Abstract
Protein engineering of cytochrome P450 monooxygenases (P450s) has been very successful in generating valuable non-natural activities and properties, allowing these powerful catalysts to be used for the synthesis of drug metabolites and in biosynthetic pathways for the production of precursors of artemisinin and paclitaxel. Collected experience indicates that the P450s are highly 'evolvable'--they are particularly robust to mutation in their active sites and readily accept new substrates and exhibit new selectivities. Their ability to adapt to new challenges upon mutation may reflect the nonpolar nature of their active sites as well as their high degree of conformational variability.
Introduction
Cytochrome P450 monooxygenases (CYPs or P450s) are heme-containing enzymes that use molecular oxygen and the hydride donor NAD(P)H (coupled via redox partners) to effect the overall oxidative insertion of one oxygen atom into an organic substrate. Oxidation is manifested as hydroxylation, epoxidation, dealkylation, and other transformations and is carried out in a regio- and stereoselective manner. Selective C–H functionalization at unactivated carbons--one of the most challenging reactions in synthetic chemistry--is conducted under mild conditions by these impressive biocatalysts [1]. P450s are responsible for steps in the biosynthesis of valuable natural products such as the anti-cancer drug paclitaxel and the anti-malaria drug artemisinin [2]. Additionally, they are critical for metabolism of drugs and toxins [3].
P450s could have wide-ranging applications in the production of drugs and drug metabolites or as catalysts in other chemical processes; they could also serve as sensors or bioremediation agents [4–7]. Despite their unique catalytic capabilities, however, only a limited number have been exploited in preparative chemical reactions or industrial chemical processes [8]. Many natural P450s are insoluble (they are often membrane-associated), expressed at low levels, and exhibit activity insufficient for practical biocatalysis. To expand the applications and enhance the utility of P450s, it will be necessary to improvecatalytic properties: substrate scope, selectivity (regio and stereo), activity (TTN (total turnover number), kcat, KM), inhibition, and coupling efficiency (ratio of substrate reacted to NAD(P)H cofactor consumed, expressed as mole%). Particularly challenging is achieving high coupling efficiency (ideally close to 100%), which is severely compromised (often less than 10%) when P450s are presented with novel substrates. Uncoupling wastes expensive reduced cofactors and leads to generation of reactive oxygen species and enzyme inactivation. Improvements also may be sought for physical properties such as thermostability, solvent tolerance, oxidative stability, and substrate and product tolerance [8]. Furthermore, P450s may require multiple protein redox partners, and reconstituting the entire system can be tricky. These daunting challenges for optimizing P450s in new applications have been the focus of recent protein engineering efforts.
P450s can be engineered using both rational and evolutionary approaches [4–6,9•]. Rational approaches are characterized by the deliberate mutation of one or more amino acids based on mechanistic or structural information. Obtaining detailed information for P450s, however, may be very difficult, since these enzymes are notoriously hard to crystallize and furthermore require additional protein partners and cofactors for activity. Directed evolution has thus become a valuable complementary tool in P450 engineering [10]. Here, mutations are introduced in a random or semi-random manner, e.g. by site-saturation mutagenesis at residues thought to be important for the desired property, and the resulting mutant P450s are screened for enhancement of that property or set of properties. Although most random mutations are either neutral or deleterious, a small percentage (the specific value depends on the protein and property targeted) may be advantageous [9•]. These mutations are accumulated in an iterative process or by recombination until the functional goal is met (or not).
In this review, we examine examples of P450 engineering with emphasis on the past two years. One goal is to illustrate how readily these enzymes are able to adopt new functions, such as the ability to accept a new substrate. Another is to offer some ideas as to why these particular enzymes are so adaptable. We initially focus on P450 BM3 (also known as CYP102A1) from Bacillus megaterium, as it has been the target of the most engineering efforts. We then examine selected studies of other P450s, which have been engineered for a variety of biocatalytic applications using mutagenesis or by generating self-sufficient P450s that mimic the natural fusion of the hydroxylase and reductase domains in P450 BM3. Some engineered P450s have been incorporated into metabolic pathways for in vivo synthesis of natural products.
Properties of the P450s that facilitate their ability to adapt
It appears that some proteins are more easily endowed with new functions in the laboratory than others; these have been called more “evolvable”. Evolvable proteins include immunoglobulins, HIV proteases, chaperone proteins, and enzymes in the alkaline phosphate and glutathione transferase superfamilies [11,12]. One thing these proteins all share is natural functional diversity: nature discovered an adaptable framework and diversified it widely through mutation and selection. The cytochrome P450s are a functionally diverse enzyme family with more than 11,000 known members (Box 1) that contribute to catabolism of a wide range of xenobiotics and production of large numbers of secondary metabolites including terpenes, fatty acids, and alkaloids. It is not surprising therefore that P450s adapt readily and take on new functions in the laboratory. Although functional diversity in the natural enzyme family is a useful rule-of-thumb for predicting evolvability, the structural basis and mechanisms of evolvability are still largely speculative. Important for evolvability, however, is the ability to accept mutations in the first place (mutational robustness) [13]. Even though P450s are not exceptionally stable enzymes, their active sites are highly accepting of mutations with respect to maintaining both the folded structure and catalytic competence. This mutational robustness reflects the central role of the iron-heme prosthetic group in enzyme reactivity, and likely also reflects the nonpolar nature of the active site and/or its unusual conformational variability.
Online collections of data relevant to P450 engineering
http://drnelson.uthsc.edu/cytochromeP450.html “The Cytochrome P450 Homepage” compiled by David R Nelson includes sequences and collections of presentations and publications. As of the latest statistics update on August 20, 2009, the site listed 11,294 known P450 sequences, and sequences continue to be listed at a rapid pace.
http://www.cyped.uni-stuttgart.de “The CYP450 Engineering Database” developed at the University of Stuttgart includes sequence and structure data for numerous P450s. Downloadable PDB files are available for structures of wild-type and mutant enzymes [49].
www.muteindb.org “MuteinDB” developed at Graz University of Technology is a database of wild-type and mutant enzymes (muteins) that includes experimental data on substrates, kinetic parameters, and other properties of interest for applications. This database includes extensive datasets for P450 BM3, CYP2D6, and CYP3A4.
P450 active sites possess a greater degree of conformational variability than the active sites of other enzymes [14•], and this property may aid in the ability of the P450s to tolerate more mutation in this region. Active site conformational variability is well established in the P450s, which exhibit major conformational changes [15] and repositioning of active site residues upon substrate binding [16,17]. Examples are CYP3A4 and CYP2B4, whose substrate-binding-induced conformational changes have been investigated by X-ray crystallography [16]. Conformational variability enables structural changes to take place in the active site upon substrate binding, molecular oxygen binding, and reduction of the heme-bound iron atom during catalysis. This malleable P450 active site could also experience less mutational disruption than a more rigid structure.
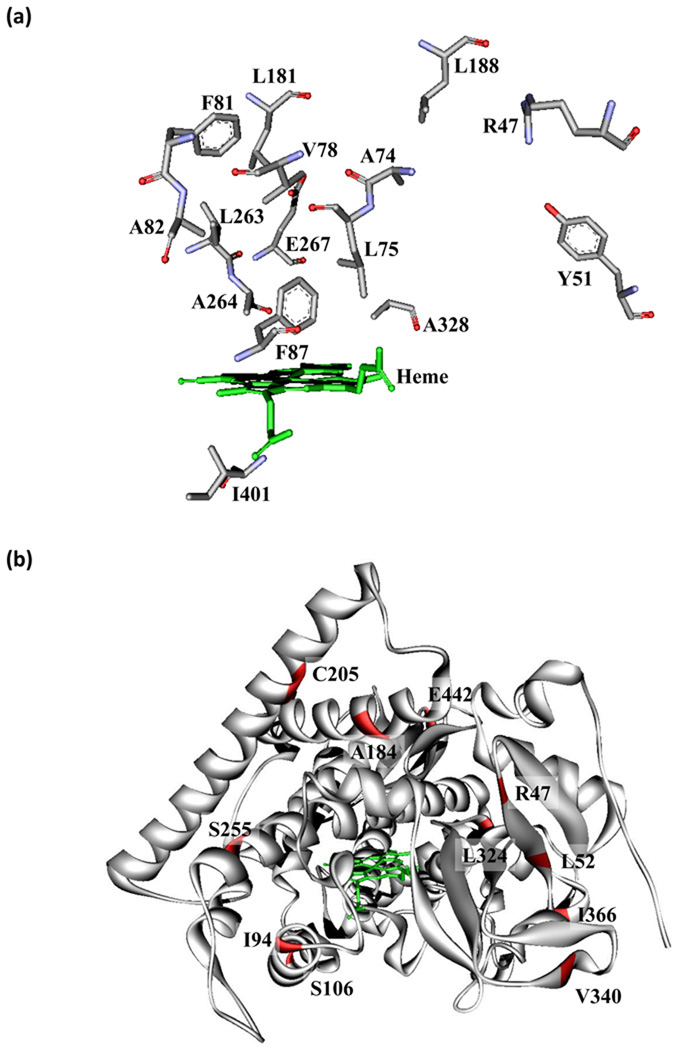
The ability to tolerate conformational and mutational changes may also be a result of the non-polar nature of the amino acids in the active site. We visualize the P450 active site as consisting of the reactive iron-heme and a greasy distal cavity, the majority of whose residues make up a relatively amorphous, conformationally variable agglomeration. Because it is relatively weak van der Waals interactions that tend to be disturbed when various conformations are accessed rather than stronger and more directional bonds such as hydrogen bonds or salt bridges, conformational changes readily occur. These weak interactions may also facilitate the enzyme’s ability to accept mutations in residues in the substrate binding pocket. If one of the non-polar amino acids is replaced with a similar non-polar amino acid, the interactions that are disturbed tend to be weak ones that are less likely to disrupt global protein stability. Figure 2(a) shows the active site residues that tolerate mutation in P450 BM3: most of the non-polar ones have been individually mutated in active variants, typically to other non-polar amino acids.
Figure 2.
Crystal structure showing the residues of P450 BM3 altering substrate recognition and thermostability. (a) Stick representations are shown of 15 active site residues distal to the heme which can undergo mutation and alter substrate scope and selectivity. (b) Residues that improve thermostability are colored red on the crystal structure (PDB: 1BU7).
Tokuriki and Tawfik have proposed a general mechanism for why conformationally variable proteins can more easily adopt new functions [18•]. Transiently stable conformations inherent to a protein are each endowed with different substrate specificities and other properties. Mutations can alter the relative stabilities and therefore the equilibrium distributions of these transient conformations as well as provide access to new ones. Proteins with greater conformational variability are thus able to find conformations that accept new substrates or catalyze other reactions.
Engineering P450 BM3
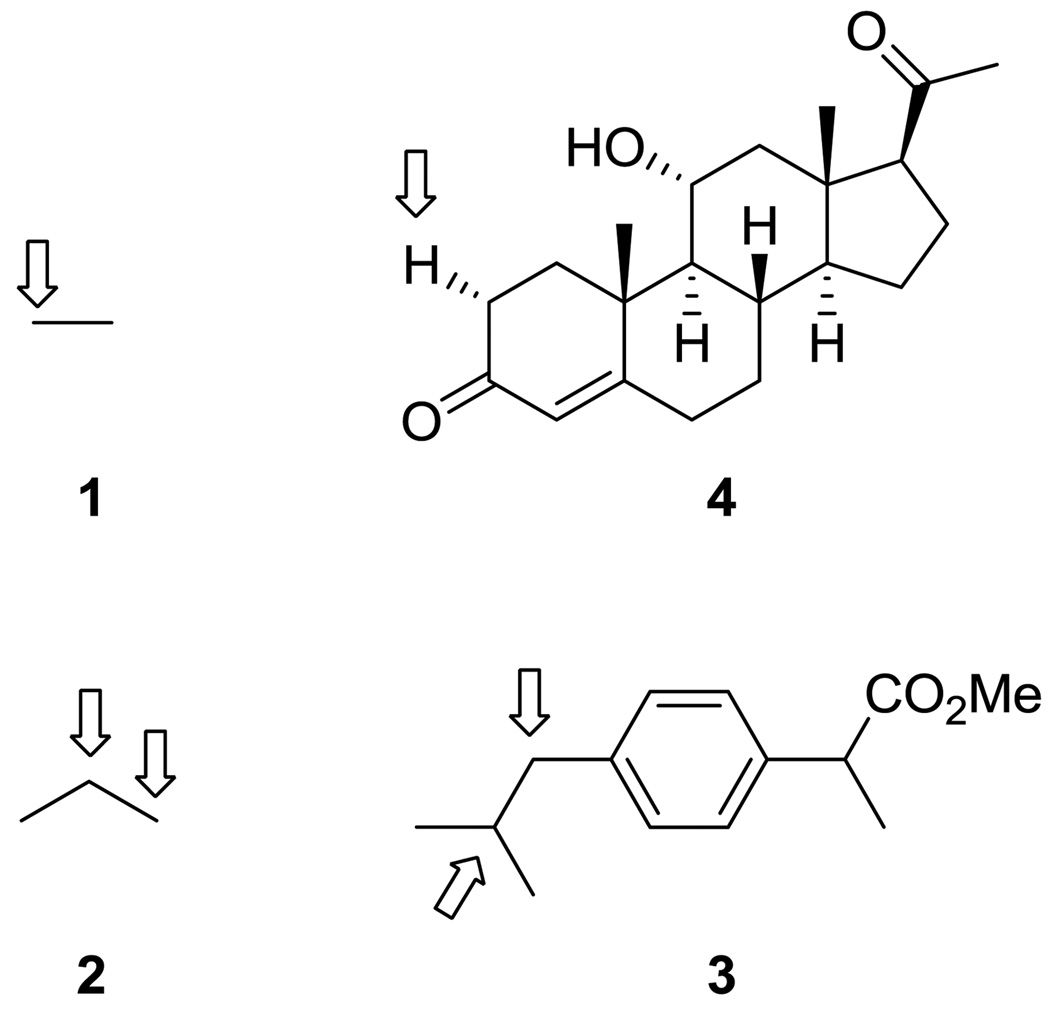
P450 BM3 possesses favorable properties that make it an attractive target for engineering [19]. This soluble bacterial enzyme is naturally fused to its reductase and expresses well in E. coli. It is also highly active: the hydroxylation of arachidonic acid catalyzed by P450 BM3 is the most rapid P450-catalyzed hydroxylation known (kcat = 17,000 min−1) [20]. Thus, P450 BM3 has been the target of many mutagenesis studies, whose combined results amply demonstrate its ability to adapt to accept new substrates (Figure 1). Protein engineering studies of P450 BM3 reveal that mutants 1) can be endowed with new and differentiated substrate scopes, 2) can exhibit regio- and enantio-selctivity on new substrates, and 3) can be engineered to be highly selective and active toward new substrates. Furthermore, increases in P450 activity are often accompanied by increased coupling efficiency and selectivity.
Figure 1.
P450 BM3 has been engineered to hydroxylate a broad spectrum of molecules of different molecular weights. Molecular weights range from 30 for ethane (1) to 330 for 11-α-hydroxyprogesterone (4). Propane (2) and ibuprofen methyl ester (3) are also readily hydroxylated. Arrows denote sites of hydroxylation.
Directed evolution of P450 BM3 for activity on non-native substrates often yields libraries of mutants that exhibit a range of new substrate specificities and selectivities. Mutagenesis targeted to the P450 BM3 active site, for example, generates variants that have few mutations but nonetheless display differentiated substrate scope. Pleiss and coworkers mutated just two amino acids in the active site, F87 and A328, to create a library of 24 mutants with non-polar amino acids (A, V, F, L, and I) at these two positions. The library included members with measurable activity on a variety of linear terpenes, cyclic monoterpenes, and cyclic sesquiterpenes [21•]. Members of this library also exhibited hydroxylation activity on cyclooctane, cyclodecane, and cyclododecane[22]. When measured as percentage conversion, a different mutant proved to be most active on each of those three substrates. In a separate study, combinatorial alanine incorporation in the enzyme’s active site created a library of P450 BM3 mutants which collectively accepted a variety of larger substrates, including steroids, opiate alkaloids, and peralkylated monosaccharides [23•]. Finally, three separate studies on a small library of 17 P450 BM3 mutants showed that various members hydroxylated and/or dealkylated many bioactive compounds including resveratrol, phenacetin, ethoxyresorufin, lovastatin, and simvastin [24–26]. Sawayama et al. described a collection of P450 BM3 mutants created by random mutagenesis, site-saturation mutagenesis, and structure-guided recombination with P450 BM3 homologs CYP102A2 and CYP102A3 that hydroxylated a variety of drugs and drug candidates [27•]. Together, members of this collection of only 120 P450 BM3 mutants produced 12 out of 13 human metabolites of the drugs verapamil and astemizole.
Although active mutants of P450s readily exhibit (generally low) activity on an expanded range of substrates, individual mutants can also possess regio- and enantioselectivity. When establishing a novel chemo-enzymatic two-step fluorination process, for example, Rentmeister et al. screened Sawayama’s collection of active P450 BM3 mutants for hydroxylation and demethylation of privileged bioactive compound classes including cyclopentenones, ibuprofen, Corey lactones, and 5-phenyloxazoline derivatives [28]. Activity was found for each of these classes, and library members exhibiting high regio- and enantioselectivity and yield were used for preparative-scale reactions without further optimization. Lewis et al. screened a similar collection of P450 BM3 mutants for demethylation activity on permethylated monosaccharide derivatives of glucose, mannose, and galactose [29]. In this case, they found individual mutants which removed one methyl in the presence of similar functionalities on several other positions, demonstrating excellent regioselectivity. They in fact identified P450 BM3 variants that could catalyze regioselective demethylation at nearly all the possible regiomeric O-methyl groups.
Mutants with activity toward a new substrate can be made more active, substrate-specific, and regio- and enantioselective upon further rounds of directed evolution. The P450 BM3 mutants that catalyzed regioselective demethylation of permethylated monosaccharides, for instance, were subjected to additional rounds of mutagenesis and screening for regioselectivity, resulting in increased regioselectivity and yield (up to 100% selectivity in the case of O-3 demethylation of β-pentamethyl galactose and 98% isolated yield) [29]. In another study, the P450 BM3 heme domain was engineered to selectively recognize dopamine for functional MRI imaging in the brains of living rats [30]. Five rounds of random mutagenesis and screening produced a P450 BM3 mutant with ~300-fold increased affinity for dopamine compared to wild-type P450 BM3 and greatly reduced affinity to the excellent substrate of wild-type P450 BM3, arachidonic acid.
We have observed that directed evolution leading to high TTN with a new substrate is accompanied by greater coupling efficiency, substrate specificity, and regioselectivity. This correlation was observed during the evolution ofP450 BM3 to hydroxylate propane [31]. As TTN on propane increased over many generations of directed evolution, coupling efficiency with this substrate increased from less than 10% to 98%. TTN is sensitive to uncoupling, which results in formation of reactive oxygen species and enzyme deactivation. Thus forcing the enzyme to increase TTN on propane also optimized cofactor utilization and reduced NADPH oxidase activity. Substrate specificity was also strongly affected by the continued evolution of propane activity. By the time the TTN on propane reached >45,000 in the newly-evolved propane hydroxylase, laurate hydroxylation was not detectable and the TTN for palmitate hydroxylation had fallen to less than 150 TTN, or < 0.5% of the value in wild-type P450 BM3. Thus substrate specificity was completely refocused from fatty acids to propane, even though the directed evolution included no negative selection against activity on fatty acids [31].
In general, evolution finds the easiest, or most probable, path to achieve a given functional goal. Thus while it may be possible to make an enzyme that is highly active on very different substrates, there are apparently more solutions to increasing activity on one at the cost of activity on others when the activity gets high enough. Conversely, because total activity as measured by TTN is related to coupling efficiency through enzyme deactivation, these two properties move hand-in-hand. Thus increasing TTN seems to be an excellent route to improving coupling efficiency.
Two recent studies demonstrate a positive correlation between increasing P450 BM3 oxidation activity (as measured by rate of product formation) and increasing coupling efficiency on non-natural substrates. Engineering of P450 BM3 for epoxidation of amorphadiene by site-directed mutagenesis (R47L and Y51F, located in the substrate access channel) increased coupling efficiency from 35% to 63% and epoxidation rate from ~8 to 30 min−1 [32•]. In another study, a proline substitution at residue 401 (I401P) was found to increase coupling efficiency for the oxidation of non-natural substrates [33,34]. For the conversion of propylbenzene, 3-methylpentane fluorene, and (+)-α-pinene, the mutation increased both coupling efficiency and activity. For example, the rate of hydroxylation of fluorene increased from 0.1 to 188 nmol per min per nmol enzyme, and was accompanied by an increase in coupling efficiency from 0.9% to 26%.
Most mutations are destabilizing, and mutations that enhance other properties not directly coupled to stability are also mostly destabilizing. Thus, if an evolving enzyme is only barely stable, nearly any mutation beneficial for the desired property would be lost due to its negative effect on folding. High stability allows an enzyme to accept a wider range of mutations while still maintaining a properly folded conformation. It has therefore proven useful to incorporate stabilizing mutations into P450 BM3 mutants before conducting additional random mutagenesis. This strategy was employed in the previously mentioned study on combinatorial alanine incorporation in the P450 BM3 active site, where six thermostabilizing mutations were introduced into an active mutant used as a starting point for directed evolution [23•]. These stabilizing mutations were known from previous directed evolution studies on P450 BM3, and were located throughout the enzyme. Their introduction increased the half-life of a promiscuous mutant of P450 BM3 called 9–10A from 3 min to 136 min at 50°C. This stabilization enabled the incorporation of an average of 3.9 alanines per mutant with 65% of clones still properly folded, which was not possible with 9–10A itself [23•]. For directed evolution of P450BM3 heme domain as a dopamine binder, a thermostabilizing mutation was also introduced prior to engineering for selective binding [30]. A summary of stabilizing mutations is presented in Figure 2(b), which also illustrates their distribution over the entire structure. Incorporation of these mutations into P450 BM3 may increase the probability of success in engineering new functions.
Engineering other P450s
Recent results demonstrate that a variety of other P450s can adopt new functions via directed evolution and other protein engineering approaches, and that these engineered enzymes are valuable in synthetic chemistry and biotechnology applications. For the enantioselective hydroxylation of N-benzylpyrrolidine to N-benzyl-3-hydroxypyrrolidine, a valuable intermediate for pharmaceutical synthesis, Zhao and coworkers developed a high throughput screening system to detect enantioselective hydroxylation catalyzed by P450 mutants [35•]. They used P450pyr from Sphingomonas species for the hydroxylation and two alcohol dehydrogenases which respectively transform (R) and (S) forms of P450pyr-mediated hydroxylated products to N-benzylpyrrolidinone to generate a colorimetric response. To generate the mutant library, 17 amino acid residues located in the active site were individually randomized by site-saturation mutagenesis. Although catalytic activity was decreased somewhat (shown by a decrease in conversion from 55% to 33% after 4 h with 5 mM substrate), it is notable that introducing a single mutation (N100S) could invert the (S)-enantioselectivity of wild-type P450pyr to (R) (42% ee). By directed evolution on the N100S variant, they further improved (R)-enantioselectivity to 83% ee. This study marks the first published high-throughput screen for enantioselective P450-mediated hydroxylation. The generality of this screening approach, however, depends on identifying appropriate dehydrogenases for quantitative detection of the enantiomeric products.
In humans, vitamin D3 (VD3) is activated to physiologically functional 1α,25(OH)2VD3 by two sequential P450-mediated hydroxylations. Vitamin D3 hydroxylase (Vdh), a CYP107 family enzyme that produces 1α,25(OH)2VD3 from VD3, was isolated from Pseudonocardia autophica, cloned and characterized. By screening a randomized Vdh library, Fujii et al. isolated a variant containing four mutations and exhibiting 21.6 fold higher VD3 hydroxylase activity than wild-type Vdh [36]. Hayashi et al. showed that a double mutant (R73V/R84V in the substrate recognition site) of another P450, CYP105A1, could transform vitamin D3 to 1α,25(OH)2VD3 with a catalytic efficiency (kcat/Km) two orders of magnitude greater than wild-type CYP105A1 [37].
A kind of structure-guided chimeragenesis has been used to create a P450 displaying combined beneficial properties of parent P450s from bacteria and insect [38•]. P450 BM3 hydroxylates farnesol to generate a mixture of 2,3-epoxyfarnesol, 10,11-epoxyfarnesol, and 9-hydroxyfarnesol. In contrast, CYP4C7 from the cockroach predominantly converts farnesol to 12-hydroxyfarnesol. A partial sequence alignment (around substrate recognition sites) between BM3 and CYP4C7 shows that these enzymes have only about 30% sequence identity in these regions. The X-ray crystal structure of P450 BM3 was aligned with a computationally modeled structure of CYP4C7, enabling the identification of residues interacting with substrate. Based on the alignment, substrate-interacting regions of P450 BM3 were replaced by those of CYP4C7. Substitution of nine amino acid residues on P450 BM3 (78–82, F87L, 328–330) with the equivalent residues from CYP4C7 gave P450 BM3 the insect terpenoid hydroxylase activity, producing 12-hydroxyfarnesol as the major product, while maintaining the high catalytic activity of wild-type P450 BM3. Remarkably, the variant (78–82, F87L, 328–330) converted farnesol with 2 fold and 100 fold increased rate (567 nmol farnesol consumed per min per nmol enzyme) compared to P450 BM3 (285 nmol per min per nmol enzyme) and CYP4C7 (4.1 nmol per min per nmol enzyme), respectively [38•].
A significant barrier to engineering and using P450s has been finding, cloning, and co-expressing the redox partners necessary for P450 activity. As each P450 requires redox partner(s) to bind NAD(P)H and transport a hydride to the heme domain, identification of the native redox partners or use of non-natural redox partners is necessary. Redox partner fusion may be an alternative strategy to finding and cloning the native redox partners. Developments through early 2009 in artificial fusion P450 constructs are covered in an excellent review [39•]. In addition to P450 BM3, another attractive native fusion of heme domain and redox partner is P450RhF isolated from Rhodococcus sp. NCIMPB 9784. Work by Sabbadin and coworkers has established a general method for rapid generation of libraries of various heme domains fused to the reductase domain of P450RhF (RhFRED) as a general redox partner [40]. The method was demonstrated on heme domains from P450cam and P450 XpIA, and the resulting enzymes were shown to be active on their original substrates, D-camphor and hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), respectively, with KM values similar to that of their native heme domains. Notably, coupling efficiency was impressively high, at 82% for the XpIA-RhFRED construct. Robin and colleagues explored the role of linker length between the reductase and heme domains when P450cam was fused to RhFRED [41]. The length was found to be optimal when extended 2 or more amino acids beyond the 22 amino acid linker in natural P450RhF, as measured by hydroxylation activity on D-camphor. The Sherman lab fused the RhFRED to macrolide P450 monooxygenase (PikC) involved in the biosynthesis of bioactive compounds pikromycin, methymycin and neomycin. Although coupling efficiency was not reported, the fusion protein (PikC-RhFRED) exhibited four-fold higher catalytic efficiency (kcat/Km) for the hydroxylation of 12- or 14-memberedring macrolide substrates compared to the reconstituted system. By the combination of RhFRED fusion, single mutation of PikC (D50N), and tethering of desosamine glycoside to the substrate for better recognition by the monooxygenase, they achieved a 31-fold improvement in catalytic efficiency (7.44 µM−1×min−1) for the 12-membered ring macrolide compared to the wild-type PikC reconstituted with reductase partners (0.24 µM−1×min−1). They subsequently used this system for regioselective hydroxylation of a variety of non-natural substrates, carbocyclic rings [42,43].
Because fusion of the heme domain and redox partners may offer a general approach to engineering P450s for biocatalysis applications, other natural fusion constructs have been investigated. Weis and coworkers expressed nine P450s from bacterial and fungal sources, all of which were natural fusion proteins of the heme domain and redox partners [44]. Although activities were low (8% and 12% isolated yield from two representative reactions that were scaled up), variants from the self-sufficient P450s could reproduce the same major metabolites generated by human P450s from the drug substrates diclofenac and chlorzoxazone (both non-steroidal anti-inflammatory agents, NSAIDS).
As an alternative to direct fusion of heme domain and redox partners in a single polypeptide, Hirakawa et al. used PCNA (proliferating cell nuclear antigen), a trimeric DNA binding protein complex, to make a heterotrimeric P450 [45]. The three components of P450cam monooxygenase system (putridaredoxin (PdR), putridaredoxin reductase (PdX), and P450cam) were fused to PCNA1, PCNA2, and PCNA3 to generate PCNA1-PdR, PCNA2-Pdx, and PCNA3-P450cam, respectively. Although the specific activity was much decreased as the protein concentration was lowered due to the quite low affinity of PCNA3 to the heterodimer (PCNA1-PdR - PCNA2-Pdx) (Kd = 270 nM), the designed complex exhibited 50-fold higher NADPH and oxygen consumption rates at 90 nM concentration when D-camphor was used as substrate compared to the reconstituted system consisting of equimolar concentrations (90 nM) of PdR, PdX, and P450cam domains. The rapid oxygen and cofactor consumption rates may indicate uncoupling and high NADPH oxidase activity, however, as no effort was made to directly measure D-camphor oxidation.
Engineered P450s are being used in metabolic pathways to produce valuable chemicals. The engineering of P450s for metabolic pathways poses new challenges in that the enzyme must now be compatible with the more complex cellular milieu. For example, P450 BM3 was engineered as part of a pathway to produce propanol from propane in vivo. Cofactor utilization was improved through engineering of reductase domain of P450 BM3 to utilize both NADH and NADPH [46]. In another application, Ajikumar et al. utilized a P450 in a process that overproduced (about 1 g/L) a cyclic intermediate for paclitaxel (taxol) synthesis in E. coli [47••]. The regioselective hydroxylation of taxadiene to taxadien-5α-ol, a critical step for the complete synthesis of paclitaxel, was performed by a P450 isolated from the Taxus species. By optimizing metabolic flux for taxadiene synthesis and using the chimeric enzyme in which the plant P450 heme domain was fused to the taxus CYP reductase, taxadien-5α-ol production was increased about 2,400 fold compared to a previously reported S. cerevisiae production system. These examples demonstrate that engineered P450s can be incorporated to improve intracellular processes through a combination of protein engineering and metabolic engineering.
Conclusions
The ability of the P450s to adopt new functions has mechanistic underpinnings that have yet to be fully elucidated, but is clearly an advantage that the iron-heme responsible for the unique P450 chemistry is retained in properly folded enzymes. The nonpolar nature and unusual conformational variability of the substrate binding pocket probably also contribute significantly to this enzyme’s ability to remodel its active site to adapt to new substrates and selectivities. The recent examples of P450 engineering presented here amply demonstrate that these enzymes can be ‘tamed’ for applications. Yet the bar for engineering these enzymes is high. Incorporation into complex metabolic pathways and commercial demands for enzymes that are functional in nonnatural environments (elevated temperature, nonnative pH, high substrate and product concentrations, organic solvents) present challenges in multi-variable optimization that have been met by P450s only partially to date. The multi-variable optimization that has been accomplished with other enzymes, such as a new transaminase engineered for the commercial production of the diabetes drug sitagliptin [48•], remains a model for biocatalysis. Given the facility with which P450s adopt new functions, this enzyme class will likely meet the challenge as well.
Table 1.
Examples of recent P450 engineering
| P450 | Substrate | Optimization goal | Ref. |
|---|---|---|---|
| P450 BM3 | Linear and cyclic terpenes, cycloalkanes | Hydroxylation | [21•,22] |
| P450 BM3 | Steroids, opiate alkaloids, peralkylated monosaccharides | Dealkylation and hydroxylation | [23•,29] |
| P450 BM3 | Lovastatin, resveratrol, phenacetin, ethyoxyresorufin, simvastin | Hydroxylation and dealkylation | [24–26] |
| P450 BM3 | Verapamil, astemizole | Production of human metabolites via hydroxylation and dealkylation | [27••] |
| P450 BM3 | Cyclopentenones, ibuprofen methyl ester, Corey lactones, 5-phenyloxazoline | Hydroxylation and demethylation | [28] |
| P450 BM3 | Dopamine | Binding for MRI contrast | [30] |
| P450 BM3 | Amorphadiene | Epoxidation | [32•] |
| P450 BM3 | Propylbenzene, 3-methylpentane, fluorene, (+)-α-pinene | Increased coupling efficiency and activity | [33,34] |
| P450pyr | N-benzylpyrrolidine | Enantioselective hydroxylation | [35•] |
| Vdh, CYP105A1 | Vitamin D3 (VD3) | Increased activity for two sequential hydroxylations to produce 1α,25(OH)2VD3 | [36,37] |
| P450 BM3 + CYP4C7 (chimera) | Farnesol | Regioselective hydroxylation | [38] |
| Taxus P450 | Taxadiene | Hydroxylation with alkene migration within a metabolic pathway | [47••] |
Acknowledgments
The authors acknowledge the support of the U.S. Department of Energy, BES grant DE-FG02-06ER15762, National Institutes of Health ARRA grant 2R01GM068664-05A1, and National Institutes of Health grant 1R01-DA028299. RL acknowledges the support of NIH fellowship 1F32GM095061-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funding agencies. We thank Eric Brustad, Philip Romero, Kersten Rabe, Mike Chen, and Indira Wu for helpful comments on various drafts of this review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors are aware of no conflicts of interest regarding the preparation and submission of this manuscript.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of interest
•• of outstanding interest
- 1.Ortiz de Montellano PR. Hydrocarbon hydroxylation by cytochrome P450 enzymes. Chem Rev. 2010;110:932–948. doi: 10.1021/cr9002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dewick PM. Medicinal Natural Products: A Biosynthetic Approach. 3rd Edition. John Wiley & Sons; 2009. [Google Scholar]
- 3.Guengerich FP. Cytochrome P450s and other enzymes in drug metabolism and toxicity. AAPS J. 2006;8:E101–E111. doi: 10.1208/aapsj080112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillam EMJ. Engineering cytochrome P450 Enzymes. Chem Res Toxicol. 2008;21:220–231. doi: 10.1021/tx7002849. [DOI] [PubMed] [Google Scholar]
- 5.Grogan G. Cytochromes P450: exploiting diversity and enabling application as biocatalysts. Curr Opin Chem Biol. 2011 doi: 10.1016/j.cbpa.2010.11.014. article in press. [DOI] [PubMed] [Google Scholar]
- 6.Kumar S. Engineering cytochrome P450 biocatalysts for biotechnology, medicine and bioremediation. Expert Opin Drug Metab Toxicol. 2010;6:115–131. doi: 10.1517/17425250903431040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabe K, Gandubert V, Spengler M, Erkelenz M, Niemeyer C. Engineering and assaying of cytochrome P450 biocatalysts. AnalBioanal Chem. 2008;392:1059–1073. doi: 10.1007/s00216-008-2248-9. [DOI] [PubMed] [Google Scholar]
- 8.Julsing MK, Cornelissen S, Bühler B, Schmid A. Heme-iron oxygenases: powerful industrial biocatalysts? Curr Opin Chem Biol. 2008;12:177–186. doi: 10.1016/j.cbpa.2008.01.029. [DOI] [PubMed] [Google Scholar]
- 9. Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol. 2009;10:866–876. doi: 10.1038/nrm2805. • This review discusses lessons learned from laboratory evolution studies and strategies for designing new directed evolution experiments.
- 10.Lewis JC, Arnold FH. Catalysts on Demand: Selective Oxidations by Laboratory-Evolved Cytochrome P450 BM3. Chimia. 2009;63:309–312. [Google Scholar]
- 11.O'Loughlin TL, Patrick WM, Matsumura I. Natural history as a predictor of protein evolvability. Protein Eng Des Sel. 2006;19:439–442. doi: 10.1093/protein/gzl029. [DOI] [PubMed] [Google Scholar]
- 12.Khersonsky O, Tawfik DS. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 13.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc Natl Acad Sci U S A. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pochapsky TC, Kazanis S, Dang M. Conformational plasticity and structure/function relationships in cytochromes P450. Antioxid Redox Signal. 2010;13:1273–1296. doi: 10.1089/ars.2010.3109. • This review examines the structural changes that several P450s undergo during catalysis, as determined by crystal structures and molecular dynamics simulations.
- 15.Ekroos M, Sjogren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci U S A. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol Cell. 2000;5:121–131. doi: 10.1016/s1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- 17.Williams PA, Cosme J, Ward A, Angove HC, Matak Vinkovic D, Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–468. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 18. Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. • The authors propose that the transiently stable conformations of proteins have different properties, and mutations alter the balance between these conformations during evolution of new functions.
- 19.Munro AW, Leys DG, McLean KJ, Marshall KR, Ost TW, Daff S, Miles CS, Chapman SK, Lysek DA, Moser CC, et al. P450 BM3: the very model of a modern flavocytochrome. Trends Biochem Sci. 2002;27:250–257. doi: 10.1016/s0968-0004(02)02086-8. [DOI] [PubMed] [Google Scholar]
- 20.Warman AJ, Roitel O, Neeli R, Givan HM, Seward HE, Murray SA, McLean KJ, Joyce MG, Toogood H, Holt RA, et al. Flavocytochrome P450 BM3: an update on structure and mechanism of a biotechnologically important enzyme. Biochem Soc Trans. 2005;33:747–753. doi: 10.1042/BST0330747. [DOI] [PubMed] [Google Scholar]
- 21. Seifert A, Vomund S, Grohmann K, Kriening S, Urlacher VB, Laschat S, Pleiss J. Rational design of a minimal and highly enriched CYP102A1 mutant library with improved regio-, stereo- and chemoselectivity. ChemBioChem. 2009;10:853–861. doi: 10.1002/cbic.200800799. • Mutations of two active site residues (F87 and A328) to selected non-polar amino acids created a library of 24 variants that displayed broad substrate scope for hydroxylation of cyclic and acyclic terpenoids.
- 22.Weber E, Seifert A, Antonovici M, Geinitz C, Pleiss J, Urlacher VB. Screening of a minimal enriched P450 BM3 mutant library for hydroxylation of cyclic and acyclic alkanes. Chem Commun. 2011;47:944–946. doi: 10.1039/c0cc02924f. [DOI] [PubMed] [Google Scholar]
- 23. Lewis JC, Mantovani SM, Fu Y, Snow CD, Komor RS, Wong C-H, Arnold FH. Combinatorial alanine substitution enables rapid optimization of cytochrome P450BM3 for selective hydroxylation of large substrates. ChemBioChem. 2010;11:2502–2505. doi: 10.1002/cbic.201000565. • To accommodate larger substrates including steroids, alkaloids and peralkylated monosaccharides, alanines were incorporated combinatorially into the substrate-binding pocket of thermostabilized P450 BM3.
- 24.Kim DH, Ahn T, Jung HC, Pan JG, Yun CH. Generation of the human metabolite piceatannol from the anticancer-preventive agent resveratrol by bacterial cytochrome P450 BM3. Drug MetabDispos. 2009;37:932–936. doi: 10.1124/dmd.108.026484. [DOI] [PubMed] [Google Scholar]
- 25.Kim DH, Kim KH, Kim D, Jung HC, Pan JG, Chi YT, Ahn T, Yun CH. Oxidation of human cytochrome P450 1A2 substrates by Bacillus megaterium cytochrome P450 BM3. J Mol Catal. 2010;63:179–187. [Google Scholar]
- 26.Yun CH, Kim KH, Kang JY, Kim DH, Park SH, Park SH, Kim D, Park KD, Lee YJ, Jung HC, et al. Generation of human chiral metabolites of simvastatin and lovastatin by bacterial CYP102A1 mutants. Drug Metab Dispos. 2011;39:140–150. doi: 10.1124/dmd.110.036392. [DOI] [PubMed] [Google Scholar]
- 27. Sawayama A, Chen M, Kulanthaivel P, Kuo MS, Hemmerle H, Arnold F. A Panel of cytochrome P450 BM3 variants to produce drug metabolites and diversify lead compounds. Chem-Eur J. 2009;15:11723–11729. doi: 10.1002/chem.200900643. • P450 BM3 mutants were shown to produce 12 of 13 human metabolites of the drugs verapamil and astemizole.
- 28.Rentmeister A, Arnold FH, Fasan R. Chemo-enzymatic fluorination of unactivated organic compounds. Nat Chem Biol. 2009;5:26–28. doi: 10.1038/nchembio.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis JC, Bastian S, Bennett CS, Fu Y, Mitsuda Y, Chen MM, Greenberg WA, Wong C-H, Arnold FH. Chemoenzymatic elaboration of monosaccharides using engineered cytochrome P450BM3 demethylases. Proc Natl Acad Sci U S A. 2009;106:16550–16555. doi: 10.1073/pnas.0908954106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shapiro MG, Westmeyer GG, Romero PA, Szablowski JO, Kuster B, Shah A, Otey CR, Langer R, Arnold FH, Jasanoff A. Directed evolution of a magnetic resonance imaging contrast agent for noninvasive imaging of dopamine. Nat Biotechnol. 2010;28:264–270. doi: 10.1038/nbt.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fasan R, Meharenna YT, Snow CD, Poulos TL, Arnold FH. Evolutionary history of a specialized P450 propane monooxygenase. J Mol Biol. 2008;383:1069–1080. doi: 10.1016/j.jmb.2008.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dietrich JA, Yoshikuni Y, Fisher KJ, Woolard FX, Ockey D, McPhee DJ, Renninger NS, Chang MC, Baker D, Keasling JD. A novel semi-biosynthetic route for artemisinin production using engineered substrate-promiscuous P450(BM3) ACS Chem Biol. 2009;4:261–267. doi: 10.1021/cb900006h. • The authors use a P450 BM3 mutant to oxidize the sesquiterpene amorphadiene to artemisinic-11S,12-epoxide at levels of 250 mg per L in E. coli. They chemically reduce the epoxide to (R)-dihydroartemisinic alcohol, which is an intermediate in the synthesis of the anti-malarial drug, artemisinin.
- 33.Whitehouse CJC, Bell SG, Yang W, Yorke JA, Blanford CF, Strong AJF, Morse EJ, Bartlam M, Rao Z, Wong L-L. A highly active single-mutation variant of P450BM3 (CYP102A1) ChemBioChem. 2009;10:1654–1656. doi: 10.1002/cbic.200900279. [DOI] [PubMed] [Google Scholar]
- 34.Whitehouse CJC, Yang W, Yorke JA, Rowlatt BC, Strong AJF, Blanford CF, Bell SG, Bartlam M, Wong L-L, Rao Z. Structural basis for the properties of two single-site proline mutants of CYP102A1 (P450BM3) ChemBioChem. 2010;11:2549–2556. doi: 10.1002/cbic.201000421. [DOI] [PubMed] [Google Scholar]
- 35. Tang WL, Li Z, Zhao H. Inverting the enantioselectivity of P450pyr monooxygenase by directed evolution. Chem Commun. 2010;46:5461–5463. doi: 10.1039/c0cc00735h. • P450pyr was used to catalyze stereoselective hydroxylation of a pyrrolidine. Production of both enantiomers was screened in a coupled enzyme high throughput assay.
- 36.Fujii Y, Kabumoto H, Nishimura K, Fujii T, Yanai S, Takeda K, Tamura N, Arisawa A, Tamura T. Purification, characterization, and directed evolution study of a vitamin D3 hydroxylase from Pseudonocardia autotrophica. Biochem Biophys Res Commun. 2009;385:170–175. doi: 10.1016/j.bbrc.2009.05.033. [DOI] [PubMed] [Google Scholar]
- 37.Hayashi K, Yasuda K, Sugimoto H, Ikushiro S, Kamakura M, Kittaka A, Horst RL, Chen TC, Ohta M, Shiro Y, et al. Three-step hydroxylation of vitamin D3 by a genetically engineered CYP105A1: enzymes and catalysis. FEBS J. 2010;277:3999–4009. doi: 10.1111/j.1742-4658.2010.07791.x. [DOI] [PubMed] [Google Scholar]
- 38. Chen CK, Berry R, Shokhireva T, Murataliev M, Zhang H, Walker F. Scanning chimeragenesis: the approach used to change the substrate selectivity of fatty acid monooxygenase CYP102A1 to that of terpene ω-hydroxylase CYP4C7. J Biol Inorg Chem. 2010;15:159–174. doi: 10.1007/s00775-009-0580-y. • Chimeras of P450 BM3 and CYP4C7 (an insect P450) were constructed to combine the high activity of P450 BM3 and the selectivity of CYP4C7.
- 39. Hlavica P. Assembly of non-natural electron transfer conduits in the cytochrome P450 system: A critical assessment and update of artificial redox constructs amenable to exploitation in biotechnological areas. Biotechnol Adv. 2009;27:103–121. doi: 10.1016/j.biotechadv.2008.10.001. • This review discusses the characteristics of redox partners for P450 heme domains, including fusion constructs.
- 40.Sabbadin F, Hyde R, Robin A, Hilgarth EM, Delenne M, Flitsch S, Turner N, Grogan G, Bruce NC. LICRED: a versatile drop-in vector for rapid generation of redox-self-sufficient cytochrome P450s. ChemBioChem. 2010;11:987–994. doi: 10.1002/cbic.201000104. [DOI] [PubMed] [Google Scholar]
- 41.Robin A, Roberts GA, Kisch J, Sabbadin F, Grogan G, Bruce N, Turner NJ, Flitsch SL. Engineering and improvement of the efficiency of a chimeric [P450cam-RhFRed reductase domain] enzyme. Chem Commun. 2009;18:2478–2480. doi: 10.1039/b901716j. [DOI] [PubMed] [Google Scholar]
- 42.Li S, Chaulagain MR, Knauff AR, Podust LM, Montgomery J, Sherman DH. Selective oxidation of carbolide C-H bonds by an engineered macrolide P450 mono-oxygenase. Proc Natl Acad Sci U S A. 2009;106:18463–18468. doi: 10.1073/pnas.0907203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Podust LM, Sherman DH. Engineering and analysis of a self-sufficient biosynthetic cytochrome P450 PikC fused to the RhFRED reductase domain. J Am Chem Soc. 2007;129:12940–12941. doi: 10.1021/ja075842d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weis R, Winkler M, Schittmayer M, Kambourakis S, Vink M, Rozzell JD, Glieder A. A diversified library of bacterial and fungal bifunctional cytochrome P450 enzymes for drug metabolite synthesis. Adv SynthCatal. 2009;351:2140–2146. [Google Scholar]
- 45.Hirakawa H, Nagamune T. Molecular assembly of P450 with ferredoxin and ferredoxin reductase by fusion to PCNA. ChemBioChem. 2010;11:1517–1520. doi: 10.1002/cbic.201000226. [DOI] [PubMed] [Google Scholar]
- 46.Fasan R, Crook NC, Peters MW, Meinhold P, Buelter T, Landwehr M, Cirino PC, Arnold FH. Improved product-per-glucose yields in P450-dependent propane biotransformations using engineered E. coli. Biotechnol Bioeng. 2011;108:500–510. doi: 10.1002/bit.22984. [DOI] [PubMed] [Google Scholar]
- 47. Ajikumar PK, Xiao WH, Tyo KEJ, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. •• A plant P450 was engineered into a cellular pathway to produce a paclitaxel intermediate. The plant P450 was expressed as a fusion construct with a different reductase domain, producing higher than native activity.
- 48. Savile CK, Janey JM, Mundorff EC, Moore JC, Tam S, Jarvis WR, Colbeck JC, Krebber A, Fleitz FJ, Brands J, et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science. 2010;329:305–309. doi: 10.1126/science.1188934. • Directed evolution was conducted on a transaminase used to install a chiral amine on a precursor of the diabetes drug sitagliptin. Multiple properties including organic solvent tolerance and substrate tolerance were optimized for the now-commercial process.
- 49.Sirim D, Wagner F, Lisitsa A, Pleiss J. The cytochrome P450 engineering database: Integration of biochemical properties. BMC Biochem. 2009;10:27. doi: 10.1186/1471-2091-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]