Synopsis
Ubiquitination is a post-translational modification pathway involved in myriad cellular regulation and disease pathways. The ubiquitin (Ub) transfer cascade requires three enzyme activities: a Ub-activating (E1) enzyme, a Ub-conjugating (E2) enzyme, and a Ub ligase (E3). Because the E2 is responsible both for E3 selection and substrate modification, E2s function at the heart of the Ub transfer pathway and are responsible for much of the diversity of Ub cellular signaling. There are currently over ninety three-dimensional structures of E2s, both alone and in complex with protein binding partners, providing a wealth of information regarding how E2s are recognized by a wide variety of proteins. In this review, we describe the prototypical E2/E3 interface and discuss limitations of current methods to identify cognate E2/E3 partners. We present non-canonical E2-protein interactions and highlight the economy of E2s in their ability to facilitate many protein-protein interactions at nearly every surface on their relatively small, compact catalytic domain. Lastly, we compare the structures of conjugated E2~Ub species, their unique protein interactions, and the mechanistic insights provided by species that are poised to transfer Ub.
Keywords: ubiquitin, ubiquitin-conjugating enzyme, ubiquitin ligase enzyme, ubiquitination, ubiquitylation
Introduction
In 1983, while studying the regulation of protein degradation, Hershko and co-workers discovered three enzymatic activities required for the ATP-dependent addition of the small modifier protein ubiquitin (Ub) to protein substrates [1]. Over the last three decades, much has been learned about the structures and activities of the E1 Ub-activating enzymes, E2 Ub-conjugating enzymes, and E3 Ub ligases that comprise the signaling pathway. From this body of work a general paradigm for the stepwise transfer of activated Ub from the E1 to a lysine side chain on a protein substrate has been developed, as depicted in Figure 1. For simplicity and clarity, we illustrate only the canonical pathways of RING and HECT E3s in the scheme. These will be elaborated in the text to describe the emerging, more complicated details of the Ub system. Fundamental questions remain: How do E2s and E3s function in a concerted fashion to target specific proteins for modification? What structural factors of E2s, E3s, and Ub govern how a protein is modified and which specific Ub signals are generated? A central player in Ub transfer choreography is the E2. Once thought to serve merely as carriers of Ub from E1 to an E3/substrate complex, it has become increasingly clear that E2s must perform multiple functions. An E2 must interact with an E1, form a covalent thioester conjugate to the C-terminal tail of Ub, and function with a variety of E3s. Recent work has also demonstrated that E2s play an important role in dictating the final product, be it mono-Ub or a poly-Ub chain of specific lysine linkage[2–4]. This realization demands that the simple model for E2 function illustrated in the cartoon of Figure 1 be modified and expanded. The fast pace of research in the general area of protein ubiquitination and, in particular, Ub-conjugating enzymes is evidenced by an abundance of recent review articles [5–10]. In hopes of avoiding redundancy, we present newly emerging themes in E2 structure, function, and mechanism not covered in the two most recent reviews on E2s [9,10]. In particular, we focus on new non-canonical ways in which E2s interact with other proteins and what additional insights these provide about the mechanism of their activity. While we focus this review on Ub E2s, when illustrative we draw parallels between the related Ubl SUMO pathway to better elucidate emerging patterns in the Ub system.
Figure 1.
The 1, 2, 3's of protein ubiquitination. Simplified schematic shows the three enzymatic activities associated with the central paradigm of protein ubiquitination: E1, the Ub-activating enzyme; E2, the Ub-conjugating enzyme; and E3, the Ub ligase. Mechanistically, there are two types of E3s, the RING/UBox-type E3s that effect transfer of Ub directly from the active site of an E2 to a lysine residue of a substrate, and the HECT-type E3s that facilitate Ub transfer from an E2 to substrate via a thioester intermediate on the E3. Auto-ubiquitination of the E3 is also observed in some cases and can be used as a proxy signal to assay the activity of an E2/E3 pair in the absence of a substrate. The best studied fate of poly-ubiquitinated substrates is degradation by the proteasome (not depicted).
The sheer number of E2s and the fact that many are well-behaved, small, soluble proteins have combined to generate a large amount of structural information. There are currently ~100 structures of E2 enzymes in the Protein Data Bank, over half of which are human E2s (www.rcsb.org/pdb) (see [11], for a nearly up to date list of all E2 PDB entries). Most of the structures are of an individual E2, or more specifically, the catalytic domain of an E2 (referred to as “Ubc”). There are five structures of E1/E2 complexes [12–16], and approximately a dozen structures of E2s in complex with an E3 [17–28]. There are nine structures of E2s in complex with other proteins, including “Ubiquitin Enzyme Variants” (UEVs) [29–32] or non-covalently bound Ub or Ub-like proteins (Ubls) such as SUMO [33–37]. Until recently, structures of the activated form of E2, the E2~Ub conjugate, have been missing [26, 38–41]. As discussed below, the growing structural information regarding this mechanistically crucial species is providing new insights into the mechanisms by which E2/E3 pairs transfer Ub. Overall, the wealth of available structural information reveals that E2 Ubc domain structures from yeast to human are remarkably similar. While these similarities have provided many general insights, a wide array of functional diversity within this enzyme family demands a closer inspection of the distinctive features of E2s and how they interact with other proteins.
E2 Morphology
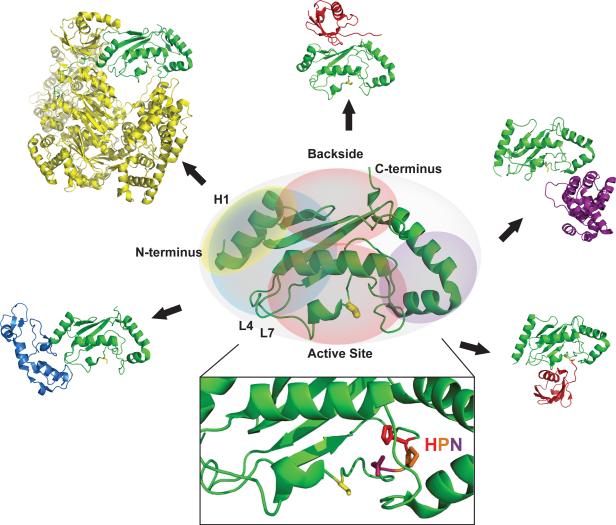
E2 enzymes are easily recognized from their evolutionarily conserved ~150 residue catalytic `Ubc' domains. Ubcs have a compact structure shaped essentially like a prolate ellipsoid, comprised of four α-helices, a short 310 helix near the active site, and a four-stranded anti-parallel β-sheet. On the “bottom” surface (in the view shown in Fig. 2), the active site Cys to which Ub (or Ubl) becomes conjugated is nestled in a catalytic groove surrounded by highly conserved amino acids that mediate both thioester formation (reception of Ub from the E1 Ub- activating enzyme) and substrate ubiquitination (isopeptide bond formation with a substrate lysine). Most notable is a trio of residues known as the `HPN' motif, usually found ten residues to the N-terminal side of the active site Cys. The His is thought to play a structural role in forming the E2 active site, whereas the Asn residue is important for mediating the catalysis of an isopeptide between Ub and a substrate lysine [42, 16]. As the general mechanism for ubiquitination suggests, an E2 must interact with an E1 and an E3; both interactions use partly overlapping surfaces on one pole of the Ubc ellipsoid (Fig. 2). The E1/E3 interaction site is comprised of residues on the N-terminal helix, or helix 1 (H1), and loops 4 and 7 (L4 and L7; sometimes referred to as L1 and L2). The shared surface implies that an E2 must be free of the E3 before becoming re-loaded with Ub [43].
Figure 2.
Structure of a Ubc domain and its interaction surfaces. Shown in the center is the E2, Ubc13 in ribbon structure (PDB code 2GMI), with interaction surfaces described in the text colored. The E2 in each interacting pair is shown as green. Clockwise from lower left corner: the E1/E3-binding surface as seen in complex of Ubc13 with the RING E3 Traf6 (blue) (PDB code 3HCT) and in Ubc12 in complex with the NEDD8-activating E1 (yellow) (PDB code 2NVU); backside-binding surface as seen in the E2/Ub complex, UbcH5c/Ub (red) (PDB code 2FUH); substrate-binding surface as seen in the SUMO E2 Ubc9 in complex with its substrate RANGAP1(purple) (PDB code 1Z5S); activated Ub/Ubl surface as seen in UbcH5b~Ub complex (PDB code 3A33). The inset shows the highly conserved HPN motif at the active site, with the active site Cys shown as yellow stick representation.
Many E2s are characterized by additional segments located within and flanking the Ubc domain. For example, Cdc34 has a 12-residue acidic loop near the 310-helix of its Ubc that is important for catalyzing poly-Ub chain formation [44, 45]. More common are E2s that contain variable N- or C-terminal extensions appended to the Ubc domain. Thus historically, the E2 family has been divided into four classes according to the presence of these additional protein segments: Class I, Ubc domain only; Class II, Ubc domain plus a C-terminal extension; Class III, Ubc plus an N-terminal extension; Class IV, Ubc plus both N- and C-terminal extensions. However, while the topology of the Ubc domain is highly conserved, N- and C-terminal extensions are widely variable in both size and structure. Sizes of extensions vary from a ~50-residue C-terminal extension in the class III E2 Ube2k to a greater than 4000-residue N-terminal extension in the Class IV E2 Birc6 (see [10] for review of extension-containing E2s). Although a tidy way to group E2s, the classifications are not predictive for functionality, such as E3s with which a given E2 will interact, or whether an E2 transfers mono-Ub or is capable of building a poly-Ub chain. In a recent effort to classify E2s, a phylogenetic analysis of similarities within the Ubc domains yielded 17 different families of E2s [46]. In this scheme, five of the ~45 Human E2s are assigned to the family that includes the UbcH5 isoforms while other families have only one or two members. Assigning functional significance to each family may have limited utility when there are half as many families as E2s, and additional biochemical and functional characterization will be required to lend predictive value to this fine-grained classification.
Canonical E2–E3 interactions: UbcH7-E6AP/cCbl and other prototypical E2–E3 complexes
How does an E2 bind a particular E3? Crystal structures of the human E2 UbcH7 in complex with a HECT-type (E6AP)[17] and RING-type (cCbl)[18] ligase have long provided the framework for our basic understanding of E2–E3 interactions (Figure 3). Although mechanistically distinct, HECT- and RING-type ligases bind a similar surface on the E2 Ubc domain, suggesting that regardless of the Ub destination, be it a HECT active-site Cys or a substrate lysine, E3 binding at this region of an E2 may somehow help to optimally position the activated E2 for transfer of its Ub.
Figure 3.
RING- and HECT-type E3s recognize the same surface on the E2 UbcH7. Shown are the archetypes for canonical E2/E3 interactions: A) UbcH7 in complex with the HECT-E3, E6AP (PDB code 1C4Z); B) UbcH7 in complex with the RING-E3, cCbl (PDB code 1FBV).
A growing number of E2/E3 structures are available for comparison. Structures that contain the same E2 bound to different E3s allow for both generalizations and distinctions to be drawn. In general, the E2 residues observed to contact E3s include polar and charged residues on H1, a highly conserved hydrophobic residue in L4, usually a phenylalanine (Phe63 in UbcH7; Phe62 in UbcH5c), and hydrophobic residues in L7 (Pro97 and Ala98 in UbcH7). The L4 phenylalanine is required for E2s to interact with HECT-type ligases 47], while the analogous residue appears unimportant in interactions involving isoforms of UbcH5 and the RINGs of BRCA1 [2] or CNOT4 [19]. L7 residues contribute to interactions with both HECT and RING type ligases. In the case of BRCA1 and its cadre of ten interacting E2s, the L7 alanine (Ala96 in UbcH5c) is a defining feature of the set of interacting E2s, as its mutation to aspartic acid disrupts the ability of E2s to continue to make productive interactions [2].
Comparison of structures of a single E3 in complex with different E2s reveals plasticity in the E2/E3 interface. Structures of the homodimeric U-box ligase, CHIP, in complex with Ubc13 [21] or UbcH5a [23] illustrate how an E3 can accommodate different E2s. The CHIP-interacting surface of UbcH5a includes residues in the N-terminal helix and hydrophobic residues in L4 (Pro61, Phe62) and in L7 (Pro95, Ala96), with Phe62 protruding into a hydrophobic groove on CHIP [23]. In Ubc13, the structurally homologous residue to Phe62 is Met64. However, while mutation of UbcH5a residue Phe62 disrupts CHIP-UbcH5a interactions, substitution of Met64 in Ubc13 does not disrupt the complex [23]. In contrast, the mutation M64A disrupts the interaction between Ubc13 and RING TRAF6 in a Y2H system [48], as well as its interaction with Rad5, another RING type ligase in yeast [49]. There are salt bridge interactions between Ubc13 and CHIP not observed in the CHIP/UbcH5a complex and these may account for the lowered importance of the L4 hydrophobic residue [21, 23]). A salt bridge interaction plays a key role in the complex of CNOT4 RING domain and UbcH5 loop L4 residue, Lys63 [50, 19]. Although yet to be exploited, the ability to manipulate charge-charge interactions to yield altered specificity pairs could be a powerful approach for in vivo studies [51, 52]. In summary, the E2/E3 structures solved to date identify a modest number of E2 residues that are involved in binding a variety of E3s. It is important to note, however, that there are currently E2/E3 structures for only a small number of Ub E2s, namely, isoforms of UbcH5, UbcH7, and Ubc13. As these E2s are quite similar in their E3-binding loops, it may be ill-advised to extrapolate lessons learned from them to other more distantly related E2s.
E2/E3 Interactions within the Ubc domain: Which with Which?
There are at least two compelling reasons for studying E2/E3 interactions. First, knowledge of active E2/E3 pairs can guide and inform biological studies. A given E3 may interact with numerous E2s, and the ultimate outcome of the reaction (i.e., type of product) depends on which E2 is involved [2–4]. Second, structural information on E2/E3 complexes may provide insight regarding the mechanism and products of Ub-transfer reactions. Therefore, knowledge of the complete cadre of E2s for a given E3 is critical.
The question of how to best identify active E2/E3 pairs remains a thorny one, with different pathways (i.e., Ub, SUMO, etc.) posing different challenges. In the case of the Ubls, including SUMO, the E3s are not as easily recognized as the Ub E3s, which all contain either a RING/U-box or a HECT domain that can be identified through sequence conservation [53]. For Ub transfer, a genome will contain dozens of E2s and hundreds of E3s. Given the relative ease with which E2s and E3s can be recognized directly from protein sequence information and numerous structural studies of E2/E3 complexes, the goal of defining the E2/E3 interactome might seem straightforward. However, sequence analysis alone has had limited power for predicting uncharacterized E2/E3 pairs. Part of the problem arises from the functional necessity that E2s use overlapping surfaces for binding and recognition of both E1s and E3s, requiring that these surfaces have broadly conserved features (See Figure 2.) [43].
The modest affinity and transient nature of E2/E3 complexes pose additional technical challenges for the identification of E2/E3 pairs. Standard techniques such as co-purification, pull-down, or co-immunoprecipitation rarely succeed due to the weak affinity of the complexes. Expression analysis in which substrate degradation is monitored when certain E2s or E3s are over-expressed or depleted from cell extracts can be successful in identifying E2/E3 pairs [54,55]. Depletion of the APC-specific E2 Ube2c (UbcH10) from cellular extracts did not completely stabilize APC substrates such as securin, leading to the discovery of another APC-interacting E2, Ube2s [55]. Such approaches may suffer from the pleiotropic or compensatory nature of some E2s/E3s, and their success requires a targeted approach that may not be feasible for large scale de novo determination of interacting E2–E3 pairs.
Directed yeast two-hybrid (Y2H) approaches have been somewhat more successful in identifying E2/E3 pairs, presumably because a positive read-out can be obtained even for transient interactions. During the past year, two large-scale Y2H screens for E2 partners have been reported [56, 57]. A screen that utilized full-length E2s as bait against circa 150 RING-protein preys yielded putative partners for all but two of 39 E2s (Cdc34 and Birc6) and for approximately 90 of the E3 preys [56]. However, a screen of over 250 RING domain preys with 36 E2 Ubc domain baits failed to identify a binding partner for ten E2s that are known to conjugate Ub and fewer than half the RING domains returned a positive E2 interaction [57]. The different outcomes in the screens may be due in part to the use of full-length versus specific E2 and E3 domains, consistent with emerging evidence for non-canonical E2/E3 interactions. There is growing recognition that some RING E3s, such as Rad18 and gp78, utilize regions outside the canonical RING domain for binding to Ubc domains [58, 25, 27]. In addition, the requirement of some RING E3s to exist as heterodimeric or multi-component complexes may further affect the attainable yield in a Y2H screen. For example, in a targeted Y2H screen aimed at identifying the human E2s that interact with the heterodimeric RING E3, BRCA1/BARD1, a bait construct in which the RING domains of BRCA1 and BARD1 were fused into a single polypeptide that folds correctly into the E3-active structure identified ten E2s that interact with BRCA1/BARD1 [2]. Screens using baits comprised solely of the BRCA1 RING failed to identify any E2s that have been shown biochemically to transfer Ub [2, 56, 57]. It is reasonable to expect similar behavior from other heterodimeric E3s, implying that the use of single RING domains as bait is likely to fail for this class of E3. Furthermore, in the same screens, using the individual RINGs from the heterodimeric E3 RING1/Bmi, only a small subset of the known interacting E2s could be identified [56, 57]. Although it is tempting to predict that the RING constructs for which no interacting E2s were identified in the screens reported to date are either heterodimeric or multi-components E3s, many things can conspire to give a negative Y2H result, so caution should be exercised. Within a Y2H screen, the E2 may or may not be conjugated to Ub, depending on whether the endogenous yeast E1 is able to charge the E2 of interest among other factors. Therefore, an issue that may contribute to a failure to identify E2s for an E3 in a Y2H experiment is that Y2H studies may only screen for interactions between an E3 and a free E2, although the functionally relevant interaction involves the E2~Ub conjugate. There are examples of E3s that show detectable binding only to an E2~Ub. For example, SspH2 (a bacterial protein with E3 ligase activity) binds only to an activated E2~Ub conjugate and not to the individual components [59]. Attempts to identify this interaction using Y2H screens with E2s as prey uniformly failed (P. Brzovic & R. Klevit, unpublished observation). It remains to be seen if this feature will be unique to bacterial E3s that have evolved via convergent evolution to work with host E2 enzymes or whether there are eukaryotic E3s that only bind to E2~Ub species. In any case, these examples demonstrate the complicated and context-dependent nature of E2/E3 interactions that confound the ability to identify them.
E2–E3 Interactions Expanded: the E2 “backside”
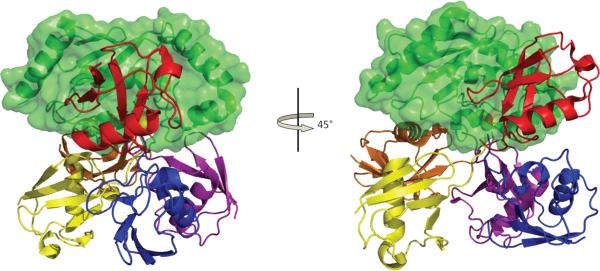
An emerging interaction surface used in E2s is the so-called “backside” (the top in the view shown in Figure 2)—a surface comprised primarily of the E2 β-sheet. First identified as a site for non-covalent Ub and SUMO binding with the E2s UbcH5c [33] and Ubc9 [35,37] respectively, the list of E2s that use the backside for mediating protein-protein interactions continues to grow and now includes Ube2g2 [25, 27, 60] (Figure 4).
Figure 4.
Ub/Ubl and non-canonical E3 binding to the E2 backside. Top, non-covalent complexes of E2s with Ub/Ubl (red) bound on the backside. Bottom, non-canonical E3 (blue) binding uses the same E2 surface. PDB codes are (left to right from the top: 2EKE, 2UYZ, 2FUH, 3A4S, 3H8K, 1Z5S).
There are several examples of E3s that utilize the E2 backside surface in addition to the canonical E3-binding surface (H1, L4, and L7). Some E3s contain domains that are structural mimics for Ubls, as seen with the human RENi protein Nip45 and its yeast counterpart Rad60 (Figure 4) [61, 62]. Nip45 uses “SUMO-Like Domains” (SLDs) to bind the backside of Ubc9 with similar affinity as SUMO, using conserved electrostatic interactions [62]. The biochemical function of Nip45 remains to be determined; current suggestions are that Nip45 may function as a SUMO E3 ligase or as a target for a SUMO-targeted Ub E3 ligase [62, 63]. In another example, the SUMO E3 Nup358/RanBP2 contacts the Ubc9 backside using a domain that recapitulates the contacts of SUMO in a structurally unique way (Figure 4) [20]. In addition to the canonical Ubc9 E3-binding interface comprising residues in H1 and L4, the E3 Nup358/RanBP2 makes additional contacts with the first 3 β-strands on the backside of Ubc9--the same surface that binds SUMO. Notably, in contrast to Ub E2s, residues of Ubc9 L7 do not participate in this E2–E3 interaction.
Similar to RanBP2, the Ub E3 ligase gp78 uses a distinct structural domain to interact with the backside of the E2 Ube2g2 [25, 27]. In addition to binding Ube2g2 via a RING domain, the E3 gp78 interacts with the backside of Ube2g2 through a short region called the “Ube2G2 Binding Region” (G2BR) [25, 27]. However, unlike the SUMO E3 that uses a contiguous surface to contact the Ubc9 canonical E3 binding surface (H1/L4/L7) and the backside [20], the gp78 G2BR is located almost 200 residues away from the RING domain [64]. Unstructured in its apo form [25], G2BR adopts an α-helical structure that contacts the Ube2g2 backside surface through mostly hydrophobic interactions bolstered by additional charged contacts [25,27]. Binding of G2BR, even in trans, results in a ~50 fold increase in affinity for the RING domain of gp78 [25]. Functionally, Ube2g2 showed slower rates of thioester formation with Ub in the presence of G2BR [25], as well as an enhanced ability to build chains [25, 27]. Although a structure of gp78's RING bound to Ube2g2 does not yet exist, it is reasonable to expect that it will show primarily canonical E2/E3 interactions. Thus, the mode of binding for this Ub E2/E3 pair bears a striking resemblance to the mode used by the SUMO E2/E3, using a mix of canonical E3-binding surface and the backside of the E2. Although no structural information yet exists, a small auxiliary E2-binding module that is 300 residues away from the RING of the E3, Rad18 was identified from Y2H analysis with the E2, Rad6 (yeast) [58]. It seems reasonable to expect that additional variations on this theme will be discovered for other E3/E2 pairs in the future. In summary, similar to the shared E1/E3-binding surface, E2s appear to use their backside surfaces to provide a multi-functional binding surface for E3s and Ub/Ubls, and possibly, other binding partners.
Interestingly, Ube2g2 binds Ub on its backside surface, although with much weaker affinity than for G2BR [25]. A functional significance, if any for this non-covalent Ub interaction has yet to be established for Ube2g2. This begs the question, could the binding of Ub via the `backside' reported for the other E2s be a harbinger for non-canonical E3 binding, or is the E2 using overlapping surfaces to manage multiple functionalities? Given the overlap in the E3 and E1 binding surfaces on the E2, the E2 backside may also serve multiple functions binding both Ub and an E3. As functionally significant Ub binding occurs on the backside surfaces of several E2-variant proteins, including the UEVs Mms2, Tsg101, and Vps23 [31, 32, 34], it remains a possibility that E2 Ub binding on the backside surface may be biologically and mechanistically important.
E2/E3 Interactions Expanded: roles for non-Ubc extensions of E2s
Available structures of most Class II, III, and IV E2s are for the Ubc portion only. In a few cases, extensions were included in the crystallization, but their structure could not be defined, indicating that they are disordered, at least in the absence of a binding partner [65, Ube2T, PDB code 1YH2). With the exception of yeast Ubc1 and its human counterpart Ube2k (also known as E2-25K), there is very little structural information regarding the non-Ubc portions of E2s. Ubc1/Ube2k are unique in that their C-terminal extensions are comprised of an identifiable, structurally well-characterized domain, the UBiquitin-Associated domain (UBA). UBA domains appear in a wide variety of proteins and are known to function as Ub-binding domains. Paradoxically, the function of the UBA of Ube2k (and yeast Ubc1), the only non-Ubc domain for which structural information is available, remains enigmatic [66, 67]. Although known to bind Ub and poly-Ub chains, the UBA domain is dispensable for the generation of poly-Ub chains, and does not seem to be important for determining chain linkage specificity [3].
Current knowledge points to a role for non-Ubc extensions of E2s in mediating interactions with E3s or substrates. Y2H analysis of the interaction between the yeast E2 Ubc2 and the E3 ligase, Ubr1, identified acidic residues in the C-terminal extension of the E2 that interact with a basic region in the E3 [68]. Removal of the extension resulted in some loss of Y2H signal when presenting the RING H2 region alone. The 37-residue C-terminal extension of Cdc34, which contains a high density of acidic residues helps mediate the interaction with the multi-protein SCF E3 complex through a “basic canyon” on a surface located in the C-terminal domain of the Cul1 subunit [69]. Substitution of basic residues flanking the canyon with acidic residues disrupts ubiquitination mediated by Cdc34, but not by other E2s such as UbcH5c that lack acidic tails. Removal of Cdc34's acidic tail decreases its activity with the SCF, as does addition of the tail in trans [70]. The interaction of the E3 with the Cdc34 Ubc domain and with its tail are each quite weak, but when presented within a single E2 moiety, the avidity effect results in orders-of-magnitude increase in Km relative to the Ubc domain alone. In addition to binding E3s, the C-terminal tail of Cdc34 has recently been shown to contain two non-covalent ubiquitin binding sites composed primarily of two islands of hydrophobic residues in the otherwise acidic tail [71]. Mutations in one of these Ub-binding sites result in defects in SCF-mediated substrate ubiquitination. How Cdc34 coordinates Ub binding and E3 binding via the same 37-residue stretch remains to be established. Combined, these observations underscore an emerging theme of dual functionality for E2 regions exhibiting non-covalent Ub-binding.
The unique 30-residue N-terminal extension of Ube2c (UbcH10) is thought to provide this E2 with specificity towards substrates [72]. Deletion of the N-terminus of Ube2c makes the enzyme more active but less specific for particular lysines and degradation signals when transferring Ub with the APC E3 ligase. In contrast to the Cdc34 tail which provides additional contacts to the E3, the Ube2c N-terminus probably mediates an interaction with substrate, using its Ubc domain as the primary E3-binding component. Confirmation of this proposal must await further structural characterization of Ube2c in complex with APC and/or a substrate. The N-terminal extensions of members of the Ube2e family (also known as UbcH6, Ube2e2, and UbcM2) are implicated in novel interactions with cullin adapter complexes based on the observation that mutations in the canonical E3 binding region of the E2s do not disrupt the interaction [73]. The functional consequences of these interactions have yet to be resolved. In summary, non-Ubc regions of E2s appear to play a variety of roles, not all yet defined, through interactions with E3s, substrates, adaptors, and Ub itself.
Activated E2~Ub: their structures and recognition
The conjugated E2~Ub is the active form of an E2 enzyme. The in vivo steady-state equilibrium of the human E2 Ubc2b favors the conjugated form and this is likely a general feature of the protein ubiquitination system [74]. Thus, cellular proteins are more likely to encounter E2~Ubs than free E2s. However, due to the relative ease with which free E2s can be studied structurally and the technical challenges posed by generating sufficient quantities of purified E2~Ub species, a vast majority of current structural information on E2s are for free E2s. The recent emergence of several E2~Ub structures provides an excellent foundation to begin to understand this functionally important species.
E2~Ub conjugates present a technical challenge for structure determination because the thioester formed between E2 and Ub is inherently unstable—a chemical property that is necessary for catalysis. Manipulation of the unstable thioester bond has enabled structural studies of several E2~Ubs. For many (but not all) E2s, mutation of the active site Cys to a Ser can allow formation of a longer lived oxy-ester, which is more amenable to structural determination [26, 39, 41, 59, 75]. The Cys-to-Ser mutation differs by only a single atom in the resulting ester. A disulfide-linked mimic of a UbcH8~Ub (Ube2L6) species has been generated by linking Ub with a Cys residue added to its C-terminus to the E2 active site Cys [40]. At the writing of this review, there are 5 structures of E2~Ub-like species: Ubc13~Ub (PDB code 2GMI); Ubc1~Ub (PDB code 1FXT); UbcH8~Ub (PDB code 2KJH); and UbcH5b~Ub (PDB codes 3A33 and 3JVZ). Structural information from NMR studies of Ube2b (Rad6b)~Ub is also available, but without a PDB entry [75]. These structures provide the first glimpse of E2~Ubs and the possibility to better understand the mechanism of Ub transfer. Another informative structure representing an E2/E3/product complex in the SUMO system (Ubc9/Nup358/SUMO-RanGAP1) adds to the growing picture of how Ub or a Ubl is transferred from an E2 to a substrate lysine [40]. As the Ubc9 active site Cys is directed at the isopeptide bond between the RanGAP1 substrate and SUMO, certain features of this structure may be pertinent to a Ubc9~SUMO conjugate.
The first general conclusion that can be drawn from comparison of the structures of the conjugated E2s and their free E2 counterparts is that neither the E2 nor Ub undergoes significant conformational change upon conjugation [26, 38–41, 59]. The comparison of free Ubc13 (PDB code 1JAT) with conjugated Ubc13~Ub (PDB code 2GMI) shows only 1.6 Å r.m.s. deviation over all Cα positions [39]. Movements of side chains in or near E2 active sites have been noted, but such changes should be interpreted with care, as the positions of side chains in free E2 structures can be as variable as the differences being noted [76, 41]. For example, the location of proposed catalytic residue, Asn77 of the HPN motif, differs by nearly 2 Å in four different free UbcH5b structures depending on whether the structure was solved crystallographically or in solution (PDB codes 2C4O, 2ESK, 2CLW, and 1W4U) and dynamics of Asn77's side chain in solution is apparent in the NMR ensemble (PDB code 1W4U) [77]. In some cases the resolution of the crystal structure is on par or lower than the 2Å changes noted above and therefore lacks sufficient precision to draw conclusions about the position and role of Asn77 (PDB codes 3A33, 2C4O, and 2CLW). Furthermore, comparison of the conformation of Asn77 in a variety of E2 structures does not provide a unified theme: Asn77 is in the gauche conformation, pointing away from the active site Cys in free Ube2g2 (PDB code 2CYX), free Ubc2 (yeast Rad6) (PDB code 1AYZ), and in Ubc13~Ub (PDB code 2GMI), and is in the trans conformation, pointing towards the Cys and presumably in a better conformation to stabilize the oxyanion intermediate in the solution state structure of Ube2g2 [78].
Given that neither component undergoes conformational change upon conjugation, it is instructive to compare the available E2~Ub structures as a group. Figure 5 shows five structures with the E2 components superposed. A striking feature of this display is that Ub/Ubls adopt many positions relative to the E2. It has been proposed that differences in the positioning of Ub in an E2~Ub conjugate may explain the variety of activity observed for the various E2 family members although this remains to be tested experimentally [76]. Observed Ub positions may be caused in part by either crystal packing effects or additional protein-protein interactions in a given crystal. For example, in the crystal of Ubc13~Ub/Mms2, the UEV protein Mms2 binds the hydrophobic patch on the surface of Ub centered about Leu8, Ile44, and Val70 of a Ub from another Ubc13~Ub moiety and positions its Lys63 side chain towards the thioester bond of Ubc13~Ub [39]. This non-covalent interaction involving the ~Ub may contribute to its ultimate position in the crystal lattice. Similarly, in the structure of UbcH5b~Ub, a Ub moiety from one E2~Ub is bound non-covalently on the backside site of the E2 of another E2~Ub moiety [41]. Thus, these “secondary” interactions may dictate the ~Ub positions observed in each crystal. In two cases, Ubc1~Ub and UbcH8~Ub, the position of the conjugated Ub is not influenced by a third protein [38, 40]. In both cases, a single E2~Ub conformation is reported, despite evidence that the Ub takes many positions in solution in the E2~Ub conjugates [76].
Figure 5.
Ub conjugated to an E2 assumes multiple orientations relative to the E2. Shown in green, the E2 Ubc of each structure has been superimposed; active site Cys is shown in yellow spheres. Ub moiety in each structure is shown in a different color: Red, Ubc1~Ub (PDB code 1FXT); Blue, UbcH5b~Ub in complex with Nedd4L (PDB code 3JVZ); Yellow, Ubc13~Ub (PDB code 2GMI); Orange, UbcH5b~Ub (PDB code 3A33); Purple, UbcH8~Ub (PDB code 2KJH). View on the left shows the same E2 orientation as in Figures 2, 3, and 4; view on the right is rotated 45 degrees about the vertical axis.
The array of Ub positions observed among the available structures may be taken as indication that E2~Ub species are dynamic in solution and that each structure represents a snapshot along some trajectory or within an ensemble. NMR studies indicate that conjugated Ub is indeed flexible relative to the E2 and that the two proteins behave as two loosely connected entities rather than one globular protein complex [59, 75]. Further investigations of E2~Ub species in solution are needed to more fully understand the nature and relevance of the observed and inferred dynamics.
The Ubc9/Nup358/SUMO-RanGAP1 structure is presumably a snapshot of an E2/E3/product complex [20]. Here, the E3 is seen to contact the β-sheet surface of SUMO that is now attached to product. The interaction between the ligase and SUMO may provide a hint into the role an E3 plays to catalyze Ub/Ubl transfer. The SUMO is in a unique position relative to the E2 in the admittedly small number of E2~Ub structures currently available and may represent the orientation of an E2 and Ub just after formation of the isopeptide bond.
Ub itself binds to a remarkable array of proteins, doing so mainly via a hydrophobic surface centered around residue Ile44. Thus, when an E2 is conjugated with Ub, there is the potential to utilize this domain as a site for protein-protein interactions. The ability to distinguish between free E2s and E2~Ub conjugates would seem to offer a functional advantage. Recently several E3s have been shown to possess domains that bind Ubls [79–81, 59, 26] in some cases using this site to specifically recruit E2~Ubls [26,59], thereby enhancing productive associations.
Most E3s appear to bind both free and conjugated forms of E2s. Although there is anecdotal evidence in support of the general expectation that binding is stronger with an E2~Ub, this property has only been quantitatively demonstrated in a few instances. A disulfide-linked mimic of activated UbcH7~Ub showed no increased affinity for the HECT E3 E6AP compared to the affinity measured for free E2 [82]. In contrast, a kinetic analysis of the yeast ligase E3α and the E2, Ubc2 revealed that binding to Ubc2~Ub is ten times stronger than to free Ubc2 [74]. Similarly, kinetic analysis showed that Cdc34~Ub binds Cul1-Rbx1 about twice as tightly as free Cdc34 [83]. These relatively modest differences in binding energy make the measurement technically challenging; kinetic measurements in which KI is compared for a free E2 and an oxyester form of E2~Ub are a better choice than thermodynamically-based approaches.
Although most E3s appear to bind a free E2 with low, but detectable affinity, it is the E2~Ub/E3 complex that represents an enzyme poised to react. The first structure of an E3/E2~Ub complex was recently reported for a HECT E3, NEDD4L, and UbcH5bUb (Fig. 6) [26]. The ability to capture this important species in a crystal was made possible by inclusion of several mutations in the E2: an active site Cys-to-Ser mutation allowed formation of the oxyester with Ub, and mutations were made in the E3 binding region to create a stronger-binding complex amenable to structural investigation [26, 84]. The structure is characterized by a combination of canonical and novel binding surfaces on the E2 and on Ub. The N-terminal lobe (N-lobe) of the HECT domain binds the canonical E3-binding site on UbcH5 much as previous structures have revealed. However, the HECT domain C-terminal lobe (C-lobe), which contains the E3 active site Cys 922, is in a dramatically different position relative to previous structures [17, 26]. Contact between the C-lobe and ~Ub moiety closes the distance between the E2 and E3 active sites to about 8 Å, much closer than distances observed in any previous structures of HECT E3s in complex with an E2 [17, 26]. NEDD4 C-lobe residues Leu916 and Met943 make hydrophobic contacts with Ub residues Ile36, Leu71, Leu73. Mutations in the E3 or Ub at this interface decreased the ability of the E2 to transfer Ub to the HECT active site Cys. Based on this structure, it is reasonable to propose that HECT E3s can be described in general as consisting of an E2-binding domain (the N-lobe) and a ~Ub-binding domain (the C-lobe), even though an interaction between the C-lobe and free Ub has never been detected [80]. However, with only a single E3/E2~Ub structure currently available, it is wise to avoid the temptation to draw many generalizations. There are indications that various active E3/E2~Ub complexes interact in different ways--mutations in Ub I36D and L71A decrease formation of a HECT~Ub in NEDD4L and other family members, such as RSP5 and ITCH, but the HECT E3, E6AP, is not impaired by the same mutations [26]. We can therefore expect that even among HECT-E3s, the details of how they recognize and bind to E2~Ubs will be distinct.
Figure 6.
Structure of an E3/E2~Ub complex: HECT-E3, Nedd4L (blue), in complex with UbcH5b~Ub (green/red). Left, HECT N-lobe contacts E1/E3 surface on UbcH5, as observed in previous structures (see Figure 3) and HECT C-lobe makes contacts with the ~Ub moiety. The NEDD4L active site is shown in yellow (PDB code 3JVZ). Right, view rotated by 180 degrees to show NEDD4L residues analogous to those observed in Rsp5 and SMURF2 that are important for non-covalent Ub binding in the N-lobe, shown as orange spheres.
In contrast to eukaryotic E3s, the bacterial HECT-type E3, SspH2, binds only to E2~Ub, with no detectable binding to the individual components [59]. The E3-interacting surface mapped by NMR studies only modestly overlaps with the canonical binding surface of eukaryotic E3s, as binding on helix H1 and loop L4 of the E2 were not detected. Instead, the interaction involves a surface on UbcH5c not previously observed in E2/E3 complexes, with the SspH2-binding site comprised of residues in regions around the active site, the cross-over helix, and loop L7. In addition, the E3 makes contacts to the hydrophobic Ile44 surface of the Ub moiety in the E2~Ub. Mutation of the loop L4 phenylalanine of UbcH5, known to be essential for HECT E3 binding [47], does not disrupt the interaction with SspH2 [59], consistent with the non-overlapping nature of the SspH2-binding site relative to other HECT-E3s. Thus, this effector protein from a pathogenic bacteria has evolved by convergent evolution to recognize specifically the active form of a critical host enzyme, an E2~Ub, and accomplishes the task using a surface on the E2 different from host E3s. Whether this novel binding surface is used by other bacterial effectors that hijack host E2s is an open question.
The SUMO E3, Pc2, was recently shown to contain a cryptic SUMO-Interacting Motif (SIM) that is distal to its Ubc9-binding domain [79]. Point mutants in the Pc2 SIM fail to recruit Ubc9 to subnuclear foci and a Ubc9 active-site mutant incapable of conjugating SUMO also fails to be localized by Pc2. These observations are consistent with the E3 preferentially binding Ubc9~SUMO. Interestingly, Ubc9 mutants defective for non-covalent “backside” SUMO-binding also fail to be relocalized by Pc2, suggesting Pc2 may bind both activated and noncovalently-bound SUMO on the E2. Whether these interactions have mechanistic consequences in addition to their demonstrated sub-cellular localization functions remains to be explored.
There is a growing list of proteins that do not contain canonical RING, UBox, or HECT domains that exhibit preferential binding of E2~Ub over free E2. The nuclear import protein, importin-11, interacts with the conjugated UbcM2~Ub (Ube2e3), but not with the free E2 [85]. Thus, preferential binding of conjugated E2~Ub results in specific subcellular localization of an E2. UbcM2~Ub is restricted to the nucleus, whereas free UbcM2 cannot enter the nucleus, a mechanism that may spatially regulate the activity of UbcM2. Likewise, the kinase OspG from Shigella bacteria exclusively binds human UbcH5b~Ub, but not free UbcH5b or Ub alone [86]. OspG binding to UbcH5b~Ub presumably disrupts Ub transfer activity in the host as observed by decreased Ub-dependent degradation of IκBα.
In summary, the observed range of Ub positions relative to the E2 in E2~Ub complexes may play a variety of roles. Certain orientations may help to recruit specific binding proteins. Or the dynamics of conjugated Ub itself may play a role in catalysis by sampling numerous states which could alter the environment of the active site and modulate reactivity. Additional structural information regarding E2~Ubs alone and in complex with E3s will help to fill in the blanks in the not-too-distant future.
Additional Interactions among E2, E3, and Ub/Ubl
Several HECT E3s contain an additional Ub-binding site in their N-terminal lobe, distinct from the E2 binding region [80, 81]. Ub binding to the N-lobe of the HECT E3 Rsp5 was detected by GST pull-downs and scanning alanine mutagenesis [80]. A similar site has been identified in HECTs NEDD4 [80] and SMURF2 [81], but not in Tom1 [80], again suggesting that not all HECT-E3s recognize the components of ubiquitination in the same fashion. The canonical hydrophobic Ile44 Ub surface is responsible for the HECT N-lobe/Ub non-covalent interaction, different from the surface used by the C-lobe of NEDD4L (Fig 6) [81]. The functional significance of HECT N-lobe/Ub-binding has yet to be settled unambiguously. Rsp5 mutants with decreased affinity for Ub appear to synthesize Ub chains more efficiently than the wild-type enzyme [80]. In contrast, SMURF2 mutants deficient for Ub binding have disrupted polyUb chain synthesis [81]. SMURF2 can bind its substrate more efficiently when the substrate is mono-ubiquitinated, and mutations in the E3 Ub-binding site preclude this interaction [80]. In neither case was a role for the N-terminal Ub-binding domain in recruiting E2~Ub demonstrated [80, 81]. As interactions required to recruit a ubiquitinated substrate and a Ub-laden E2 may be quite similar, E2~Ub recruitment remains a formal possibility.
From structures to mechanism
Considering the wealth of structural and biochemical information regarding E2s, it is remarkable how much remains unclear about the mechanism by which Ub is transferred from an E2~Ub. Fundamental questions such as “How does an E3 catalyze transfer?” are not yet answered. Studies with the SUMO E2 Ubc9 show that the `natural' hydrolysis (in absence of any other nucleophile or E3) is slow, suggesting that thioesters in native E2s are relatively stable, and must be coaxed into discharging their Ub to substrate [87]. Hypotheses invoking allosteric effects on the E2 are pervasive but neither well-documented nor supported. Studies utilizing E2~Ub conjugates (or their mimics) are likely to finally shed light on Ub transfer mechanisms.
To understand the mechanism of Ub transfer, it is sensible to focus on the E2 active site and its surroundings. The current paradigm for an E2 active site includes the Cys residue to which Ub/Ubl will be conjugated and the conserved HPN motif (see above). However, these residues have been insufficient to completely explain the mechanism, suggesting that we must look further afield for clues. In the following section, we suggest some additional features that may play direct roles in E2 function.
Dynamic behavior of E2s may play a role in catalysis and mechanism. Some E2s have flexible loops near their active sites: for example, there are two flexible loops (residues 95–107 and 130–135) surrounding the active site of the E2 Ube2g2, identified by comparison of solution and crystal structures [78]. Similar loops are present in yeast Ubc7, human and yeast Cdc34, but not in the large UbcH5 family. Two acidic residues in the Ube2g2 loops (Asp98, Asp99) are required for activity [78]. Current hypotheses regarding the role of the acidic loop residues include: 1) that they polarize the histidine of the conserved HPN motif to create a general base [78], or 2) that they guide a lysine into the active site [78]. Either case requires a fixed orientation of the acidic residues in question and therefore predicts that the flexible loops become more ordered either as a result of formation of the E2~Ub conjugate or as a result of E3 binding. Further investigations will be required to sort out these possibilities.
The position of conjugated Ub within an E2~Ub conjugate likely plays a role in catalysis, perhaps by influencing the approach of the nucleophilic substrate lysine. Early studies demonstrated specificity for Ub transfer to lysine over other amines, indicating that the E2~Ub protects its thioester from other reactive species [88]. The reactivity of the E2~Ub thioester is significantly lower than that of a free thioester in solution, which may be due to limited access to the thioester within the active site [88]. In the structure of the oxyester mimic of Ubc13~Ub/Mms2, the substrate Ub's Lys63 is seen poised to attack the ester and the direction of nucleophilic attack is defined by the geometry of the E2~Ub Ub/UEV complex [39]. The ability to form such a structure may explain Ubc13/Mms2`s ability to build free Lys63-linked-poly-Ub chains in the absence of an E3. Intriguingly, in the structure of the E3/E2/product mimic, Ubc9 Nup38/Ubc9/SUMO-RanGAP1 (PDB code 1Z5S) the now reacted lysine is observed at nearly the same angle as the about-to-be-reacted Lys63 just discussed (Figure 6) [20]. Whether the direction for substrate lysine approach for other E2s will be the same as the two examples currently available cannot be predicted, but it is tempting to speculate that the orientation observed represents one of perhaps several that allow for optimal reactivity. Access of substrate to the optimal approach path might also be modulated by E2~Ub dynamics in which the preferred route is blocked much of the time. In such a scenario, E3 binding could either limit the available E2~Ub conformations and/or stabilize the most reactive conformer, thereby effecting catalysis.
Another factor that may control thioester reactivity is the ability of the E2~Ub active site to favorably position the atoms of the tetrahedral intermediate—formation of which is likely the rate-limiting step of the transfer reaction. Computational calculations of transition state models of thioesters indicate that electron delocalization and the dihedral angles of the tetrahedral oxyanion intermediate contribute significantly to activation energies and hence thioester reactivity [89]. These factors have yet to be considered in the context of an E2 active site cradling a thioester, but it is likely that the geometry of the active site plays a role in the reactivity of Ub transfer by influencing the dihedral angles of the tetrahedral oxyanion intermediate. Since every E2~Ub structure is a snapshot of the reactant ester before lysine attack, it is virtually impossible to test the implications of steric dihedral angle regulation of the oxyanion intermediate by the E2 active site. Trapping the Ub transfer reaction at the oxyanion intermediate structurally will be challenging, but computational simulations of the intermediate in the context of an E2 active site may well provide insights.
Concluding Remarks & Future Perspectives
Over the past few years, investigation of the roles E2s play in the Ub transfer pathway have increased appreciation for their importance. We are beginning to understand the functional diversity the E2 family provides--a remarkable feat considering the high homology within this class of enzyme. In this review, we have highlighted the economy of E2s in their ability to facilitate many protein-protein interactions at nearly every surface on their compact ellipsoid Ubc domain. In fact, several surfaces are used for more than one protein-protein interaction including the E3/E1 binding site, as well as the backside Ub/E3 binding site. We have focused on non-canonical E2-protein interactions, and predict that new ones will be discovered, possibly involving the few remaining Ubc surfaces yet to be identified as mediating protein binding. Lastly, we stressed the importance of studying the conjugated E2~Ub, the active form of the enzyme, to fully inform function.
In the future, several discoveries could quickly advance the understanding of Ub transfer. Mechanistically, a structure of a RING E3/E2~Ub will be highly informative as such a structure may provide insight into the catalytic role of the RING E3, beyond its ability to recruit target substrates to an E2~Ub. As ~95% of E3s in the human genome are RING/UBox-type E3s (including multi-subunit ligases such as APC and SCF) [53], advancements in our understanding of RING-mediated Ub transfer stands to impact almost every field of biology. Second, there is a need to develop approaches that incorporate non-canonical E2/E3 interactions in efforts to comprehensively identify E2/E3 partners. Third, since traditional Y2H screens for E3/E2 pairs cannot parse novel interactions with the conjugated E2~Ub versus the free E2, new techniques are needed to uncover E2~Ub-specific interactions. The ability to identify binding partners for E2~Ubs (or a reasonable mimic of the conjugate) will likely result in the discovery of different and potentially more relevant E3 partners than have been found by screening with an E2 alone. Fourth, identification of non-Ub-pathway binding partners for E2s may provide important new breakthroughs in the future. A large-scale Y2H screen in which 39 full-length E2s were used as bait against high complexity prey libraries identified 229 total interactions, only 30% of which involved known E3 ligase domains [56]. Although the ~160 E2/non-E3 interactions remain to be confirmed, it is unlikely that such a large percentage of hits all correspond to false positives, suggesting that there are even more E2-binding partners than we might have been led to expect. The nature of these proteins and of their interactions with E2s is likely to be one of the newly emerging themes of E2 investigation in coming years.
E2s function at the center of the Ub transfer pathway, and provide much of the diversity the Ub signal transmits in the cell. From E3 selection to the type of Ub transfer (mono, poly, and the type of lysine linkage), the E2 is at the crux of substrate regulation, and arguably, many disease pathways. Considering this responsibility, it is perhaps not surprising that the human genome contains in excess of thirty-five E2 genes. Many of these have yet to be characterized even at a rudimentary level. There are sure to be more surprises on the horizon as new insights into the mechanisms of E2-catalyzed Ub transfer and a complete description of the cellular roles of E2s emerge.
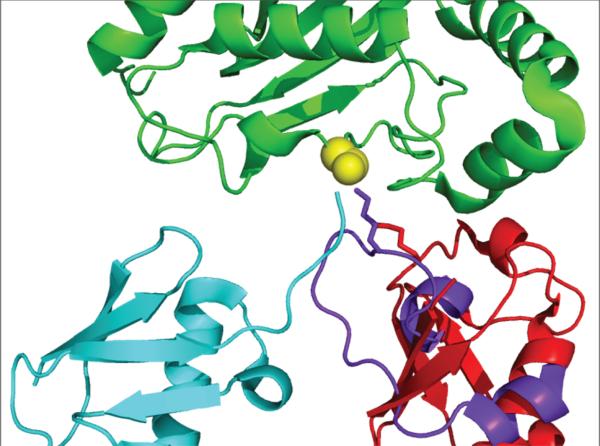
Figure 7.
Position of substrate lysine approach into the E2 active site. Lys63 of a putative substrate Ub in the formation of a Lys63-linked Ub chain observed in a structure of Ubc13~Ub/Mms2 (PDB code 2GMI) is shown in red stick representation. Substrate lysine residue involved in isopeptide linkage to Ubl (SUMO) observed in a structure of E2/E3/product (Ubc9/Nup358/SUMO-RanGAP1; PDB code 1Z5S) is shown in purple stick representation. The E2s of each structure were superimposed for this figure and shown in green, with active site Cys in yellow spheres (conjugated Ub in Ubc13~Ub is shown in cyan).
Acknowledgements
We thank P. Brzovic, N.Zheng, R.Gardner, and members of the Klevit lab for careful reading of this manuscript and many helpful comments.
Funding: We acknowledge support from the National Institute of General Medical Sciences in the form of 5R01 GM088055 (R. E. K.), T32 GM008268 (K. E. S.), and T32 GM07270 (D. M. W.), and the National Science Foundation in the form of MCB0615632 (R. E. K.)
Abbreviations used
- APC
Anaphase Promoting Complex
- BARD1
BRCA1-Associated RING domain
- BRCA1
Breast Cancer susceptibility protein 1
- Birc6
Baculoviral IAP repeat-containing protein 6
- Bmi1
B-cell-specific Moloney murine leukaemia virus integration site 1
- cCbl
Casitas B cell lymphoma
- CDC34
cell division cycle 34
- CHIP
carboxy terminus of Hsp70p-interacting protein
- CNOT4
CCR4-NOT transcription complex, subunit 4
- Cul1-Rbx1
cullin-1 ring-box 1
- E1
ubiquitin activating enzyme
- E2
ubiquitin conjugating enzyme
- E2~Ub
E2 conjugated to Ub
- E3
ubiquitin ligase
- E6AP
ubiquitin protein ligase E3A or Ube3A
- gp78
endoplasmic reticulum membrane spanning RING E3 ligase
- G2BR
Ube2g2 Binding Region
- HECT
Homologous to E6AP Carboxy Terminus ligase
- IκBα
Inhibitor of Kappa Light Chain Gene Enhancer in B cells alpha
- ITCH
itchy E3 ubiquitin protein ligase homolog
- Mms2
Methyl methanesulfonate sensitive 2
- NEDD4L
neural precursor cell expressed, developmentally down-regulated 4
- NIP45
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein
- NMR
nuclear magnetic resonance
- Nup358/RanBP2
RAN binding protein 2
- OspG
secreted by the Mxi-Spa secretion machinery
- PC2
chromobox homolog 4
- PDB
Protein Data Bank
- Rad5
Radiation sensitivity protein 5
- Rad6
Radiation sensitivity protein 6
- Rad18
Radiation sensitivity protein 18
- Rad60
Radiation sensitivity protein 60
- RENi'
protein family composed of Rad60, Esc2 and mouse NIp45
- RING
Really Interesting New Gene
- RSP5
E3 ubiquitin ligase of the NEDD4 family
- SCF
Cullin RING ubiquitin ligase
- SIM
SUMO-Interacting Motif
- SLD
SUMO-Like Domain
- SMURF2
SMAD specific E3 ubiquitin protein ligase 2
- SspH2
Salmonella secreted effector protein
- SUMO
Small Ubiquitin Like Modifier
- Tom1
target of myb1 E3 ubiquitin ligase of the HECT-domain class
- TRAF6
TNF-receptor-associated factor 6
- Tsg101
Tumor susceptibility protein 101
- Ub
ubiquitin
- UBA
Ubiquitin-Associated domain
- Ubl
Ubiquitin-like protein
- U-box
domain that is similar to a RING, but does not coordinate Zn
- Ubr1
Ubiquitin protein ligase E3 component n-recognin-1
- UEV
Ubiquitin conjugating Enzyme Variant
- Vps23
vacuolar protein sorting-associated protein 23
- Y2H
Yeast 2-Hybrid
References
- 1.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. J. Biol. Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 2.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 3.Roderigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–39. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 4.Windheim M, Peggie M, Cohen P. Two different classes of E2 ubiquitinconjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology. Biochem J. 2008;409:723–9. doi: 10.1042/BJ20071338. [DOI] [PubMed] [Google Scholar]
- 5.Tang Z, Hecker CM, Scheschonka A, Betz H. Protein interactions in the sumoylation cascade: lessons from X-ray structures. FEBS J. 2008;275:3003–15. doi: 10.1111/j.1742-4658.2008.06459.x. [DOI] [PubMed] [Google Scholar]
- 6.Christensen DE, Klevit RE. Dynamic interactions of proteins in complex networks: identifying the complete set of interacting E2s for functional investigation of E3-dependent protein ubiquitination. FEBS J. 2009;276:5381–9. doi: 10.1111/j.1742-4658.2009.07249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deshaies RJ, Joazerio CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 8.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–31. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye Y, Rape M. Building Ubiquitin Chains: E2 enzymes at work. Nature. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Wijk SJL, Timmers HTM. The family of ubiquitin-conjugating enzymes (E2s): deciding between live and death of proteins. FASEB J. 2010;24:981–993. doi: 10.1096/fj.09-136259. [DOI] [PubMed] [Google Scholar]
- 11.Burroughs AM, Jaffee M, Iyer LM, Aravind L. Anatomy of the E2 ligase fold: implications for enzymology and evolution of ubiquitin/Ub-like protein conjugation. J. Struct. Biol. 2008;162(2):205–218. doi: 10.1016/j.jsb.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang DT, Miller DW, Mathew R, Cassell R, Holton JM, Roussel MF, Schulman BA. A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat. Struct. Mol. Biol. 2004;10:927–935. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1. Mol. Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 14.Huang DT, Hunt HW, Zhuang M, Ohi MD, Holton JM, Schulman BA. Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature. 2007;45:394–398. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Hu W, Cai S, Lee B, Song J, Chen Y. The intrinsic affinity between E2 and the Cys domain of E1 in ubiquitin-like modifications. Mol. Cell. 2007;27:228–37. doi: 10.1016/j.molcel.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, Schulman BA. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 18.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 19.Dominguez C, Bonvin AM, Winkler GS, van Schaik FM, Timmers HT, Boelens R. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure. 2004;12:633–644. doi: 10.1016/j.str.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMORanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 22.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat. Struct. Mol. Biol. 2006;13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 23.Xu Z, Kohli E, Devlin KI, Bold M, Nix JC, Misra S. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct. Biol. 2008;8:26. doi: 10.1186/1472-6807-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mace PD, Linke K, Feltham R, Schumacher FR, Smith CA, Vaux DL, Silke J, Day CL. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J. Biol Chem. 2008;283:31633–31640. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- 25.Das R, Mariano J, Tsai YC, Kalathur RC, Kostova Z, Li J, Tarasov SG, McFeeters RL, Altieri AS, Ji X, Byrd RA, Weissman AM. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol. Cell. 2009;34:674–685. doi: 10.1016/j.molcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamadurai HB, Souphron J, Scott DC, Duda DM, Miller DJ, Stringer D, Piper RC, Schulman BA. Insights into Ubiquitin Transfer Cascades from a Structure of UbcH5B~Ubiquitin-HECTNEDD4L Complex. Mol. Cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, Tu D, Li L, Wollert T, Ghirlando R, Brunger AT, Ye Y. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3722–3727. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, Zheng L, Rich RL, Campos AD, Myszka DG, Lenardo MJ, Darnay BG, Wu H. E2 interaction and dimerization in the crystal structure of TRAF6. Nat. Struct. Mol. Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moraes TF, Edwards RA, McKenna S, Pastushok L, Xiao W, Glover JN, Ellison MJ. Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13. Nat. Struct. Biol. 2001;8:669–673. doi: 10.1038/90373. [DOI] [PubMed] [Google Scholar]
- 30.Van Demark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell. 2001;105:711–720. doi: 10.1016/s0092-8674(01)00387-7. [DOI] [PubMed] [Google Scholar]
- 31.Sundquist WI, Schubert HL, Kelly BN, Hill GC, Holton JM, Hill CP. Ubiquitin recognition by the human TSG101 protein. Mol. Cell. 2004;13:783–789. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- 32.Teo H, Veprintsev DB, Williams RL. Structural insights into endosomal sorting complex required for transport (ESCRT-I) recognition of ubiquitinated proteins. J. Biol. Chem. 2004;279:28689–96. doi: 10.1074/jbc.M400023200. [DOI] [PubMed] [Google Scholar]
- 33.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/Ub Noncovalent Complex Is Required for Processive BRCA1-Directed Ubiquitination. Mol. Cell. 2006;21:873–80. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Lewis MJ, Saltibus LF, Hau DD, Xiao W, Spyracopoulos L. Structural basis for non-covalent interaction between ubiquitin and the ubiquitin conjugating enzyme variant human MMS2. J. Biomol. NMR. 2006;34:89–100. doi: 10.1007/s10858-005-5583-6. [DOI] [PubMed] [Google Scholar]
- 35.Capili AD, Lima CD. Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J. Mol. Biol. 2007;369:608–618. doi: 10.1016/j.jmb.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duda DM, van Waardenburg RC, Borg LA, McGarity S, Nourse A, Waddell MB, Bjornsti MA, Schulman BA. Structure of a SUMO-binding-motif mimic bound to Smt3p-Ubc9p: conservation of a non-covalent ubiquitin-like protein-E2 complex as a platform for selective interactions within a SUMO pathway. J Mol Biol. 2007;369:619–630. doi: 10.1016/j.jmb.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 2007;26:2797–2807. doi: 10.1038/sj.emboj.7601711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamilton KS, Ellison MJ, Barber KR, Williams RS, Huzil JT, McKenna S, Ptak C, Glover M, Shaw GS. Structure of a Conjugating Enzyme-Ubiquitin thioester Intermediate Reveals a Novel Role for the Ubiquitin Tail. Structure. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 39.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 40.Serniwka SA, Shaw GS. The Structure of the UbcH8-Ubiquitin Complex Shows a Unique Ubiquitin Interaction Site. Biochemistry. 2009;4:12169–79. doi: 10.1021/bi901686j. [DOI] [PubMed] [Google Scholar]
- 41.Sakata E, Satoh T, Yamamoto S, Yamaguchi Y, Yagi-Utsumi M, Kurimoto E, Tanaka K, Wakatsuki S, Kato K. Crystal Structure of UbcH5b~Ubiqutin Intermediate: Insight into the Formation of the Self-Assembled E2~Ub Conjugates. Structure. 2010;18:138–147. doi: 10.1016/j.str.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Wu PY, Hanlon M, Eddins M, Tsui C, Rogers RS, Jensen JP, Matunis MJ, Weissman AM, Wolberger C, Pickart CM. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 2003;22:5241–50. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat. Struct. Mol. Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- 44.Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SF-Cdc34. Cell. 2005;123:1107–20. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 45.Gazdoiu S, Yamoah K, Wu K, Pan ZQ. Human Cdc34 employs distinct sites to coordinate attachment of ubiquitin to a substrate and assembly of polyubiquitin chains. Mol Cell Biol. 2007;27:7041–52. doi: 10.1128/MCB.00812-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michelle C, Vourc'h P, Mignon L, Andres CR. What Was the Set of Ubiquitin and Ubiquitin-Like Conjugating Enzymes in the Eukaryotic Common Ancestor? J. Mol. Evol. 2009;68:616–628. doi: 10.1007/s00239-009-9225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nuber U, Scheffner M. Identification of determinants in E2 ubiquitinconjugating enzymes required for hect E3 ubiquitin-protein ligase interaction. J. Biol. Chem. 1999;274:7576–82. doi: 10.1074/jbc.274.11.7576. [DOI] [PubMed] [Google Scholar]
- 48.Wooff J, Pastushok L, Hanna M, Fu Y, Xiao W. The TRAF6 RING finger domain mediates physical interaction with Ubc13. FEBS Lett. 2004;566:229–33. doi: 10.1016/j.febslet.2004.04.038. [DOI] [PubMed] [Google Scholar]
- 49.Ulrich HD. Protein-protein interactions within an E2-RING finger complex. Implications for ubiquitin-dependent DNA damage repair. J Biol Chem. 2003;278:7051–8. doi: 10.1074/jbc.M212195200. [DOI] [PubMed] [Google Scholar]
- 50.Albert TK, Hanzawa H, Legtenberg YI, de Ruwe MJ, van den Heuvel FA, Collart MA, Boelens R, Timmers HT. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 2002;337:157–65. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winkler GS, Albert TK, Dominguez C, Legtenberg YI, Boelens R, Timmers HT. An altered specificity ubiquitin-conjugating enzyme/ubiquitin-protein ligase pair. J Mol Bio. 2004;337:157–65. doi: 10.1016/j.jmb.2004.01.031. [DOI] [PubMed] [Google Scholar]
- 52.Winkler GS, Timmers HT. Structure-based approaches to create new E2–E3 enzyme pairs. Methods Enzymol. 2005;399:355–66. doi: 10.1016/S0076-6879(05)99024-1. [DOI] [PubMed] [Google Scholar]
- 53.Li W, Bengston MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazerio CA. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krämer OH, Zhu P, Ostendorff HP, Golebiewski M, Tiefenbach J, Peters MA, Brill B, Groner B, Bach I, Heinzel T, Göttlicher M. Histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22:3411–20. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc. Natl. Acad. Sci. 2009;106:18213–8. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Markson G, Kiel C, Hyde R, Brown S, Charalabous P, Bremm A, Semple J, Woodsmith J, Duley S, Salehi-Ashtiani K, Vidal M, Komander D, Serrano L, Lehner P, Sanderson CM. Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res. 2009;19:1905–11. doi: 10.1101/gr.093963.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Wijk SJ, de Vries SJ, Kemmermen P, Huang A, Boelens R, Bonvin AM, Timmers HT. A comprehensive framework of E2-RING E3 interactions of the human ubiquitin-proteasome system. Mol. Syst. Biol. 2009;5:1–16. doi: 10.1038/msb.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bailly V, Prakish S, Prakish L. Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enzyme and Rad18 DNA binding protein. Mol Cell Bio. 1997;17:4536–4543. doi: 10.1128/mcb.17.8.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levin I, Eakin C, Blanc MP, Klevit RE, Miller SI, Brzovic PS. Identification of an unconventional E3 binding surface on the UbcH5~Ub conjugate recognized by a pathogenic bacterial E3 ligase. Proc. Natl. Acad. Sci. U.S.A. 2010;107:2848–2853. doi: 10.1073/pnas.0914821107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi YS, Jeon YH, Ryu KS, Cheong C. 60th residues of ubiquitin and Nedd8 are located out of E2-binding surfaces, but are important for K48 ubiquitin-linkage. FEBS Lett. 2009;583:3323–8. doi: 10.1016/j.febslet.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 61.Prudden J, Perry JJ, Arvai AS, Tainer JA, Boddy MN. Molecular mimicry of SUMO promotes DNA repair. Nat. Struct. Mol. Biol. 2009;16:509–16. doi: 10.1038/nsmb.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sekiyama N, Arita K, Ikeda Y, Hashiguchi K, Ariyoshi M, Tochio H, Saitoh H, Shirakawa M. Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45. Proteins. 2010;78:1491–502. doi: 10.1002/prot.22667. [DOI] [PubMed] [Google Scholar]
- 63.Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc. Natl. Acad. Sci. U.S.A. 2006;103:341–6. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin Y, Hwang WC, Basavappa R. Structural and functional analysis of the human mitotic-specific ubiquitin-conjugating enzyme, UbcH10. J Biol Chem. 2002;277:21913–21. doi: 10.1074/jbc.M109398200. [DOI] [PubMed] [Google Scholar]
- 66.Merkley N, Shaw GS. Solution structure of the flexible class II ubiquitinconjugating enzyme Ubc1 provides insights for polyubiquitin chain assembly. J Biol Chem. 2004;279:47139–47. doi: 10.1074/jbc.M409576200. [DOI] [PubMed] [Google Scholar]
- 67.Wilson RC, Hughes RC, Flatt JW, Meehan EJ, Ng JD, Twigg PD. Structure of full-length ubiquitin-conjugating enzyme E2-25K (huntingtin-interacting protein 2) Acta Cryst. Sect. F Struct. Biol. Cryst. Commun. 2009;65:440–444. doi: 10.1107/S1744309109011117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie Y, Varshavsky A. The E2–E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 1999;18:6832–44. doi: 10.1093/emboj/18.23.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2–E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139:957–68. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kleiger G, Hao B, Mohl DA, Deshaies RJ. The acidic tail of Cdc34 ubiquitin-conjugating enzyme functions in both binding to and catalysis with ubiquitin ligase SCFCdc4. J Biol Chem. 2009;284:36012–23. doi: 10.1074/jbc.M109.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Choi YS, Wu K, Jeong K, Lee D, Jeon YH, Choi BS, Pan ZQ, Ryu KS, Cheong C. The human Cdc34 carboxyl terminus contains a non-covalent ubiquitin binding activity that contributes to SCF-dependent ubiquitination. J. Biol. Chem. 2010;285:17754–62. doi: 10.1074/jbc.M109.090621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Summers MK, Pan B, Mukhyala K, Jackson PK. The unique N terminus of the UbcH10 E2 enzyme cotnrols the threshold for APC activation and enhances checkpoint regulation of the APC. Mol Cell. 2008;31:544–56. doi: 10.1016/j.molcel.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Plafker KS, Singer JD, Pflafker SM. The ubiquitin conjugating enzyme, UbcM2, engages in novel interactions with components of cullin-3 based E3 ligases. Biochemistry. 2009;48:3527–37. doi: 10.1021/bi801971m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Siepmann TJ, Bohnsack RN, Tokgoz Z, Baboshina OV, Hass AL. Protein Interactions within the N-end Rule Ubiquitin Ligation Pathway. J. Biol. Chem. 2003;278:9448–9457. doi: 10.1074/jbc.M211240200. [DOI] [PubMed] [Google Scholar]
- 75.Miura T, Klaus W, Gsell B, Miyamoto C. Characterization of the Binding Interface between Ubiquitin and Class I Human Ubiquitin-conjugating Enzyme 2b by Multidimensional Heteronuclear NMR Spectroscopy in Solution. J. Mol. Biol. 1999;290:213–228. doi: 10.1006/jmbi.1999.2859. [DOI] [PubMed] [Google Scholar]
- 76.McKenna S, Mores T, Pastushok L, Ptak C, Xiao W, Spyracopoulous L, Ellison MJ. An NMR-based Model of the Ubiquitin-bound Human Ubiquitin Conjugation Complex Mms2 Ubc13. J. Biol. Chem. 2003;278:13151–158. doi: 10.1074/jbc.M212353200. [DOI] [PubMed] [Google Scholar]
- 77.Houben K, Dominguez C, van Schaik FM, Timmers HT, Bonvin AM, Boelens R. Solution structure of the ubiquitin-conjugating enzyme UbcH5B. J. Mol. Biol. 2004;344:513–26. doi: 10.1016/j.jmb.2004.09.054. [DOI] [PubMed] [Google Scholar]
- 78.Ju T, Bocik W, Majumdar A, Tolman JR. Solution structure and dynamics of human ubiquitin conjugating enzyme Ube2g2. Proteins. 2009;78:1291–1301. doi: 10.1002/prot.22648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang S, Sharrocks AD. The SUMO E3 ligase activity of Pc2 is coordinated through a SUMO-interaction motif. Mol. Cell. Biol. 2010;30:2193–205. doi: 10.1128/MCB.01510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.French ME, Kretzmann BR, Hicke L. Regulation of the RSP5 ubiquitin ligase by an intrinsic ubiquitin-binding site. J Biol Chem. 2009;284:12071–9. doi: 10.1074/jbc.M901106200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ogunjimi AA, Wiesner S, Briant DJ, Varelas X, Sicheri F, Forman-Kay J, Wrana JL. The ubiquitin binding region of the Smurf HECT domain facilitates polyubiquitylation and binding of ubiquitylated substrates. J Biol Chem. 2010;285:6308–15. doi: 10.1074/jbc.M109.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Purbeck C, Eletr ZM, Kuhlman B. Kinetics of the transfer of ubiquitin from UbcH7 to E6AP. Biochemistry. 2010;49:1361–63. doi: 10.1021/bi9014693. [DOI] [PubMed] [Google Scholar]
- 83.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eletr ZM, Kuhlman B. Sequence determinants of E2-E6AP binding affinity and specificity. J Mol Biol. 2007;369:419–28. doi: 10.1016/j.jmb.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Plafker SM, Plafker KS, Weissman AM, Macara IG. Ubiquitin charging of human class III ubiqutin-conjugating enzymes triggers their nuclear import. J. Cell Biol. 2004;167:649–659. doi: 10.1083/jcb.200406001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim DW, Lenzen G, Page AL, Legrain P, Sansonetti PJ, Parsot C. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Song J, Wang J, Jozwiak A, Hu W, Swiderski PM, Chen Y. Stability of thioester intermediates in ubiquitin-like modifications. Protein Sci. 2009;18:2492–9. doi: 10.1002/pro.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pickart CM, Rose IA. Functional Heterogeneity of Ubiquitin Carrier Proteins. J. Biol. Chem. 1985;260:1573–81. [PubMed] [Google Scholar]
- 89.Yang W, Drueckhammer DG. Understanding the relative acyl-transfer reactivity of oxoesters and thioesters: computational analysis of transition state delocalization effects. J. Am. Chem. Soc. 2001;123:11004–11009. doi: 10.1021/ja010726a. [DOI] [PubMed] [Google Scholar]