008;8:813–827. doi: 10.1517/14712598.8.6.813. [DOI] [PubMed] [Google Scholar]
- Lee I, Kim DH, Sunar U, Magnitsky S, Shogen K. The therapeutic mechanisms of ranpirnase-induced enhancement of radiation response on A549 human lung cancer. In Vivo. 2007;21:721–728. [PubMed] [Google Scholar]
- Lee JE, Raines RT. Ribonucleases as novel chemotherapeutics : the ranpirnase example. BioDrugs. 2008;22:53–58. doi: 10.2165/00063030-200822010-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlejohn JE, Cao X, Miller SD, Ozvaran MK, Jupiter D, Zhang L, Rodarte C, Smythe WR. Bcl-xL antisense oligonucleotide and cisplatin combination therapy extends survival in SCID mice with established mesothelioma xenografts. Int J Cancer. 2008;123:202–208. doi: 10.1002/ijc.23452. [DOI] [PubMed] [Google Scholar]
- Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- Mikulski SM, Viera A, Darzynkiewicz Z, Shogen K. Synergism between a novel amphibian oocyte ribonuclease and lovastatin in inducing cytostatic and cytotoxic effects in human lung and pancreatic carcinoma cell lines. Br J Cancer. 1992;66:304–310. doi: 10.1038/bjc.1992.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Pass HI, Liu Z, Wali A, Bueno R, Land S, Lott D, Siddiq F, Lonardo F, Carbone M, Draghici S. Gene expression profiles predict survival and progression of pleural mesothelioma. Clin Cancer Res. 2004;10:849–859. doi: 10.1158/1078-0432.ccr-0607-3. [DOI] [PubMed] [Google Scholar]
- Pass HI, Stevens EJ, Oie H, Tsokos MG, Abati AD, Fetsch PA, Mew DJ, Pogrebniak HW, Matthews WJ. Characteristics of nine newly derived mesothelioma cell lines. Ann Thorac Surg. 1995;59:835–844. doi: 10.1016/0003-4975(95)00045-m. [DOI] [PubMed] [Google Scholar]
- Pavlakis N, Vogelzang NJ. Ranpirnase--an antitumour ribonuclease: its potential role in malignant mesothelioma. Expert Opin Biol Ther. 2006;6:391–399. doi: 10.1517/14712598.6.4.391. [DOI] [PubMed] [Google Scholar]
- Porta C, Paglino C, Mutti L. Ranpirnase and its potential for the treatment of unresectable malignant mesothelioma. Biologics. 2008;2:601–609. doi: 10.2147/btt.s2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck M, Krzakowski M, Jastrzebski D, et al. Randomized, multicenter phase III study of ranpirnase plus doxorubicin (DOX) versus DOX in patients with unresectable malignant mesothelioma (MM) J Clin Oncol. 2009;27(15s) Ref Type: Abstract. [Google Scholar]
- Robinson BW, Lake RA. Advances in malignant mesothelioma. N Engl J Med. 2005;353:1591–1603. doi: 10.1056/NEJMra050152. [DOI] [PubMed] [Google Scholar]
- Robledo R, Mossman B. Cellular and molecular mechanisms of asbestos-induced fibrosis. J Cell Physiol. 1999;180:158–166. doi: 10.1002/(SICI)1097-4652(199908)180:2<158::AID-JCP3>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Rybak SM, Pearson JW, Fogler WE, Volker K, Spence SE, Newton DL, Mikulski SM, Ardelt W, Riggs CW, Kung HF, Longo DL. Enhancement of vincristine cytotoxicity in drug-resistant cells by simultaneous treatment with onconase, an antitumor ribonuclease. J Natl Cancer Inst. 1996;88:747–753. doi: 10.1093/jnci/88.11.747. [DOI] [PubMed] [Google Scholar]
- Saxena SK, Sirdeshmukh R, Ardelt W, Mikulski SM, Shogen K, Youle RJ. Entry into cells and selective degradation of tRNAs by a cytotoxic member of the RNase A family. J Biol Chem. 2002;277:15142–15146. doi: 10.1074/jbc.M108115200. [DOI] [PubMed] [Google Scholar]
- Shi Z, Parmar S, Peng XX, Shen T, Robey RW, Bates SE, Fu LW, Shao Y, Chen YM, Zang F, Chen ZS. The epidermal growth factor tyrosine kinase inhibitor AG1478 and erlotinib reverse ABCG2-mediated drug resistance. Oncol Rep. 2009;21:483–489. [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Peng XX, Kim IW, Shukla S, Si QS, Robey RW, Bates SE, Shen T, Ashby CR, Jr, Fu LW, Ambudkar SV, Chen ZS. Erlotinib (Tarceva, OSI-774) antagonizes ATP-binding cassette subfamily B member 1 and ATP-binding cassette subfamily G member 2-mediated drug resistance. Cancer Res. 2007;67:11012–11020. doi: 10.1158/0008-5472.CAN-07-2686. [DOI] [PubMed] [Google Scholar]
- Shukla S, Robey RW, Bates SE, Ambudkar SV. Sunitinib (Sutent, SU11248), a small-molecule receptor tyrosine kinase inhibitor, blocks function of the ATP-binding cassette (ABC) transporters P-glycoprotein (ABCB1) and ABCG2. Drug Metab Dispos. 2009;37:359–365. doi: 10.1124/dmd.108.024612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh UP, Ardelt W, Saxena SK, Holloway DE, Vidunas E, Lee HS, Saxena A, Shogen K, Acharya KR. Enzymatic and structural characterisation of amphinase, a novel cytotoxic ribonuclease from Rana pipiens oocytes. J Mol Biol. 2007;371:93–111. doi: 10.1016/j.jmb.2007.04.071. [DOI] [PubMed] [Google Scholar]
- Soini Y, Jarvinen K, Kaarteenaho-Wiik R, Kinnula V. The expression of P-glycoprotein and multidrug resistance proteins 1 and 2 (MRP1 and MRP2) in human malignant mesothelioma. Ann Oncol. 2001;12:1239–1245. doi: 10.1023/a:1012292230480. [DOI] [PubMed] [Google Scholar]
- Soini Y, Kinnula V, Kaarteenaho-Wiik R, Kurttila E, Linnainmaa K, Paakko P. Apoptosis and expression of apoptosis regulating proteins bcl-2, mcl-1, bcl-X, and bax in malignant mesothelioma. Clin Cancer Res. 1999;5:3508–3515. [PubMed] [Google Scholar]
- Symanowski J, Vogelzang N, Zawel L, Atadja P, Pass H, Sharma S. A histone deacetylase inhibitor LBH589 downregulates XIAP in mesothelioma cell lines which is likely responsible for increased apoptosis with TRAIL. J Thorac Oncol. 2009;4:149–160. doi: 10.1097/JTO.0b013e318194f991. [DOI] [PubMed] [Google Scholar]
- Tsai SY, Ardelt B, Hsieh TC, Darzynkiewicz Z, Shogen K, Wu JM. Treatment of Jurkat acute T-lymphocytic leukemia cells by onconase (Ranpirnase) is accompanied by an altered nucleocytoplasmic distribution and reduced expression of transcription factor NF-kappaB. Int J Oncol. 2004;25:1745–1752. [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Reyes JL, Chua NH, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Mikulski SM, Ardelt W, Rybak SM, Youle RJ. A cytotoxic ribonuclease. Study of the mechanism of onconase cytotoxicity. J Biol Chem. 1993;268:10686–10693. [PubMed] [Google Scholar]
- Yang H, Bocchetta M, Kroczynska B, Elmishad AG, Chen Y, Liu Z, Bubici C, Mossman BT, Pass HI, Testa JR, Franzoso G, Carbone M. TNF-alpha inhibits asbestos-induced cytotoxicity via a NF-kappaB-dependent pathway, a possible mechanism for asbestos-induced oncogenesis. Proc Natl Acad Sci U S A. 2006;103:10397–10402. doi: 10.1073/pnas.0604008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Testa JR, Carbone M. Mesothelioma epidemiology, carcinogenesis, and pathogenesis. Curr Treat Options Oncol. 2008;9:147–157. doi: 10.1007/s11864-008-0067-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Ardelt B, Ardelt W, Shogen K, Darzynkiewicz Z. The cytotoxic ribonuclease onconase targets RNA interference (siRNA) Cell Cycle. 2008;7:3258–3261. doi: 10.4161/cc.7.20.6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Cell lines subjected to Onconase treatment at 20 µg/mL (A) Control proliferation has been normalized to a value of 1 for each time point. Significantly decreased proliferation compared to control is seen at 48 and 72 hours. (B) Significantly decreased invasion of MPM cell lines at both time points.
Supplemental Figure 2: Dose effects of Onconase on the proliferation of three mesothelioma cell lines. Although effects were seen with as little as 5 ug/ml, maximum effects were seen with 20ug/ml.
Supplemental Figure 3: Cell lines subjected to Onconase treatment at 20 µg/mL for 72 hours resulted in decreased expression of multidrug resistance gene ABCB. NFKB(p50) expression was also reduced at both time points.
Supplemental Figure 4: Western Blot demonstrates that cell lines subjected to Onconase treatment at 20 µg/mL for 72 hours have decreased protein for multidrug resistance genes ABCB1 and NFKB. ERK2, unaffected by Onconase, was used as a loading control.
Supplemental Figure 5: Quantification of effects of Onconase-associated microRNAs on genes downstream of NFKB. The gene expression was uniformly decreased by hsa-mir-17* and has-mir-30c inhibitor and the degree of decrease was cell line dependent.