Abstract
Mediator complex subunit 29 (MED29) is part of a large multiprotein coactivator complex that mediates regulatory signals from gene-specific activators to general transcription machinery in RNA polymerase II mediated transcription. We previously found that MED29 is amplified and overexpressed in pancreatic cancer and that MED29 silencing leads to decreased cell survival in PANC-1 pancreatic cancer cells with high MED29 expression. Here we further demonstrate decreased migration, invasion and colony formation in PANC-1 cells after MED29 silencing. Unexpectedly, lentiviral-based overexpression of MED29 led to decreased proliferation of NIH/3T3 cells as well as MIAPaCa-2 pancreatic cancer cells with low endogenous expression. More importantly, subcutaneous inoculation of the MED29-transduced pancreatic cancer cells into immuno-compromised mice resulted in dramatic tumor suppression. The mock-control mice developed large tumors, whereas the animals with MED29-xenografts showed both decreased tumor incidence and a major reduction in tumor size. Gene expression analysis in the MED29-transduced pancreatic cancer cells revealed differential expression of genes involved in control of cell cycle and cell division. The observed gene expression changes are expected to modulate the cell cycle in a way that leads to reduced cell growth, explaining the in vivo tumor suppressive phenotype. Taken together, these data implicate MED29 as an important regulator of key cellular functions in pancreatic cancer with both oncogenic and tumor suppressive characteristics. Such a dualistic role appears to be more common than previously thought and is likely to depend on the genetic background of the cancer cells and their surrounding environment.
Keywords: pancreatic cancer, MED29, mediator complex, oncogene, tumor suppression, cell cycle regulation
Introduction
Pancreatic cancer is a highly aggressive malignancy and one of the leading causes of cancer deaths. The disease is typically diagnosed at an advanced stage when curative surgery is no longer possible and other currently available treatment options are also largely ineffective. Therefore, there is an urgent need for better understanding of pancreatic cancer pathogenesis that might lead to more effective treatment strategies.
Transcriptional regulation is one of the most fundamental steps in controlling cellular growth and differentiation.1 In eukaryotes, there are three main polymerase (Pol) enzymes accounting for most DNA transcription. Pol I and III are responsible for transcription of non-coding genes, such as those encoding for tRNAs, rRNAs, and various small RNAs, whereas Pol II transcribes all coding mRNAs.2 Eukaryotic Pol II cannot initiate transcription on its own but needs general transcription factors that help to position Pol II on the gene promoter, the start site for initiation of transcription. General transcription factors including TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH assemble on all Pol II driven gene promoters and are thus necessary for Pol II mediated transcription.3 This process can be further facilitated or repressed by gene-specific activators or repressors, respectively. Gene-specific activator or repressor proteins bind distant from the transcriptional start site and mediate their signal via coregulatory complexes such as the Mediator complex.2,4
The Mediator complex is a large multiprotein coactivator that was originally identified in Saccharomyces cerevisiae when it was discovered that activators and the general transcription machinery alone were not sufficient to activate transcription in vitro.5 Mediator acts as a link between transcriptional activators and the general transcription machinery.6–7 More specifically, Mediator is recruited to gene promoters at the preinitiation stage of transcription to facilitate the formation of the preinitiation complex (PIC).8 The PIC is an inactive state of Pol II and general transcription factors that still needs an additional conformational change to initiate transcription. Mediator has been reported to be essential for both activated and basal transcription. There are supporting4,9 and opposing evidence10–11 of Mediator acting as a general transcription factor. In other words, there is no definitive consensus on the role of Mediator in transcription.
The structure and composition of the Mediator complex has been studied extensively and mammalian Mediator is known to contain up to 30 different subunits.12–17 Structural studies, mainly from Saccharomyces cerevisiae, suggest that the Mediator subunits are organized into three distinct submodules: the head, middle, and tail.16,18 The MED29 (previously known as IXL) subunit is located in the head module of the Mediator complex but its functional role is less well characterized.19 Human MED29 was originally identified from an embryonic heart cDNA library.20 It is a homolog to Drosophila melanogaster intersex which is a transcriptional regulator involved in female somatic sex determination.21 MED29 is one of the most highly conserved proteins across species and is widely expressed in humans both during embryonic development and in adult tissues,20 thereby supporting its essential role in transcriptional regulation.
We and others have previously shown that MED29 is recurrently amplified and overexpressed in pancreatic cancer.22–23 We also demonstrated that its silencing leads to decreased cell survival and increased apoptosis specifically in cells with MED29 amplification,22 implying that these cells are dependent on MED29 overexpression. In this study, we further explored the functional role of MED29 in pancreatic cancer by generating cell lines with stable MED29 overexpression. A genome-wide expression analysis was performed to systematically identify the global transcriptional effects of MED29 overexpression, and mice xenografts were created to explore the in vivo consequences of aberrant MED29 levels.
Materials and methods
Cell lines
Human pancreatic (PANC-1, SU.86.86, Hs 700T, MIA PaCa-2) and breast cancer cell lines (BT-474, MCF7), embryonic kidney cells (HEK 293T/17), and mouse embryonic fibroblasts (NIH/3T3) were obtained from the American Type Culture Collection (Manassas, VA) and grown under recommended conditions. Bacterial TOP10 Escherichia coli cells were obtained from Invitrogen (Carlsbad, CA).
siRNA transfection
ON-TARGETplus SMART siRNA pool containing a mixture of four gene-specific siRNAs for MED29 was obtained from Dharmacon (Lafayette, CO). A control siRNA targeting the firefly luciferase gene (LUC) was purchased from Sigma (St Louis, MO). siRNA transfection was performed using Interferin transfection reagent according to the manufacturer's instructions (Polyplus Transfection, San Marcos, CA). The efficiency of MED29 silencing was confirmed in each experiment using qRT-PCR.
Lentiviral constructs
MED29 cDNA clone in pT-REx-DEST31 plasmid (Invitrogen) was grown in TOP10 E. coli cells (Invitrogen) and purified using QIAfilter Plasmid Maxi Kit (Qiagen, Valencia, CA). MED29 insert was isolated by PCR amplification with two pairs of restriction primers, producing gene products of 863 bp (MED29-1) and 675 bp (MED29-2). PCR products were sequenced (ABI Prism 3100 DNA Sequencer, Applied Biosystems, Foster City, CA) and confirmed to contain the entire MED29 coding sequence. BamHI and NedI restriction sites were used to clone the inserts into the pWPI plasmid, which was co-transfected with Delta 8.9 and VSVG plasmids into HEK 293T/17 cells. The viral supernatant was harvested, filtered, and concentrated by centrifugation. Mock constructs were constructed similarly, only without the MED29 insert. The viral concentrate was diluted in polybrene to infect NIH/3T3, Hs 700T, and MIA PaCa-2 cells. A successful transduction was confirmed by visualizing GFP (included in the pWPI vector) and sustained MED29 expression was confirmed at least every two weeks by qRT-PCR. All cell lines were established from pooled clones.
Animals and tumor models
Six-week-old male athymic nu/nu mice were purchased from Harlan (the Netherlands). The total number of mice was 40. Hs700T/MED29-1, Hs700T/mock, MIAPaCa2/MED29-1, and MIAPaCa2/mock cells were inoculated subcutaneously into the flanks of the mice. Animal welfare was monitored daily for clinical signs. Tumor measurements were performed twice a week and the tumor volume was counted according to the formula of V=(π/6)(d1×d2)3/2, where d1 and d2 are perpendicular tumor diameters. Mice were weighed and photographed once a week. Hs 700T and MIA PaCa-2 tumor bearing mice were sacrificed 7 and 8 weeks after inoculation, respectively. Tumors were exposed and photographed. The final size of the tumors was measured with a caliper and the volume calculated as described.24 The animal experiments were carried out according to the European Convention for the Protection of Vertebrate Animals used for Experimental and other Scientific Purposes, Statutes 1076/85 § and 1360/90 of The Animal Protection Law in Finland, and EU Directive 86/609. The experimental procedures were reviewed by the local Ethics Committee on Animal Experimentation at the University of Turku and approved by the local Provincial State Office of Western Finland.
Quantitative RT-PCR
Gene expression analyses were performed using the Light Cycler qRT-PCR equipment (Roche Applied Science). Total cell line RNA was isolated using RNeasy Mini kit (Qiagen). Tumor RNA was extracted by mechanical lysis using MagNA Lyser instrument (Roche Diagnostics GmbH, Mannheim, Germany) and further purification by RNeasy Mini kit (Qiagen). Superscript III reverse transcriptase (Invitrogen) was used for cDNA transcription. Expression levels were normalized against housekeeping genes TBP (TATA box binding protein) or GUSB (glucuronidase beta). Primers and probe sets for MED29 and TBP were obtained from TIB MolBiol (Berlin, Germany). The Universal ProbeLibrary gene assay (Roche Applied Science) was used for GUSB.
MED29 sequencing
PANC-1 and SU.86.86 cDNA was first amplified using primers: F-cgggatccatgctgaaaagcaacgggga and R-ggaattccatatgctacagagtgcccccag. The PCR reaction was performed using 1 μl cDNA, 5 μl of 10× PCR Gold Buffer, 4 μl of 25μM MgCl2, 1 μl of dNTPs, 1 μl each of 10 μM sense and antisense primers, and 0.3 μl Amplitaq Gold DNA polymerase (Applied Biosystems, Foster City, CA). The reaction was subjected to 35 cycles of denaturation at 95°C for 30 seconds, annealing at 55°C for 1 minute, and extension at 72° for 1 minute. PCR products were purified using Qiagen QIAquick PCR Purification Kit according to the manufacturer's protocol (Qiagen Inc.). The cycle sequencing reactions were carried out using Big Dye Terminator chemistry (Applied Biosystems) according to the manufacturer's protocol and the sequencing was carried out using ABI 3100 sequencer (Applied Biosystems).
Western blot
Cells were lysed, separated by SDS-PAGE and blotted as previously described.25 Immunodetection was performed using a custom-made MED29 primary antibody (GenWayBiotech, San Diego, CA) and β-Tubulin 1 (Sigma-Aldrich Chemie GmbH) was used as a loading control. The proteins were visualized by using a BM chemiluminescence western blotting kit (Roche Diagnostics GmbH).
Cell proliferation and serum starvation assays
Proliferation assays were performed on 24-well plates and the cells were counted 24–120h after plating using the Coulter Counter (Beckman Coulter, Fullerton, CA). Each assay contained six replicates and was repeated at least twice. In serum starvation experiments, NIH3T3/MED29 cells were grown in medium containing 1% FBS. For synchronization experiments, MIAPaCa2/MED29 cells were starved in serum-free medium for 72h and their accumulation to the G1-phase of the cell cycle was confirmed using flow cytometry. Regular growth medium was then given to the cells and their growth was monitored for 96h as indicated above.
Cell migration and invasion assays
Migration was studied by using cell culture migration chambers (BD Biosciences, Bedford, MA) and invasion by using matrigel-coated invasion chambers (BD Biosciences) according to the manufacturer's instructions. Images were captured from each membrane with Aperio ScanScope® XT (software version 9; Aperio Technologies, USA) using a lossless image format as primary output. The total area of migrated cells on a single membrane was determined from four images by quantitation with the ImageJ software (http://rsb.info.nih.gov/ij/). Each assay was performed in six replicates and repeated at least twice.
Soft agar assay
Cells were grown on 0.3% agarose on 6-well plates for 14 days. Images (six images per well) were captured with Olympus IX71 microscope (Olympus Corporation, Tokyo, Japan) using Capture Pro 6.0 program and the number and total area of colonies were quantified by ImageJ software.
Statistical analyses
The Mann-Whitney test was used to statistically compare the medians of the different groups in all experiments above.
Microarray-based gene expression profiling
Total RNA was extracted from MED29-overexpressing MIA PaCa-2 and Hs 700T, and corresponding mock cells as described above. A 2100 Bioanalyzer (Agilent, Santa Clara, CA) was used to measure the quantity and integrity of the RNA. Sample (MED29) RNA was labeled with Cy5 and mock RNA with Cy3, and they were co-hybridized onto 44K whole human genome oligonucleotide microarrays (Agilent) according to manufacturer's two-color microarray based gene expression protocol. A total of six arrays (two biological replicates corresponding to the two ME29 constructs and three technical replicates) were done per cell line. The microarray slides were scanned by Agilent microarray scanner.
Microarray data analysis
Agilent Feature Extraction software (version 10.5.1.1) was used to transform the microarray image into spot intensity data for the analysis. The preprocessing and analysis of the data was done using the Limma package of Bioconductor.26–27 The raw data were preprocessed by filtering out the spots flagged by the software as controls, saturated, non-uniformity outliers or population outliers. Preprocessing of the data was continued through background correction by normexp method with offset 5028 and normalization by loess normalization method.29 The log ratio of normalized red (MED29) and green (mock) intensity values was used as the representative value for each given probe. Replicate probes representing the same mRNA were removed by taking their average. Differentially expressed genes were identified by means of empirical Bayes with Benjamini-Hochberg adjustment for P-values.30–31 The thresholds for differential expression were set at fold change of 1.5 and adjusted P-value of 0.05. Clustering of the samples by hierarchical clustering method with correlation distance and average linkage was conducted to examine the linkages between the samples. The cellular networks and canonical pathways were generated through the use of Ingenuity Pathway Analysis (Ingenuity® Systems, www.ingenuity.com). DAVID bioinformatics resources32–33 were used to find enriched gene ontologies (molecular function, biological process, and cellular component) among the differentially expressed genes.
Human Cell Cycle RT-PCR Array
Ready-to-use Profiler PCR plates (SA Biosciences, Frederick, MD) containing 84 cell cycle related genes and 5 housekeeping genes were used. RNA from the cells and mice xenografts was extracted as described above with an additional DNase treatment step. Total RNA was transcribed into cDNA and PCR amplified according to manufacturer's instructions. Experiments were performed using the Bio-Rad CFX96 equipment (Bio-Rad, Hercules, CA) and analyzed with the PCR Array data analysis protocol (SA Biosciences). Expression values for each gene were normalized using the average of the five housekeeping genes on the same array.
Results
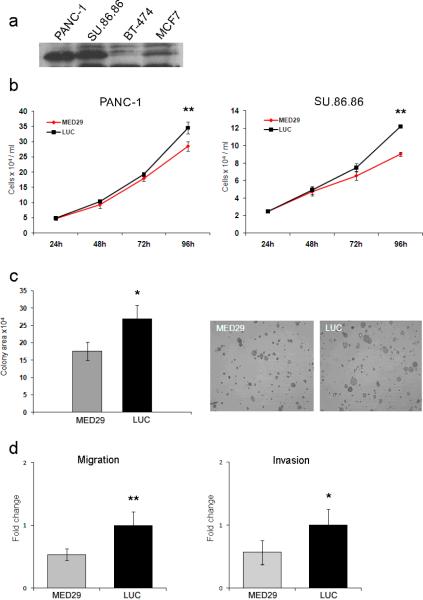
We previously reported the occurrence of MED29 amplification and concomitant high-level mRNA expression in a subset of pancreatic cancer cell lines and primary tumors.22 To extend these findings, we now demonstrate that MED29 amplification leads to elevated protein expression in PANC-1 and SU.86.86 cells compared to cells without amplification (Fig. 1a). We also searched for possible MED29 splice variants in these pancreatic cancer cells with high mRNA expression but sequencing only revealed a single MED29 transcript corresponding to that (NM_017592) in public databases. Our previous data revealed decreased survival, manifesting as a G1 cell cycle arrest and increased apoptosis, after MED29 silencing in PANC-1 cells with amplification.22 To extend these findings, we first confirmed the reduced cell growth after MED29 siRNA treatment in PANC-1 cells (an 18% average reduction in cell number as compared to control non-silencing siRNA, P=0.0022) and then showed that similar growth suppression is also evident in another MED29 amplified cell line, SU.86.86 (26% growth reduction, P=0.0022, Fig. 1b). In addition, MED29 protein levels were greatly reduced by siRNA treatment (data not shown). In addition to the altered cell growth, MED29 silencing also inhibited colony formation of PANC- 1 cells when grown in soft agar (Fig. 1c). Both the size and the number of colonies was smaller in the MED29 silenced cells compared to cells treated with control siRNA (P=0.0087). Cell migration (a 47% average reduction, P=0.0022) and invasion (a 43% average reduction, P=0.015) were also significantly compromised in PANC-1 cells after MED29 silencing (Fig. 1d). The efficacy of gene silencing was monitored by qRT-PCR during each experiment and an average of 80–85% downregulation in MED29 mRNA level was observed 48 hours after transfection, with this effect lasting at least 96 hours. Taken together, silencing of MED29 expression in pancreatic cancer cells with high endogenous expression due to gene amplification leads to attenuation of several cancer-associated phenotypic characteristics.
Figure 1.
MED29 silencing leads to reduced cell growth, colony formation, migration, and invasion in pancreatic cancer cells with high endogenous expression. (a) Western blot analysis of MED29 protein (21 kDa) levels in PANC-1 and SU.86.86 pancreatic cancer cell lines with amplification and BT-474 and MCF7 breast cancer cell lines without amplification. (b) PANC-1 and SU.86.86 cells were treated with MED29 or luciferase control (LUC) siRNAs and the number of cells (mean and s.d. of six replicates) was determined at indicated time points. (c) PANC-1 cells were first treated with MED29 or LUC siRNAs for 24h and then grown on 0.3% agarose on 6-well plates for 14 days. The colony formation was quantified by ImageJ software as the average area of the colonies per field counted from four separate images per sample. Representative images of MED29 and LUC siRNA treated cells are shown. (d) PANC-1 cells were treated with MED29 or LUC siRNA for 24h and then seeded on the upper part of migration and invasion chambers. The number of migrated (left panel) and invaded (right panel) cells was quantified at 22h and is presented as fold changes. The error bars represent s.d. of six replicates. Each experiment was repeated at least two times. *P < 0.05, **P < 0.005.
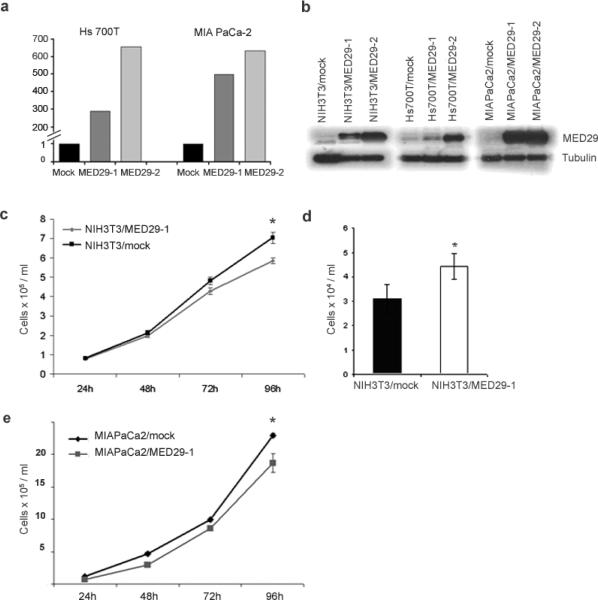
To study the effects of forced MED29 expression in cells with no or low endogenous expression, stable MED29-overexpressing cell lines were generated using a lentiviral based system. MED29 was transduced into NIH/3T3 cells (referred to as NIH3T3/MED29) as well as into Hs 700T (Hs700T/MED29-1 and Hs700T/MED29-2) and MIA PaCa-2 (MIAPaCa2/MED29-1 and MIAPaCa2/MED29-2) pancreatic cancer cells. A highly increased MED29 mRNA and protein expression was achieved compared to empty vector (mock) controls (Fig. 2a, b). The persistence of the increased expression level was monitored regularly during the maintenance of the cells and at the end of each experiment by qRT-PCR. Based on the visualization of GFP (which was expressed in line with MED29), MED29 was mainly localized in the nucleus and to a smaller extend in the cytoplasm (data not shown), similarly to what has been previously reported.20 We then followed the growth of the cells for four days and observed that MED29 expression led to decreased number of NIH/3T3 cells (up to a 17% reduction compared to mock control, P=0.0022) when cells were cultured in regular growth medium (Fig. 2c). Interestingly, in low serum (1% FBS) conditions, the NIH3T3/MED29 cells showed improved survival (30% increase in cell number, P=0.0286) compared to mock transfected cells (Fig. 2d). At first it appeared that MED29 expression did not have any consistent effect on the growth of the Hs 700T or MIA PaCa-2 cells in vitro (data not shown). However, when MIAPaCa2/MED29 cells were synchronized to G1-phase of the cell cycle prior to growth curve analysis, an up to 30% growth reduction as compared to the mock cells was observed (Fig. 2e). Unfortunately, we were unable to synchronize the Hs700T/MED29 cells. To conclude, forced MED29 expression has a context-dependent effect on the proliferation of mice fibroblasts and also influences the proliferation of human pancreatic cancer cells.
Figure 2.
MED29 expression leads to reduced growth of NIH/3T3 cells in vitro. NIH/3T3 mouse fibroblast cell line and Hs 700T and MIA PaCa-2 human pancreatic cancer cell lines were stably transfected with MED29 or an empty control vector. (a) Relative MED29 mRNA expression levels were assessed by qRT-PCR in MED29 transduced Hs 700T and MIA PaCa-2 cells and their respective empty vector (mock) control cells. MED29 expression values were normalized against a TATA-box binding protein house-keeping gene. (b) Western blot was used to detect MED29 protein (21 kDa) in stable MED29-expressing cells vs. mock control cells. β-Tubulin was used as a loading control. (c) The growth of the NIH3T3/MED29-1 and NIH3T3/mock cells was monitored at indicated time points in normal growth medium and (d) after 24h of serum starvation (1% FBS). (e) The growth of the MIAPaCa2/MED29 cells were monitored at indicated time points after release from G1-arrest. The mean +/− s.d. of six replicates are shown. The experiments were repeated three times with similar results. *P < 0.05, **P < 0.005.
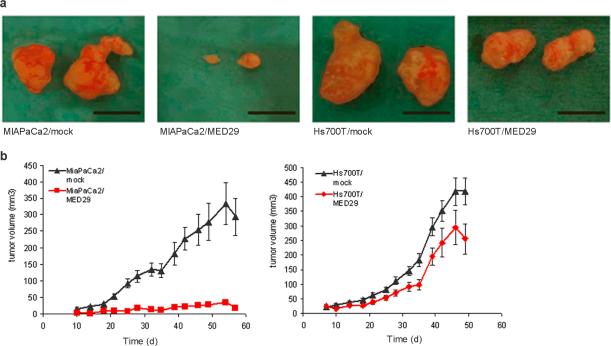
To study the possible role of MED29 in in vivo tumor formation, Hs700T/MED29-1 and MIAPaCa2/MED29-1 cells and corresponding mock cells were inoculated into the flanks of nude mice and allowed to grow for seven (Hs 700T) or eight (MIA PaCa-2) weeks. The tumor occurrence was 100%, 100%, 60%, and 90% in Hs700T/MED29, Hs700T/mock, MIAPaCa2/MED29, and MIAPaCa2/mock bearing mice, respectively. In addition, there was a striking difference in tumor size between MIAPaCa2/MED29 and MIAPaCa2/mock mice (Fig. 3a, 3b). The MIAPaCa2/mock tumors had a clear exponential growth pattern starting from day 20 of the experiment whereas MIAPaCa2/MED29 tumors showed hardly any increase in size throughout the entire observation period (Fig. 3b). At the time of sacrifice, the average volume of the MIAPaCa2/mock tumors was 13 times higher than that of the MIAPaCa2/MED29 tumors (194 mm3 vs. 15 mm3). A similar but less dramatic reduction in tumor growth was also seen in the Hs700T/MED29 mice (Fig. 3b) and the average size of the Hs700T/mock tumors at the end of the study was 1.3-fold higher compared to Hs700T/MED29 tumors (292 mm3 vs. 231 mm3). All tumors showed vascularized morphology under macroscopic examination (Fig. 3a), thereby implying the presence of functional angiogenesis. Finally, we confirmed sustained MED29 expression in the tumors derived from MED29 transduced cells (data not shown). These data indicate clearly that MED29 expression leads to tumor suppression in vivo.
Figure 3.
MED29 expression inhibits tumor growth in vivo. A total of 2.5 × 106 cells (MIAPaCa2/MED29, MIAPaCa2/mock, Hs700T/MED29, and Hs700T/mock) were s.c. inoculated into the flanks of nude mice (n=40) and allowed to grow for up to seven (Hs 700T) or eight (MIA PaCa-2) weeks. (a) Photographs of MIAPaCa2/MED29, Hs700T/MED29, and the corresponding mock tumors taken after the 7–8 weeks of growth in nude mice. The black scale bar corresponds to 1 cm. (b) Tumor diameters were measured twice a week and the growth curves of the Hs 700T and MIA PaCa-2 tumors were generated according to the formula V=(π/6)(d1×d2)3/2, where d1 and d2 are perpendicular tumor diameters. Average tumor volumes +/− s.e.m. are shown.
To get an insight into the mechanisms of MED29 action, we examined global gene expression levels in the MED29 transduced Hs 700T and MIA PaCa-2 pancreatic cancer cells compared to the mock cells using Agilent 44K whole human oligonucleotide microarrays. A total of 463 differentially expressed genes (≥1.5-fold change with adjusted P-value <0.05) were identified (Supporting Information Table S1). Overall, there were more downregulated genes (65%) than upregulated genes (35%) in the MED29 transduced cells. Unsupervised hierarchical clustering of the differentially expressed genes revealed common up- and downregulated gene clusters between MIAPaCa2/MED29 and Hs700T/MED29 cells (Supporting Information Figure S1, Table 1). As expected, the biological and technical replicates of each cell line clustered together (Supporting Information Figure S1).
Table 1.
List of genes commonly up- or downregulated in MIAPaCa2/MED29 and Hs700T/MED29 cells according to the hierarchical clustering.
| Common upregulated genes | Common downregulated genes | ||||
|---|---|---|---|---|---|
| ID | Symbol | GeneName | ID | Symbol | GeneName |
| NM_001002236 | SERPINA1 | serpin peptidase inhibitor, clade A, memeber 1 | NM_002263 | KIFC1 | kinesin family member C1 |
| NM_003641 | IFITM 1 | interferon induced transmembrane protein 1 | NM_178448 | C9orf140 | chromosome 9 open reading frame 140 |
| NM_018273 | TMEM143 | transmembrane protein 143 | NM_001255 | CDC20 | cell division cycle 20 homolog |
| XR_015904 | XR_015904 | pseudogene | NM_202002 | FOXM 1 | forkhead box M1 |
| NM_001001891 | ANO7 | anoctamin 7 | NM_022908 | NT5DC2 | 5'-nucleotidase domain containing 2 |
| ENST00000377652 | ENST00000377652 | not known | NM_004217 | AURKB | aurora kinase B |
| NM_001039876 | C19orf46 | chromosome 19 open reading frame 46 | NM_002168 | IDH2 | isocitrate dehydrogenase 2 (NADP+) |
| NM_173502 | PRSS36 | protease, serine, 36 | NM_001211 | BUB1B | budding uninhibited by benzimidazoles 1 homolog beta |
| NM_004921 | CLCA3P | chloride channel accessory 3 | NM_000674 | ADORA1 | adenosine A1 receptor |
| A_24_P486924 | A_24_P486924 | not known | XR_015516 | XR_015516 | not known |
| NM_012168 | FBXO2 | F-box protein 2 | NM_003088 | FSCN1 | fascin homolog 1, actin-bundling protein |
| NM_001080401 | PPM 1N | protein phosphatase, Mg2+/Mn2+ dependent, 1N | NM_001569 | IRAK1 | interleukin-1 receptor-associated kinase 1 |
| NM_002291 | LAMB1 | laminin, beta 1 | NM_138578 | BCL2L1 | BCL2-like 1 |
| NM_005384 | NFIL3 | nuclear factor, interleukin 3 regulated | XR_018953 | XR_018953 | not known |
| NM_133646 | ZAK | sterile alpha motif and leucine zipper containing kinase AZK | A_24_P247303 | A_24_P247303 | not known |
| NM_182734 | PLCB1 | phospholipase C, beta 1 | XR_018938 | XR_018938 | not known |
| NM_022725 | FANCF | Fanconi anemia, complementation group F | A_24_P551530 | A_24_P551530 | not known |
| NM_001032374 | ZNF226 | zinc finger protein 226 | XR_019369 | LOC652524 | similar to Keratin, type II cytoskeletal 8 |
| NM_182734 | PLCB1 | phospholipase C, beta 1 | AK091178 | AK091178 | not known |
| NM_015192 | PLCB1 | phospholipase C, beta 1 | A_24_P93111 | A_24_P93111 | not known |
| CD242823 | CD242823 | not known | NM_005550 | KIFC3 | kinesin family member C3 |
| NM_012301 | MAGI2 | membrane associated guanylate kinase | A_24_P6850 | A_24_P6850 | not known |
| NM_015469 | NIPSNAP3A | nipsnap homolog 3A | A_24_P230486 | A_24_P230486 | not known |
| NM_018245 | OGDHL | oxoglutarate dehydrogenase-like | A_24_P409420 | A_24_P409420 | not known |
| AK093508 | AK093508 | hypothetical LOC100131938 | XR_018749 | XR_018749 | not known |
| BX112111 | BX112111 | not known | XR_019186 | LOC651929 | similar to keratin, type I cytoskeletal 18 |
| NP297856 | NP297856 | not known | XR_017288 | XR_017288 | not known |
| NM_033201 | C16orf45 | chromosome 16 open reading frame 45 | XR_017211 | XR_017211 | not known |
| THC2710559 | THC2710559 | not known | NM_019841 | TRPV5 | transient receptor potential cation channel, subfamily V, member 5 |
| AI916036 | AI916036 | not known | NM_001079675 | ETV4 | ets variant 4 |
| THC2649152 | THC2649152 | not known | XR_019203 | XR_019203 | not known |
| ENST00000372127 | ENST00000372127 | not known | A_24_P844100 | A_24_P844100 | not known |
| NM_198521 | C12orf42 | chromosome 12 open reading frame 45 | A_24_P281605 | A_24_P281605 | not known |
| THC2539634 | THC2539634 | not known | XR_015430 | LOC731274 | similar to tau tubulin kinase 2 |
| NM_173484 | KLF17 | Krup pel-like factor 17 | XR_018299 | XR_018299 | not known |
| AF086329 | AF086329 | not known | XR_017139 | XR_017139 | not known |
| BX113452 | BX113452 | not known | XR_038299 | LOC100128116 | similar to hCG1748768 |
| A_32_P226468 | A_32_P226468 | not known | XR_018415 | XR_018415 | not known |
| NM_020358 | TRIM 49 | trip artite motif-containing 49 | AK127993 | FLJ46111 | keratin 8 pseudogene |
| THC2684446 | THC2684446 | not known | A_24_P315594 | A_24_P315594 | not known |
| THC2709441 | THC2709441 | not known | XR_018991 | XR_018991 | not known |
| NM_080429 | AQP10 | aquaporin 10 | NM_153824 | PYCR1 | pyrroline-5-carboxylate reductase 1 |
| THC2680667 | THC2680667 | not known | NM_003708 | RDH16 | retinol dehydrogenase 16 |
| NM_000483 | APOC2 | apolipoprotein C-II | NM_005628 | SLC1A5 | solute carrier family 1, member 5 |
| AF068286 | DSTYK | dual serine/threonine and tyrosine protein kinase | NM_015374 | SUN2 | Sad1 and UNC84 domain containing 2 |
| AF187554 | LOC147977 | hypothetical LOC147977 | NM_024671 | ZNF768 | zinc finger protein 768 |
| BC000206 | BC000206 | not known | NM_015164 | PLEKHM2 | pleckstrin homology domain containing, family M, memeber 2 |
| ENST00000391372 | ENST00000391372 | not known | |||
| A_32_P225768 | A_32_P225768 | not known | |||
| BC000228 | TLE1 | transducin-like enhancer of split 1 (E(sp1) homolog | |||
| NM_152309 | PIK3AP1 | phosphoinositide-3-kinase adaptor protein 1 | |||
| A_24_P852099 | A_24_P852099 | not known | |||
| ENST00000361624 | ENST00000361624 | not known | |||
| NM_015881 | DKK3 | dickkopf homolog 3 | |||
| NM_000746 | CHRNA7 | cholinergic receptor, nicotinic, alpha 7 | |||
| AF009267 | AF009267 | not known | |||
| CR605947 | CHRNA7 | cholinergic receptor, nicotinic, alpha 7 | |||
| AK127258 | AK127258 | not known | |||
| A_24_P799680 | A_24_P799680 | not known | |||
To study the biological meaning of the MED29-associated gene expression changes, we retrieved gene ontology data on the differentially expressed genes. Several key processes involved in cell cycle, cell division, mitosis, and chromosome segregation were enriched in MED29-expressing cells (Supporting Information Figure S2, Table S2). Cellular component analysis indicated that the differentially expressed genes showed enrichment in condensed chromosomes, kinetochore, spindle, and microtubule cytoskeleton (Supporting Information Figure S2), again implying that they have a central role in cell division. Genes that were found in these enriched categories included CCNA2, CCNB2, CDK1, GTSE1, and CDKN2B, all of which are well-known regulators of the cell cycle. CCNA2, CCNB2, CDK1, and GTSE1 are cell cycle promoting genes and were all downregulated in MED29 expressing cells, whereas CDKN2B, an inhibitory cell cycle regulator, was upregulated in MED29 cells.
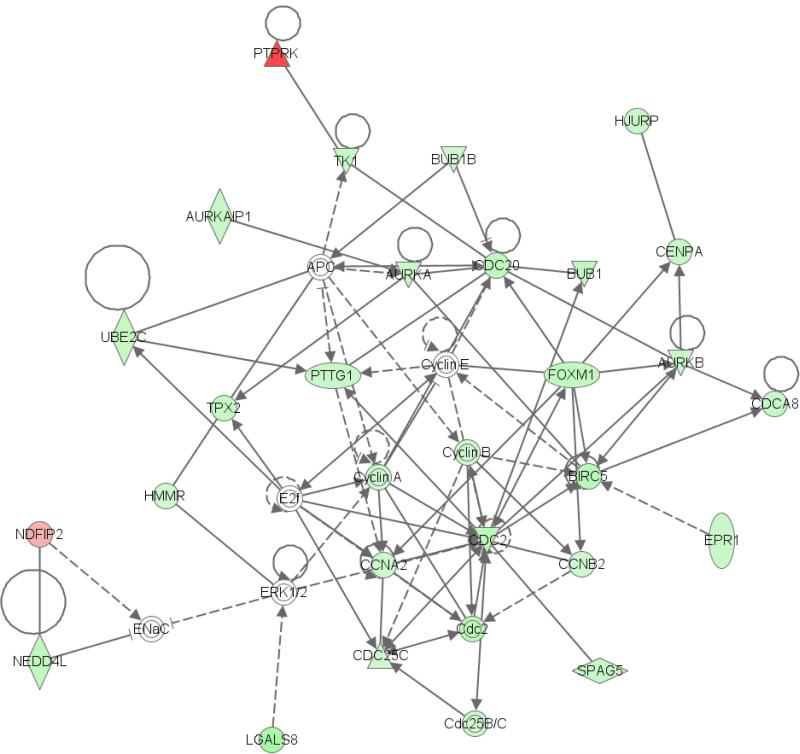
To further understand the functional role of the genes whose expression is altered after MED29 overexpression, we performed the Ingenuity Pathway Analysis on the differentially expressed genes. The results identified a network associated with cell cycle, cancer, and genetic disorder (Fig. 4). The highest enrichment among molecular and cellular functions involved cell cycle, cell death, and cellular growth and proliferation. The cell cycle was again the highest ranked among the top canonical pathways that were affected by MED29 expression. In summary, the pathway analysis on differentially expressed showed enrichment of cell cycle associated cellular networks and functions.
Figure 4.
Pathway analysis links the differentially expressed gene signatures of the MED29 transduced Hs 700T and MIAPaCa-2 cells to a cellular network involved in cell cycle, cancer, and genetic disorder. This network is characterized by involvement of several cyclins and other cell cycle related proteins, most of which are downregulated through MED29 expression. Green color indicates downregulation and red color upregulation. Lines and arrows indicate connection and dashed lines a weaker connection.
To validate the microarray data, qRT-PCR was applied to analyze the expression levels of 84 known cell cycle genes both in MIAPaCa2/MED29 and Hs700T/MED29 cells. The qRT-PCR results were in good concordance with the microarray data and highlighted the same set of cell cycle regulators (e.g., BAX, CCNF, CDC34, CDK2, CDK6, CDKN1A, GADD45, and GTF2H1) as being differentially expressed. Next we wanted to explore whether alterations in these cell cycle regulators also occurred in the xenograft tumors derived from these cell lines. In MIAPaCa2/MED29 tumors, the qRT-PCR analysis revealed 12 differentially expressed genes (at least 1.5-fold change as compared to mock tumors, Table 2). The great majority of these (11 out of 12) were downregulated. Genes that were commonly downregulated in both MIAPaCa2/MED29 xenografts and the parental cell line included CCND2, CDC16, CDK6, HERC5, and RBBP8. The only upregulated gene in the MIAPaCa2/MED29 xenografts was CDKN2B, a negative regulator of the cell cycle, and it also showed elevated expression in the parental MIAPaCa2/MED29 cells. In Hs 700T cells and xenografts, the differences in cell cycle gene expression levels between MED29 and mock cells were not as prominent. There were only five differentially expressed genes in Hs700T/MED29 tumors compared to mock tumors (Table 2). ANAPC4, ATR, CDK4, and SUMO1 were downregulated whereas HERC5 was upregulated. Taken together, the qRT-PCR analysis validated the microarray data and pinpointed crucial changes in the expression of cell cycle associated genes.
Table 2.
Differentially expressed cell cycle genes in MED29 vs. mock xenografts by qRT-PCR arrays.
| Xenograft | Gene | Fold changea |
|---|---|---|
| Hs 700T | HERC5 | 2.34 |
| Hs 700T | ANAPC4 | −1.53 |
| Hs 700T | ATR | −1.59 |
| Hs 700T | CDK4 | −1.67 |
| Hs 700T | SUMO1 | −1.67 |
| MIA PaCa-2 | CDKN2B | 3.92 |
| MIA PaCa-2 | CCNB1 | −1.62 |
| MIA PaCa-2 | CCNC | −1.75 |
| MIA PaCa-2 | CCND2 | −6.09 |
| MIA PaCa-2 | CCNG1 | −1.56 |
| MIA PaCa-2 | CDC16 | −2.23 |
| MIA PaCa-2 | CDK6 | −1.77 |
| MIA PaCa-2 | GTSE1 | −1.72 |
| MIA PaCa-2 | HERC5 | −1.68 |
| MIA PaCa-2 | MAD2L2 | −2.09 |
| MIA PaCa-2 | MCM3 | −2.46 |
| MIA PaCa-2 | RBBP8 | −1.74 |
Positive and negative values refer to up- and downregulation, respectively, in MED29 xenografts as compared to corresponding mock samples.
Discussion
Transcriptional regulation plays an important role in maintaining cellular homeostasis and diverse differentiation and developmental programs.2 This regulation involves a number of specialized proteins and protein complexes, all of which need to co-operate to ensure accurate control of expression of a given gene in a spatially and temporally regulated manner.6 Any changes in this process may lead to severe consequences in transcriptional regulation and imbalanced cellular homeostasis.
According to the current view, Mediator acts as a global regulator of transcription by communicating regulatory information between the gene-specific activators and the general transcriptional machinery.6,7,34 It plays both general and gene-specific roles in transcription and involves different compositions depending on the cellular context.35 MED29 is a component of Mediator but its exact role is currently unknown.19,36 The Drosophila melanogaster protein Intersex, an ortholog of mammalian MED29, interacts with and functions as a coactivator for the DNA-binding transcription factor Doublesex.37 Accordingly, it is likely that the mammalian MED29 also serves as a cofactor for specific transcriptional activators.
We and others have previously shown that MED29 is amplified and overexpressed in a subset of pancreatic cancers22–23 and that MED29 silencing leads to G1 arrest and increased apoptosis in PANC-1 pancreatic cancer cells with amplification but not in non-amplified cells.22 Here we extend these findings and demonstrate that in addition to reduced cell growth and survival MED29 silencing also leads to significantly decreased colony formation, cell migration, and invasion in PANC-1 cells. These data imply that increased expression of MED29 provides beneficial, oncogene-like properties to the cancer cells. This view fits well in the picture of Mediator acting as a transcriptional activator.38
To better understand the role of MED29 in pancreatic cancer, we used the lentiviral approach to induce MED29 expression first in NIH/3T3 cells and then in Hs 700T and MIA PaCa-2 pancreatic cancer cells, both of which have low endogenous MED29 levels. To our surprise, MED29 expression led to decreased proliferation of NIH/3T3 and MIA PaCa-2 cells. It is possible that this phenotype could be caused by induction of senescence, similar to what has been shown with known oncogenes such as KRAS and RAF.39–40 In low serum conditions, an opposite growth effect was seen in NIH/3T3 cells and might be explained by altered transcriptional activation mechanisms during a stress situation. Indeed, reduced association of Mediator to Pol II has been observed in response to stress conditions.10 It is important to note that the MED29 induced growth inhibition in MIA PaCa-2 was only seen after the cells were synchronized and thus the effect was not very dramatic. Unfortunately, we failed to replicate these results in Hs 700T cells as we were unable to synchronize them. Furthermore, one has to keep in mind that both MIA PaCa-2 and Hs 700T cell lines are highly aggressive and have intrinsically rapid growth rates and thereby manipulation of the expression of a single gene does not necessarily have a great impact on their phenotype.41
The MED29-expressing HS 700T and MIA PaCa-2 cells were then injected subcutaneously into nude mice to explore the possible effects of MED29 in in vivo tumor formation. All mock transfected animals developed large tumors whereas the growth of the MED29 expressing xenografts was severely compromised. Especially in the MIAPaCa2/MED29 mice, the tumor incidence was highly reduced and the tumors that did develop showed a dramatic 13-fold reduction in size compared to corresponding mock controls. In fact, the MIAPaCa2/MED29 tumors did not show exponential growth but rather remained small throughout the observation period. Similar but less extensive reduction in tumor size was also seen in Hs700T/MED29 mice but there was no change in tumor incidence compared to mock controls. Notably, all xenograft tumors were identically vascularized, implying to sufficient angiogenesis. Neovascularization is generally needed for tumor progression and is greatly influenced by the tumor microenvironment.42 The tumor microenvironment also has an effect on tumor growth and thereby an orthotopic tumor model, that more faithfully mimics the formation of pancreatic adenocarcinoma in appropriate microenvironment, is likely to provide additional information on the role of ME29 in pancreatic cancer pathogenesis. Nevertheless, these in vivo data presented here firmly suggest that MED29 also has tumor suppressive properties.
Taken together, both silencing of high endogenous MED29 expression and forced overexpression of MED29 resulted in suppression of cell growth in pancreatic cancer cells. These results were quite unforeseen and it is possible that they reflect essential differences between in vitro and in vivo assay conditions. However, it is more plausible that these differences are caused by dissimilarities in the genetic background of the cell lines. All pancreatic cancer cell lines used in this study are highly evolved and carry multiple genetic and epigenetic aberrations41, 43–44 that are expected to influence their behavior. A third possibility is that MED29 has a dualistic role in cancer, depending on the surroundings where it is expressed. There are a few examples of proteins that possess such a dualistic role in cancer. Of these, the transforming growth factor beta (TGFβ) is perhaps the most well known. TGFβ has tumor suppressive properties that cells must escape for malignant transformation to occur but, on the other hand, during cancer progression it has immunosuppressive and proangiogenic properties and also contributes to the metastatic process.45 Other examples include the macrophage migration inhibitory factor (MIF) in breast cancer46 and vski sarcoma viral oncogene homolog in pancreatic cancer.47 In a similar fashion, MED29 might have a dualistic role in cancer, depending on the surrounding milieu and other proteins it interacts with.
To examine the underlying mechanisms related to the growth regulatory role of MED29, we utilized genome-wide microarrays to identify a total of 463 differentially expressed genes between the MED29 and mock transfected pancreatic cancer cells. In general, downregulation was more prevalent than upregulation among the differentially expressed genes, implying that MED29 functions mainly as a transcriptional repressor in these cells. Even though microarray hybridization-based technology is the most widespread method to analyze the transcriptome, it suffers from certain limitations, such as restricted focus on only those transcripts included in the array and inability to detect rare splice variants, unknown transcripts, and noncoding transcripts.48–49 There is a wide range of other methods for gene expression profiling50 that have been developed to overcome these problems, including serial analysis of gene expression and massively parallel signature sequencing. Nevertheless, in order to obtain as accurate data as possible, we used both biological and technical replicates combined with careful statistical analyses. To conclude, our data provides important information on MED29-induced genome-wide gene expression changes but it is possible that there are additional changes that our method was unable to detect.
We then used two different types of approaches, the Gene Ontology database and the Ingenuity Pathway Analysis, to try to understand the biological function and nature of the differentially expressed genes. Both analyses strongly indicated that the differentially expressed genes encode for proteins involved in the regulation of cell cycle, cell division, cell growth and proliferation, and cell death. Among these, the association with cell cycle regulation was most prominent. For example, well-known cell cycle promoting genes such as CCNA2, CCNB2, CDK1, and GTSE1 were downregulated in MED29-overexpressing cells, whereas CDKN2B, an inhibitory regulator of cell cycle, was induced, thus leading to an overall attenuation of cell cycle progression.
The data shown above imply that MED29 overexpression in MIA PaCa-2 and Hs 700T cells leads to transcriptional changes that culminate in inhibition of the cell cycle. This result is in excellent concordance with our in vivo data, which demonstrated both reduced tumor incidence and reduced growth of MED29 expressing xenografts. However, further studies are needed to reveal the mechanistic details on how MED29 overexpression induces the observed transcriptional changes that lead to growth regulation.
In conclusion, silencing of high endogenous MED29 expression leads to inhibition of tumor cell-associated phenotypes, cell growth, migration, invasion, and colony formation, and thus substantiates the oncogenic role of MED29 in pancreatic cancer. In contrast, the in vivo data of forced MED29 overexpression provides new evidence for a tumor suppressive role of MED29 that is largely mediated by alterations in cell cycle regulatory genes. Therefore, MED29 seems to have a dualistic role in pancreatic cancer pathogenesis that is likely to be influenced by the genetic background of the cancer cells and their surrounding milieu.
Supplementary Material
Global expression survey reveals common up- and downregulated genes after forced expression of MED29 in Hs 700T and MIA PaCa-2 pancreatic cancer cells. The expression analysis was performed using Agilent 44K whole human oligonucleotide microarrays and two biological (MED29-1 and MED29-2) and three experimental (A, B, C) replicates were done for each cell line, resulting in twelve microarray experiments in total. Differentially expressed genes (at least 1.5-fold change with adjusted P-value less than 0.05) between the MED29 transduced and mock cell lines were identified by means of empirical Bayes with Benjamini-Hochberg adjustment for P-values. Unsupervised hierarchical clustering of the 463 differentially expressed genes was done using correlation distance and average linkage. Cell lines are presented in the columns and genes in the rows. Upregulated genes are shown in red and downregulated in green, as indicated in the color code at the left side of the dendogram. Numbers are logarithmic fold change values (MED29 vs. mock). Common up- and downregulated gene clusters between the cell lines are illustrated as red and green tree views on the left.
DAVID bioinformatics resources were used to identify enriched gene ontologies (molecular function, biological process, and cellular component) among the differentially expressed genes. Enriched gene ontologies (adjusted P-value < 0.05) involving (a) biological processes and (b) cellular components are illustrated, no significantly enriched categories were found in the molecular function ontology. The main category is shown at the top of the Fig.. White boxes represent enriched gene categories. The number of enriched genes (n) in each category is shown in the box together with the adjusted P-value for enrichment.
Differentially expressed genes between the MED29 and the mock transduced Hs 700T and MIA PaCa-2 cells were identified by means of empirical Bayes with Benjamini-Hochberg adjustment for P-values. The 463 differentially expressed genes (fold change > 1.5 with an adjusted P-value < 0.05) are listed.
List of enriched gene ontologies (molecular function, biological process, and cellular component) obtained by DAVID bioinformatics resources among the 463 differentially expressed genes. Only those with an adjusted P-value (FDR) less than 0.05 are considered significant.
Acknowledgements
We thank Kati Rouhento and Iida Leppälä for skilful technical assistance.
Grant sponsors: Academy of Finland grants no. 122440, 129657 and 134117, NIH grant PO1 CA109552, and the Sigrid Juselius Foundation.
References
- 1.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat Struct Mol Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koch F, Jourquin F, Ferrier P, Andrau JC. Genome-wide RNA polymerase II: Not genes only! Trends Biochem Sci. 2008;33:265–273. doi: 10.1016/j.tibs.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 4.Ansari SA, He Q, Morse RH. Mediator complex association with constitutively transcribed genes in yeast. Proc Natl Acad Sci U S A. 2009;106:16734–16739. doi: 10.1073/pnas.0905103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flanagan PM, Kelleher RJ, 3rd, Sayre MH, Tschochner H, Kornberg RD. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 6.Taatjes DJ, Marr MT, Tjian R. Regulatory diversity among metazoan co-activator complexes. Nat Rev Mol Cell Biol. 2004;5:403–410. doi: 10.1038/nrm1369. [DOI] [PubMed] [Google Scholar]
- 7.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 8.Han SJ, Lee YC, Gim BS, Ryu GH, Park SJ, Lane WS, Kim YJ. Activator-specific requirement of yeast mediator proteins for RNA polymerase II transcriptional activation. Mol Cell Biol. 1999;19:979–988. doi: 10.1128/mcb.19.2.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takagi Y, Kornberg RD. Mediator as a general transcription factor. J Biol Chem. 2006;281:80–89. doi: 10.1074/jbc.M508253200. [DOI] [PubMed] [Google Scholar]
- 10.Fan X, Chou DM, Struhl K. Activator-specific recruitment of mediator in vivo. Nat Struct Mol Biol. 2006;13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 11.Thiaville MM, Dudenhausen EE, Awad KS, Gjymishka A, Zhong C, Kilberg MS. Activated transcription via mammalian amino acid response elements does not require enhanced recruitment of the mediator complex. Nucleic Acids Res. 2008;36:5571–5580. doi: 10.1093/nar/gkn538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rienzo M, Nagel J, Casamassimi A, Giovane A, Dietzel S, Napoli C. Mediator subunits: gene expression pattern, a novel transcript identification and nuclear localization in human endothelial progenitor cells. Biochim Biophys Acta. 2010;1799:487–495. doi: 10.1016/j.bbagrm.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 14.Boube M, Joulia L, Cribbs DL, Bourbon HM. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell. 2002;110:143–151. doi: 10.1016/s0092-8674(02)00830-9. [DOI] [PubMed] [Google Scholar]
- 15.Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, Conaway JW, Conaway RC. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Chadick JZ, Asturias FJ. Structure of eukaryotic mediator complexes. Trends Biochem Sci. 2005;30:264–271. doi: 10.1016/j.tibs.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. Quantitative proteomic analysis of distinct mammalian mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci U S A. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dotson MR, Yuan CX, Roeder RG, Myers LC, Gustafsson CM, Jiang YW, Li Y, Kornberg RD, Asturias FJ. Structural organization of yeast and mammalian mediator complexes. Proc Natl Acad Sci U S A. 2000;97:14307–14310. doi: 10.1073/pnas.260489497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian mediator complex and its role in transcriptional regulation. Trends Biochem Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Li Y, Zeng W, Zhu C, Xiao J, Yuan W, Wang Y, Cai Z, Zhou J, Liu M, Wu X. IXL, a new subunit of the mammalian mediator complex, functions as a transcriptional suppressor. Biochem Biophys Res Commun. 2004;325:1330–1338. doi: 10.1016/j.bbrc.2004.10.159. [DOI] [PubMed] [Google Scholar]
- 21.Garrett-Engele CM, Siegal ML, Manoli DS, Williams BC, Li H, Baker BS. Intersex, a gene required for female sexual development in drosophila, is expressed in both sexes and functions together with doublesex to regulate terminal differentiation. Development. 2002;129:4661–4675. doi: 10.1242/dev.129.20.4661. [DOI] [PubMed] [Google Scholar]
- 22.Kuuselo R, Savinainen K, Azorsa DO, Basu GD, Karhu R, Tuzmen S, Mousses S, Kallioniemi A. Intersex-like (IXL) is a cell survival regulator in pancreatic cancer with 19q13 amplification. Cancer Res. 2007;67:1943–1949. doi: 10.1158/0008-5472.CAN-06-3387. [DOI] [PubMed] [Google Scholar]
- 23.Chen S, Auletta T, Dovirak O, Hutter C, Kuntz K, El-ftesi S, Kendall J, Han H, Von Hoff DD, Ashfaq R, Maitra A, Iacobuzio-Donahue CA, et al. Copy number alterations in pancreatic cancer identify recurrent PAK4 amplification. Cancer Biol Ther. 2008;7:1793–1802. doi: 10.4161/cbt.7.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janik P, Briand P, Hartmann NR. The effect of estrone-progesterone treatment on cell proliferation kinetics of hormone-dependent GR mouse mammary tumors. Cancer Res. 1975;35:3698–3704. [PubMed] [Google Scholar]
- 25.Alarmo EL, Parssinen J, Ketolainen JM, Savinainen K, Karhu R, Kallioniemi A. BMP7 influences proliferation, migration, and invasion of breast cancer cells. Cancer Lett. 2009;275:35–43. doi: 10.1016/j.canlet.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 26.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 28.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. A comparison of background correction methods for two-colour microarrays Export. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 29.Smyth GK, Speed TP. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 31.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 32.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 33.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Casamassimi A, Napoli C. Mediator complexes and eukaryotic transcription regulation: An overview. Biochimie. 2007;89:1439–1446. doi: 10.1016/j.biochi.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 35.Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Chase BA, Baker BS. A genetic analysis of intersex, a gene regulating sexual differentiation in drosophila melanogaster females. Genetics. 1995;139:1649–1661. doi: 10.1093/genetics/139.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuras L, Borggrefe T, Kornberg RD. Association of the mediator complex with enhancers of active genes. Proc Natl Acad Sci U S A. 2003;100:13887–13891. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 40.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deer EL, González-Hernández J, Coursen JD, Shea JE, Ngatia J, Scaife CL, Firpo MA, Mulvihill SJ. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–435. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D, Frese KK, Denicola G, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahlamäki EH, Bärlund M, Tanner M, Gorunova L, Höglund M, Karhu R, Kallioniemi A. Frequent amplification of 8q24, 11q, 17q, and 20q-specific genes in pancreatic cancer. Genes Chromosomes Cancer. 2002;35:353–358. doi: 10.1002/gcc.10122. [DOI] [PubMed] [Google Scholar]
- 44.Sato N, Maitra A, Fukushima N, van Heek NT, Matsubayashi H, Iacobuzio-Donahue CA, Rosty C, Goggins M. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003;63:4158–4166. [PubMed] [Google Scholar]
- 45.Massague J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verjans E, Noetzel E, Bektas N, Schütz AK, Lue H, Lennartz B, Hartmann A, Dahl E, Bernhagen J. Dual role of macrophage migration inhibitory factor (MIF) in human breast cancer. BMC Cancer. 2009;9:230. doi: 10.1186/1471-2407-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang P, Chen Z, Meng ZQ, Fan J, Luo JM, Liang W, Lin JH, Zhou ZH, Chen H, Wang K, Shen YH, Xu ZD, et al. Dual role of Ski in pancreatic cancer cells: tumor-promoting versus metastasis-suppressive function. Carcinogenesis. 2009;30:1497–1506. doi: 10.1093/carcin/bgp154. [DOI] [PubMed] [Google Scholar]
- 48.Napoli C, Lerman LO, Sica V, Lerman A, Tajana G, de Nigris F. Microarray analysis: a novel research tool for cardiovascular scientists and physicians. Heart. 2003;89:597–604. doi: 10.1136/heart.89.6.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 50.Harbers M, Carninci P. Tag-based approaches for transcriptome research and genome annotation. Nat Methods. 2005;2:495–502. doi: 10.1038/nmeth768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Global expression survey reveals common up- and downregulated genes after forced expression of MED29 in Hs 700T and MIA PaCa-2 pancreatic cancer cells. The expression analysis was performed using Agilent 44K whole human oligonucleotide microarrays and two biological (MED29-1 and MED29-2) and three experimental (A, B, C) replicates were done for each cell line, resulting in twelve microarray experiments in total. Differentially expressed genes (at least 1.5-fold change with adjusted P-value less than 0.05) between the MED29 transduced and mock cell lines were identified by means of empirical Bayes with Benjamini-Hochberg adjustment for P-values. Unsupervised hierarchical clustering of the 463 differentially expressed genes was done using correlation distance and average linkage. Cell lines are presented in the columns and genes in the rows. Upregulated genes are shown in red and downregulated in green, as indicated in the color code at the left side of the dendogram. Numbers are logarithmic fold change values (MED29 vs. mock). Common up- and downregulated gene clusters between the cell lines are illustrated as red and green tree views on the left.
DAVID bioinformatics resources were used to identify enriched gene ontologies (molecular function, biological process, and cellular component) among the differentially expressed genes. Enriched gene ontologies (adjusted P-value < 0.05) involving (a) biological processes and (b) cellular components are illustrated, no significantly enriched categories were found in the molecular function ontology. The main category is shown at the top of the Fig.. White boxes represent enriched gene categories. The number of enriched genes (n) in each category is shown in the box together with the adjusted P-value for enrichment.
Differentially expressed genes between the MED29 and the mock transduced Hs 700T and MIA PaCa-2 cells were identified by means of empirical Bayes with Benjamini-Hochberg adjustment for P-values. The 463 differentially expressed genes (fold change > 1.5 with an adjusted P-value < 0.05) are listed.
List of enriched gene ontologies (molecular function, biological process, and cellular component) obtained by DAVID bioinformatics resources among the 463 differentially expressed genes. Only those with an adjusted P-value (FDR) less than 0.05 are considered significant.