Abstract
We present an interactive program called HistoStitcher© for accurate and rapid reassembly of histology fragments into a pseudo-whole digitized histological section. HistoStitcher© provides both an intuitive graphical interface to assist the operator in performing the stitch of adjacent histology fragments by selecting pairs of anatomical landmarks, and a set of computational routines for determining and applying an optimal linear transformation to generate the stitched image. Reconstruction of whole histological sections from images of slides containing smaller fragments is required in applications where preparation of whole sections of large tissue specimens is not feasible or efficient, and such whole mounts are required to facilitate (a) disease annotation and (b) image registration with radiological images. Unlike manual reassembly of image fragments in a general purpose image editing program (such as Photoshop), HistoStitcher© provides memory efficient operation on high resolution digitized histology images and a highly flexible stitching process capable of producing more accurate results in less time. Further, by parameterizing the series of transformations determined by the stitching process, the stitching parameters can be saved, loaded at a later time, refined, or reapplied to multi-resolution scans, or quickly transmitted to another site. In this paper, we describe in detail the design of HistoStitcher© and the mathematical routines used for calculating the optimal image transformation, and demonstrate its operation for stitching high resolution histology quadrants of a prostate specimen to form a digitally reassembled whole histology section, for 8 different patient studies. To evaluate stitching quality, a 6 point scoring scheme, which assesses the alignment and continuity of anatomical structures important for disease annotation, is employed by three independent expert pathologists. For 6 studies compared with this scheme, reconstructed sections generated via HistoStitcher scored higher than reconstructions generated by an expert pathologist using Photoshop.
Keywords: whole mount histology, prostate cancer, reconstruction, reassembly, HistoStitcher, digital pathology
1. Introduction
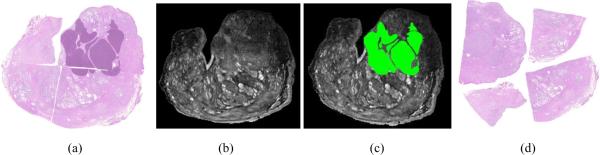
Histological sections of tissue specimens obtained via biopsy or surgical excision, such as lumpectomy or radical prostatectomy, are used for identifying presence and extent of disease, and if resolution is sufficient, the grade and malignancy of the disease [1–3]. In cases where the patient is scheduled for surgical resection of the diseased tissue or gland, it may be valuable to map disease extent from ex vivo histology sections onto in vivo radiological imaging, which is commonly acquired prior to excision, to discover signatures for disease on in vivo imaging [4–6]. For example, men with prostate cancer confirmed by biopsy and who are scheduled to undergo radical prostatectomy may receive pre-operative MRI of the prostate for identifying the presence of extra capsular spread. By registering these pre-operative MRI with corresponding ex vivo histology sections obtained from the radical prostatectomy specimen (see Figures 1(a) and (b)), pathological regions on histology (purple regions in Figures 1(a)) are then mapped onto the registered MRI (shown in green on Figure 1(c)). To achieve the required histology annotation and multi-modal registration steps described above, whole histological sections of entire slices of the specimen rather than fragmented sections of tissue are required.
Figure 1.
(a) Reconstructed pseudo-whole-mount histology from a prostate specimen with cancer (delineated in dark purple) may be used to identify signatures for disease on pre-operative in vivo imagery, similar to the MRI image shown in (b), by registering the two images. (c) The registered MRI slice that is in alignment with whole histology in (a) allows mapping of cancer onto MRI (green). (d) Quadrants comprising the whole histology section in (a) must be first reconstructed into a pseudowhole-mount section to facilitate improved annotation across cuts and registration with MRI in (b).
Obtaining whole-mount histological sections of large specimens, such as an enlarged prostate gland, while maintaining tissue integrity is not always feasible. For large specimens, a single glass slide is often too small to mount the entire contiguous tissue section, necessitating the cutting of the section into smaller fragments and mounting them onto multiple slides. Further, it is difficult to slice large specimens thin enough to achieve the very high spatial resolution required for accurate annotation without the use of specialized procedures and microtomes. For example, obtaining a whole slide for the specimen shown in Figure 1(b) with a thickness of only 4μm is difficult, prone to specimen damage, and inefficient, requiring exorbitant amounts of preparation time. As a result of these challenges, it is often preferable to adopt the much simpler approach of cutting large tissue specimens into smaller fragments and preparing multiple slides for separate analysis, such as quadrant sections shown in Figures 1(d).
Tissue fragments spread across multiple slides presents a significant hindrance for both (1) digital annotation of disease extent by a pathologist, which requires integration of visual cues across disjoint section boundaries, and (2) image processing tasks, such as registration of whole histological sections with in vivo imaging modalities. Annotation of multiple slides by a pathologist may be performed separately, however when the targeted pathology of interest crosses the boundary between slides, the process is complicated as the pathologist must repeatedly alternate between adjacent slides. As with any strenuous manual task, the annotations obtained in this manner may be inaccurate or unreliable. In addition to tissue annotation, whole histology slides are valuable in facilitating correlation with in vivo imagery by image registration, and much work has been done on registration of whole-mount histology with in vivo radiological imaging [7–10]. On the other hand, registration of smaller histologic image fragments to a sub-image or region of the in vivo data has not been widely investigated and is likely to be a more challenging registration task. Thus, in order to streamline and improve pathologist slide annotation, and facilitate image processing tasks requiring whole histological sections, it becomes useful to reconstruct a pseudo-whole-mount histological section from multiple individual fragments [6].
With the spread and growing acceptance of digital pathology [1–3], it is feasible for high resolution whole-mount sections to be digitally reconstructed from the images of the smaller fragments. Digitally reconstructed whole-mount histological sections would not only facilitate a variety of image processing tasks, but if pathologists can perform digital annotation of disease extent on high resolution histology images on computer monitors, annotations would be greatly improved compared to manual labeling of slides with felt pen or drawing on standardized examination sheets. Annotation could be improved in terms of both accuracy and efficiency using digital images by allowing labeling at any level of detail and for editing and revision of the markup. For example, in Figure 1(a) are the digitized histology quadrants of a section from a prostatectomy specimen with cancer. The closest corresponding MRI slice is shown in Figure 1(b). By digitally combining, or stitching, the quadrants a pseudo-whole-mount section can be generated (see Figure 1(c)), upon which efficient pathologist annotation can be performed on a high resolution computer monitor. The resulting cancer label established by analysis of the reconstructed pseudo-while-mount section is shown in purple, and can be seen to cross the cut between the original quadrants. Finally, having a reconstructed whole histology section, registration with corresponding in vivo imagery, such as MRI, can be performed to achieve mapping of spatial extent of disease (in this case prostate cancer) from the annotated whole histology onto corresponding radiological in vivo imaging. For example, the aligned MRI slice obtained in a previous study [4] is shown in Figure 1(d) with the mapped histopathologic cancer label shown in green. In this paper, we present a software utility called HistoStitcher© for computerized reconstruction of a whole histological section from digital images of the multiple slides of tissue fragments. While we demonstrate the utility of HistoStitcher© in the context of prostate sections in this work, the program is just as readily applicable to other domains and applications such as reassembly of whole histology sections of breast lumpectomy or mastectomy.
The rest of the paper is organized as follows. In Section 2 we discuss the challenges and requirements associated with digital reconstruction of high resolution histopathologic images. In Section 3.1 we present an overview of the HistoStitcher© software, including the interface and workflow. In Section 3.3 we detail the mathematical methods used to determine the optimal image transformation based on user-defined control points. In Section 3.4 we describe the application of the transformation to the full resolution histology and the process of stitching to create a larger image. In Section 3.6 we describe the features of HistoStitcher© that facilitate operation on very large images. In Section 4 we present formalized evaluation criteria and a 6 point scoring scheme for comparing and assessing the quality digitally reconstructed histology sections. In Section 5 we demonstrate the use of HistoStitcher© for reassembly of quadrants of prostate histology sections, and compare the results to manually reassembled (using Photoshop) sections and to block face photographs of the gland taken prior to cutting into quadrants. The reconstruction evaluation scheme is applied to compare the quality of stitches generated using HistoStitcher© to corresponding stitches generated using Photoshop for 6 prostate additional prostate studies. In Section 6 we present concluding remarks and enumerate further applications of HistoStitcher©.
2. Challenges in Digital Reconstruction of High Resolution Whole Histology
At institutions where high resolution digitization of excised tissue specimens are performed, an imaging software like Photoshop©1 is typically employed to digitally reconstruct a whole histology image when the tissue is fragmented across multiple slides. The use of conventional image editing programs for reassembly of high-resolution histology images is both inefficient and inadequate on several accounts. Firstly, these programs do not allow for simultaneous rotation, translation and scaling, which makes the digital stitching process using these programs tedious and the resulting transformation difficult to parameterize and record. Secondly, operating on high resolution (10× magnification) histology is extremely memory intensive. For example, a single digital histology image, which can be larger than 15,000 × 15,000 RGB pixels or 675 MBytes, necessitates careful memory handling to perform even basic image manipulations. The size of the images underscores the need for efficient parameterization of the transformations, which must be easily stored and applied to the full size images in chunks at a later time.
Despite the shortcomings of commercially available image editing programs for the task, digital reassembly of the fragments of whole histological sections requires expert interaction to ensure accurate reconstruction, where the corresponding edges of adjacent sections are aligned or “stitched” to preserve the continuity of anatomical structures of the tissue. Common automatic techniques for image stitching generally rely on significant overlap between adjacent images, such as with photograph [11–13] or microscopy [14–17] mosaic generation. However no such overlap exists between adjacent histology fragments. Automated edge matching techniques [18–21], which are generally designed for rigid objects such as puzzles and broken tiles, break down on account of uneven distortion and loss of tissue along the edges of histological fragments, leading to highly dissimilar contours of the two edges that were originally adjacent in the contiguous section. For example, the prostate histology quadrants shown in Figure 1(d) do not posses distinctive curvatures or other features on the contours of the adjacent edges, either at a macro or micro scale, which can be reliably used to align the edges. Many large macroscopic structures within the tissue are however visible that cross between quadrants. Thus, instead of trying to characterize overall edge similarity, the pathologist must observe the organization of tissue architecture and infer the continuity of structures across the cut. At these points along the cut where structural continuity is clear, the corresponding points along the two adjacent contours can simply be marked and used to perform the stitching.
HistoStitcher©, our interactive software package presented in this paper, adopts this approach based on identification of pairs of control points since (1) it provides a simple way for a pathologist to drive the reassembly process, (2) as few as three pairs of control points are sufficient to obtain a transformation, and (3) the transformation is easily parameterized, stored, and applied to high resolution images. While the underlying techniques employed by HistoStitcher© for image transformation using control points are not novel per se, their application to interactive digital reassembly of histology fragments and subsequent visualization of whole sections is. The design and operation of HistoStitcher© software is described in Section 3 below.
3. Features and Operation of HistoStitcher
3.1. Software Overview and Workflow
The HistoStitcher© software is comprised of an intuitive graphical user interface (GUI) and a set of computational routines for reassembly of a single stitched histological section from two images containing the adjacent fragments. A screenshot of the main HistoStitcher© GUI with two prostate histology quadrants loaded is shown in Figure 2. The operation of the program for stitching two adjacent histology fragments involves the following steps,
Load two adjacent images: the stationary image (left image) and the moving image (right image) that is to be transformed so that common edges are in the best possible alignment.
Select pairs of corresponding anatomical landmarks (points) that exist along the cut separating two adjacent sections. (Add Points button).
Specify constraints on the image transformation, including reflection, scale, and scale isotropy. (Tick boxes at bottom).
Automatically calculate the optimal coordinate transformation to minimize the error (in the least-squares sense) between pairs of control points. (Calculate Transform button).
Transform the moving image, bringing both images into a common coordinate frame.
Combine the images to generate the stitched image, expanding the image canvas as necessary. (Preview Stitch button).
Return the parameterized linear coordinate transformation and the full-resolution stitched image. (Done button).
Continue to Step 1 with the next adjacent fragment and the newly stitched image.
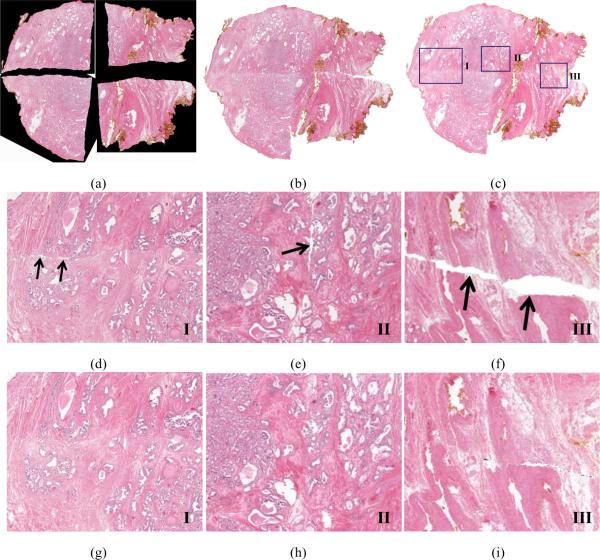
This general approach to reassembling a whole histology section from several fragments, such as the four quadrants of the prostate histology section shown in Figure 1(d), involves the cumulative stitching of two images at a time until all fragments have been combined. The process of stitching these quadrants, shown in Figures 3(a)–(d), proceeds as follows. First, two adjacent fragments (Figures 3(a) and (b)) are stitched using the steps above to generate a larger histology image (Figure 3(e)). The newly assembled semi-circular histology section is then stitched with the next histology fragment (Figure 3(c)) to generate a larger histology image (Figure 3(f)) comprising three quarters of the original whole section. Finally, the last histology fragment is stitched with the three-quarters section (Figure 3(f)) to generate the pseudo-whole prostate histology section, as shown in Figure 3(g).
Figure 2.
The HistoStitcher© graphical user interface where two histology pieces from the same section are digitally combined by selecting control point pairs along common boundaries. An optimal transformation of one fragment onto the the other is automatically determined based on user specified constraints.
Figure 3.
(a)–(d) Histology slide images of fragments (quadrants) of a whole section of a prostate specimen are reassembled using HistoStitcher©. (e) Half prostate histology section reconstructed by stitching quadrants in (a) and (b). (f) Three-quarters prostate histology section from stitching of quadrant in (c) with half in (e). (g) Final reconstructed pseudo-whole prostate histology section following stitching of quadrant in (d) with (e).
3.2. Notation
The pairs of anatomical landmarks identified using the HistoStitcher© GUI (shown as blue and green stars in Figure 2) correspond to two sets of N control points, denoted by the homogeneous coordinate matrices X = [x; y; 1] (moving) and U = [u; v; 1] (stationary), where x, y, u, v, 1 are row vectors of length N. We define the stationary and moving histology fragment images as and , where fA(c) and fB(c) are the image intensity or RGB values at each pixel c in rectangular coordinate sets CA and CB, respectively. We denote the transformation of as , defined by a new coordinate set Cβ and intensity function fβ(c) for each coordinate c ∈ Cβ. The stitched image comprising and β is denoted and defined on the coordinate frame CS. A comprehensive list of the main notations employed in this paper is given in Table 1.
Table 1.
List of notation and symbols used in this paper.
| Symbol | Description | Symbol | Description |
|---|---|---|---|
| X | Control point matrix on moving image | U | Control point matrix on stationary image |
| x,y | X- and Y-axis components of X | u, v | X- and Y-axis components of U |
| Mean-centered X | Mean-centered U | ||
| Normalized X | Normalized U | ||
| T | Linear coordinate transformation | R | Orthonormal rotation matrix |
| t | Translation vector | s | Scale factor |
| Mean cordinate of X | Mean cordinate of U | ||
| Stationary image on HistoStitcher© left | Moving image on HistoStitcher© right | ||
| CA | Coordinate set defined for | CB | Coordinate set defined for |
| fA(c) | Value of image at pixel c ∈ CA | fB(c) | Value of image at pixel c ∈ CB |
| β | Image transformed by T | C β | Coordinate set for β |
| fβ(c) | Value of image β at pixel c ∈ CB | α | Transparency value for background pixels |
| Stitched image comprising and β | Cs | Coordinate set for s.t. | |
| fS(c) | Value of image at pixel c ∈ CS | N | Number of control points |
3.3. Determining the Optimal Transformation from Control Point Pairs
The goal of this section is to determine a linear coordinate transformation T that minimizes the error e in the transformation of X via U = TX + e, subject to variable constraints on scaling (anisotropic, isotropic, none) and reflection (allowed, forced, none). To evaluate the accuracy of control point placement, the residual error in the mapping of X onto U is calculated as ‖TX – U‖2 and displayed on the GUI (shown beneath the “Calculate Transform” button in Figure 2).
Depending on the handling of the histology sections during creation and imaging of the slides, different constraints on the number of degrees of freedom (DOF) in the linear coordinate transformation, which is ultimately applied to the entire moving image, may be necessary. It is often necessary to reflect (mirror image) a section if the slice was flipped upside-down when it was scanned. Scaling may be necessary to correct for variable degrees of tissue shrinkage between fragments, or if the magnification used to acquire all sections was not exactly the same. Further anisotropic scaling (two independent scale factors on orthogonal axes) may be necessary when the slicing process causes deformations that are not equal in each direction. However, to prevent overfitting of the coordinates, especially when few or collinear landmarks were identified, it may be necessary to reduce the degrees of freedom by constraining or disallowing scaling and/or reflection. For example, when only one pair of edges are being stitched, the control points are mainly collinear and isotropic scaling should be used to prevent overfitting. When two or more pairs of edges are being stitched, the points are not collinear and anisotropic scaling can be safely used. However, if the object is truly rigid, such as if bone were being stitched, anisotropic scaling would never be used. The mathematical techniques used to find the optimal transformation T subject to the various constraints are described in Sections 3.3.1–3.3.5.
3.3.1. Unconstrained Affine: Anisotropic Scale Allowing Reflection
Maximal flexibility in the transformation of the moving image (shown in the right side of the GUI) is provided by allowing two independent scale parameters (anisotropic scaling) and reflection, in addition to rotation and translation, to align the selected control points. In the HistoStitcher© interface, this configuration corresponds to selecting the “Allow Reflection” and “Allow Scaling” check boxes, and unselecting the “Isotropic Scaling” checkbox. To achieve an alignment with these transformation possibilities, an unconstrained affine transformation Ta of X is defined as,
| (1) |
where a11, a12, a13, a21, a22, a23 are the 6 independent parameters that comprise Ta. The four parameters a11, a12, a21, a22 collectively control the scale, rotations and any reflection. The parameters a13 and a23 represent the translations of the moving image that occur after any scale, rotation and reflection operations.
Following selection of the control point pairs U and X and selecting the appropriate constraints, the user can then click the “Calculate Transform” button, directing HistoStitcher© to solve for Ta. HistoStitcher© finds the optimal transformation that minimizes the error between U and TaX,
| (2) |
where ∥·∥ is the Frobenius norm. Equation (2) can be minimized by expanding in terms of a11, a12, a13, a21, a22, a23 as,
| (3) |
and differentiating E(Ta) with respect to each of the six parameters and setting the resulting equations to zero. This produces the following linear system,
| (4) |
where Σ represents the sum over each element i (and j). Solving the system for [a11 a12 a13 a21 a22 a23]T provides the parameters of the optimal transformation Ta, which is then displayed in the HistoStitcher© GUI in the “Transformation” box beneath the stationary image on the left. To allow the operator to assess the accuracy of control point placement, the residual error of the minimization (calculated as ∥TaX − U∥2) is displayed immediately below the “Calculate Transform” button.
3.3.2. Isotropic Scaling with No Reflection
It may often be appropriate to constrain the transformation of the moving image by allowing only a single scale parameter (isotropic scaling), and disallowing any reflection (mirroring). To select this configuration, the operator checks the box for “Isotropic Scaling” and unchecks the box for “Allow Reflection”. Constraining the linear transformation to a single isotropic scaling factor and no reflection involves parameterizing the transformation Ts as,
| (5) |
where a1, a2, a3, a4 are the 4 independent parameters that comprise Ts. The parameters a1 and a2 collectively control the rotation and scale, while the negative sign prohibits reflecting the coordinates. The parameters a3 and a4 represent the translations. The error term may be formulated as,
| (6) |
| (7) |
and differentiated with respect to each of the four parameters a1, a2, a3 and a4. Setting the four resulting equations to zero provides the following matrix formulation,
| (8) |
When the “Calculate Transform” button is clicked, HistoStitcher© solves the system in Equation 8 for [a1 a2 a3 a4]T, yielding the optimal transformation Ts. This transformation is then displayed in the “Transformation” box.
3.3.3. Isotropic Scaling allowing Reflection
Similar isotropic scaling constrains may be enforced while allowing for reflection. Such constraints may be required if a the mirror image of a slide is loaded. To select this configuration, the operator checks the boxes for both “Isotropic Scaling” and “Allow Reflection”. When reflection is allowed, but not enforced, we seek a transformation Tp,s using Procrustes analysis [22] to determine a constituent 2-by-2 orthonormal transformation matrix R, a single scaling factor s and a translation vector t = [tx; ty]. The major steps of the method employed by HistoStitcher© to solve for Tp,s are as follows:
- Center x and v such that their mean corresponds to the origin, obtaining the following coordinate matrices,
(9)
where the elements of and are,(10)
and the elements of and are, - The centered coordinates and are then scaled to unit norm by,
(11)
where ∥·∥ is the Frobenius matrix norm.(12) Compute the rotation matrix R = QPT where PΣQT is the singular value decomposition (SVD) [23] of . The SVD PΣQT is comprised of two orthogonal matrixes P and Q corresponding to the eigenvectors of and respectively, and a diagonal matrix Σ of the square roots of the eigenvalues.
Compute the scale factor .
Compute the translation vector , where and are the mean coordinates of X and U.
- Construct the homogeneous transformation matrix,
(13)
When the “Calculate Transform” button is clicked, HistoStitcher© executes the above step to solve for To,s, which is then displayed in the “Transformation” box.
3.3.4. Rotation and Translation: No Scaling
If the images were acquired at exactly the same magnification and stored at the same spatial resolution, no scaling is necessary. In such an event only a transformation comprising pure rotation and translation would be required to stitch the fragments. To set up this configuration, the operator unchecks both the boxes for “Allow Scaling” and “Allow Reflection”. As described in [22], the rotation matrix R obtained via Procrustes analysis (steps 1–3 Section 3.3.3) is also used when no scaling factor is employed. The translation however is now calculated as . Finally, the homogeneous transformation matrix is constructed with the new translations,
| (14) |
HistoStitcher© calculates To,n as described above when the “Calculate Transform” button is clicked.
3.3.5. Enforcing Reflection
For any of the configurations described above (anisotropic scale, isotropic scale, no scale, allowed reflection), HistoStitcher© provides a mechanism to reflect (horizontally and/or vertically) the histology fragment in the moving image shown on the right. The toggle buttons shown to the right of the moving image in the HistoStitcher© GUI are used to reflect both the image and any previously selected control points. A manually specified horizontal or vertical reflection is combined into the transformation by TMx or TMy, where,
are the X and Y axis homogeneous reflection matrices. If both X and Y reflections are selected, a 180 degree rotation of the moving fragment is applied.
3.4. Transformation of Moving Image
On both images, background pixels not representing the tissue mounted on the slide are assigned a value α, a background intensity or color specified by a command line argument. The moving fragment image is transformed via T determined from the control points as described in Section 3.3, obtaining , defined by a new coordinate set and intensity function fβ(c) for each coordinate c ∈ Cβ. Cubic spline interpolation on fB is used to obtain the new image values fβ(c) at each c ∈ Cβ, while a value of α is assigned to coordinates originating outside the original image domain CB.
3.5. Image Reconstruction
The stationary and transformed fragment images and β are combined to create the reassembled histology section contained within a larger image by creating a continuous rectangular coordinate frame CS that is large enough to encompass both CA and Cβ (i.e. (CA ∪ Cβ) ⊂ CS). The value fS(c) of each pixel c ∈ CS in is defined by,
| (15) |
so that blending of fA and fβ only occurs at pixels c representing tissue on both and β. Note that the blending of fA and fβ will occur for few pixels in an accurately stitched pair of images, however it is necessary to address overlap that results from jagged features along the edges and other edge irregularities due to slicing and slide preparation. Thus, this area of blending will be minimal in a correct stitch and is only performed for regions where tissue deformation prevents perfect boundary alignment.
To facilitate this blending-with-transparency approach, a simple preprocessing tool is included in HistoStitcher© to identify the boundary of the tissue on the slide and hence the background. Any value of α may be selected to represent the slide background on the stitched image, such as values representing white or black.
3.6. Considerations for High Resolution Images
As previously mentioned, operations on high resolution histology images require special handling to avoid exhausting computer system memory. For example, the 10× magnification scan of the single prostate histology quadrant in Figure 3(b) contains 30k × 18k pixels, each pixel requiring 3 bytes to encode the RGB values, for a total of 1.6 GBytes of uncompressed data. Operations such as affine transformation on an image this size require excessive computation time and necessitate operating in tiles or chunks of the image when sufficient memory is not available to load the entire image. Therefore, HistoStitcher© has two important features to facilitate large images; these are (1) loading of any resolution within the image pyramid stored in an Aperio's SVS or multi-page TIFF file format and (2) down-sampling the two images for both display in the GUI and for previewing the stitch when the “Preview Overlay” button is pressed. By operating on a lower resolution pyramid level while placing control points and previewing the transformation, and by saving the obtain transformation, the full resolution images can be reassembled offline with the saved transformation using a system with more memory or using a routine that operates in blocks or tiles.
3.7. Reference Stitching: Manual Alignment of Fragments in Photoshop
In previous studies [4, 6, 10] that utilized reconstructed whole prostate histological sections for correlation with MR images, an expert pathologist performed stitching by manual manipulation of lower resolution quadrants in Photoshop. In this paper manual stitching of histology fragment images in Photoshop is compared to use of HistoStitcher©. Manipulation of the images in general purpose image editing software such as Photoshop involves performing any rotations, translations, and stretching (scaling) steps sequentially and thus independently. By contrast, HistoStitcher© simultaneously determines the single transformation that contains the optimal sequence of rotations, scale, and translation. Further, to closely stitch the edges of two histology fragments in a general image editing program, it is also necessary to specify a transparency layer by carefully delineating the boundary of the tissue on the slides.
4. Criteria for Stitching Quality and Evaluation by Multiple Expert Consensus
The criteria for assessment of the quality of a resconstructed histology section is defined to reflect how well the resulting pathology image facilitates the disease annotation process. When a pathologist reviews a histopathological section for prostate cancer, the pathologist first analyzes the tissue architecture at a low power magnification, at which a differential diagnosis is often made, and will zoom into higher power for confirmation. Thus, good stitching quality is most critical for the low power assessment where first impressions and often the diagnosis, which is of crucial importance in prostate cancer, are made. The disease annotation process involves inspecting salient anatomical structures (e.g. ducts, BPH nodules, urethra, capsule), which may span multiple quadrants. Therefore, with a well reconstructed section the assessment at low power is more efficient and accurate because a pathologist will not be required to perform complicated mental image transformations of the individual quadrants in order to follow the salient features across the cuts/boundaries.
In consort with our collaborating pathologists, we have defined stitch quality in terms of (1) continuity of tissue across the stitch boundaries, such that gaps and misalignment of anatomical structures are minimized, and (2) utility of the stitch for annotation of disease extent by a pathologist. A better stitch, as defined by this criterion, will facilitate a pathologist's ability to follow anatomical structures across the boundaries and, given the low to high power assessment approach employed by pathologists, perform annotation of disease extent more accurately and efficiently.
A quantitative 6 point scoring scheme, proposed by our collaborating and co-authoring pathologists, is employed to assess stitching quality. Under the 6 point scoring scheme, each reconstructed section receives a score from 0 to 6, with up to 2 points awarded for alignment of each of the following three anatomical features:
Capsule: continuity across the quadrants
Urethra: ability to visualize in its entirety
Glands: boundary and structural preservation of glands and other histological instances across the cut
Each anatomical feature receives either 0 points for no alignment, 1 point for partial alignment, or 2 points for full alignment. Therefore, even with some remaining gaps, points are awarded for good alignment of the edges of the anatomical structures.
5. Experimental Results and Discussion
5.1. Experiment 1: Reconstructing High Resolution Whole Prostate Sections from Quadrants
To demonstrate the operation of HistoStitcher© and evaluate both the accuracy of the reassembled histology sections and the efficiency of its use, we reconstruct high resolution whole histology sections of two di erent prostate specimens by stitching digitized slide images of the quadrants which compose each section. In the first prostate specimen, HistoStitcher's© operation is demonstrated in the context of the reassembly of a whole histology section from the very high resolution scans of the slides of the four fragments, or quadrants (Figures 3(a)–(d)). The slides were digitized using an Aperio slide scanner at 20× apparent magnification for approximately 0.5 microns per pixel. In HistoStitcher ©, image pyramid level 1 containing the 10× resolution images were loaded. The pixel dimensions of these four images shown in Figure 3(a)–(d) are approximately: 25k × 20k, 30k × 18k, 29k × 22k and 18k × 17k. To evaluate the efficacy of HistoStitcher© in terms of both the quality of the reassembled sections and the efficiency of stitching process, the same fragments were also stitched at a lower resolution (approximately 0.5× magnification or 20 microns per pixel) by an expert pathologist using Photoshop to manipulate the positions of the quadrants, as described in Section 3.7. To establish a baseline for stitching quality, we also acquired an image of the gross pathology section with a digital camera prior to cutting the section into quadrants, and staining and mounting each quadrant on slides. While this photograph shown in Figure 4(a) is not useful for diagnostic purposes, it shows the original configuration of the quadrants prior to cutting.
Figure 4.
(a) Photograph of specimen face prior to cutting into quadrants and mounting on slides. (b) Manually reassembled whole histology section obtained using Photoshop with low resolution images (final stitched image is 2,796 × 2,358 pixels). (c) HistoStitcher© reassembled whole histology section from high resolution images (final stitched image is 48k × 41k pixels). Both stitches in (b) and (c) are validated by the photograph of the section taken prior to cutting. (d)–(g) Close up views of regions I–IV over the seams of the HistoStitcher©-reassembled image in (c). (h)–(k) Close up views of the regions nearest to I–IV on the manually-reassembled image in (b). The seams in (d)–(g) contain smaller gaps and better continuity of internal tissue structures compared to the seams in (h)–(k).
As described in Section 3.1, the stitching of multiple fragments is cumulative and begins by stitching any two originally adjacent fragments. In this case, the quadrants in Figures 3(a) and (b) were stitched by placing corresponding pairs of control points along their common edge, automatic transformation of the moving image (the quadrant in Figure 3(a)), followed by reassembly of the larger image shown in Figure 3(e). Next, the quadrant in Figure 3(c) was stitched to the reassembled half in Figure 3(e) to generate the image in Figure 3(f). Finally, the quadrant in Figure 3(d) was stitched to the section in Figure 3(f) to generate the pseudo-whole mount section in Figure 3(g). The total time required to reassemble the final WMH section from the four quadrants was approximately 6 minutes. This included any time required to (i) load the images, (ii) select control points, (iii) preview the stitch, (iv) refine and/or add landmarks, (v) select appropriate transformation constraints, and (vi) perform the full resolution image transformation.
The reassembled whole-mount section generated using HistoStitcher© on the high resolution (10× apparent magnification) histology quadrants is shown in Figure 4(c), while the result of the manual stitching using Photoshop (see Section 3.7) is shown in Figure 4(b) for much lower resolution (0.5× magnification) images. To validate the accuracy of reassembled sections generated manually and using HistoStitcher ©, a block face photograph of the whole section prior to slicing into quadrants is shown in Figure 4(a). The HistoStitcher© result is shown in Figure 4(c) with four rectangular regions along the stitched boundaries highlighted. Close-up views of each of regions I–IV delineated in Figure 4(c) are shown in Figures 4(d)–(g), illustrating the continuity and smoothness of the stitched boundaries between the quadrants on the HistoStitcher© -reassembled section. For the manually stitched section in Figure 4(b), close-up views of approximately the same regions I–IV are shown in Figures 4(h)–(k), illustrating significantly larger gaps between the adjacent quadrants and lack of continuity of internal structures compared with the HistoStitcher© result. The full 10× reconstruction had dimensions of 48k × 41k pixels (approximately 5.9GB) and was obtained using a 64-bit workstation with 8GB RAM. To perform stitching at resolutions higher than 10× magnification would require either a server machine with greater amounts of memory, or computational methods to process and save the stitched image in blocks, or tiles. Note however that if the parameterized linear transformations for each of the three stitching steps were saved (Save Points button), they may easily be reloaded into HistoStitcher© (via Load Points button), which automatically adjusts the translation for the resolution difference.
5.2. Experiment 2: Comparison with Manual Stitching of High Resolution Sections
On a second high resolution study, manual stitching was performed at 1× magnification (approximately 7MB per quadrant), and also attempted at approximately 4× apparent magnification (approximately 110MB per quadrant). HistoStitcher© was again used to reassemble the quadrants of this second study on 8× resolution images (approximately 420 MB per quadrant). For this study, we compare both the (a) feasibility of manually stitching very high resolution images without a tool such as HistoStitcher ©, and (b) the efficiency of the stitching process in terms of total processing time. In both of these experiments, we use the background mask obtained by HistoStitcher© to establish the transparency layer used in the manual stitching process. This speeds up the manual stitching process and provides for a more fair comparison of stitching quality between the two methods.
The results of attempted manual stitching using Photoshop at 4× apparent magnification, and successful stitching at 1× magnification are shown in Figures 5(a) and (b), while the HistoStitcher© result using 8× magnification images is shown in 5(c). The obvious failure of Photoshop-based manual stitching at the full resolution (Figure 5(a)) is a result of insufficient memory to accommodate the necessary transparency layers and all three color planes. Without the transparency layer, the quadrants could not be brought any closer, as clearly shown in the resulting stitch in Figure 5(a). Therefore, the manual stitching task was performed again at a lower resolution of 1× apparent magnification. The reassembled section obtained by manipulation of these low resolution quadrants is shown in Figure 5(b). The machine used for manual stitching with Photoshop had 2GB of RAM, illustrating that this approach is clear memory inefficient. The high resolution reconstruction obtained using HistoStitcher© is shown in Figure 5(c). The 8× reconstruction the whole section had dimensions of about 22k × 18k pixels (approximately 1.2GB), and was obtained on a desktop computer with 6GB of RAM, although peak memory usage during stitching was less than 3GB, suggesting better memory efficiency.
Figure 5.
Comparison of whole histology reassembly via Photoshop vs. HistoStitcher©. (a) Inadequate stitching is obtained using Photoshop on 4× resolution images (final image is 13975 × 13675 pixels), as computer system memory of 2GB was insufficient to introduce the transparency layers required to bring the images closer. (b) Stitching results using Photoshop on low resolution (about 1× magnification) images (final image was 2706 × 2244 pixels). (c) HistoStitcher© result on high resolution (8× magnification) images (final image is about 22k × 18k). (d)–(f) Zoomed regions I-III of manually stitched image (b), compared to (g)–(i) same regions of HistoStitcher© result (c), demonstrating both more contiguous stitching with minimal gaps and improved continuity of tissue structures across the stitch using HistoStitcher©.
A summary of these stitching results is described in Table 2, which also lists the operation times required to generate each result. The close-up views of the manually stitched section in Figure 5(b) are shown in Figures (d)–(f), while the same regions on the HistoStitcher© result in Figure 5(c) are shown in Figures 5(g)–(i). Continuity of the gland architecture and tissue morphology is clearly visible across the stitched edges in Figures 5(g)–(i). The reconstructed section obtained using HistoStitcher© not only has smoother alignment of adjacent boundaries and better continuity of internal structures compared to the manually reconstructed section, but was obtained on the full resolution images in approximately the same amount of time required to perform a manual alignment on low resolution images. Using HistoStitcher ©, the reconstructed sections can be generated faster and/or more accurately than using an improvised approach in general purpose software, as evidenced by the results in Figure 5 and Table 2. Note in Table 2 the excessive time (56 minutes) required to operate on the medium resolution quadrants in Photoshop compared with the much shorter time (6 minutes) to combine the high resolution quadrants via HistoStitcher© . Only using drastically down-sized images was it possible to obtain a comparable stitch in a reasonable amount of time.
Table 2.
Comparison of HistoStitcher© to manipulation via Photoshop for the task of reassembling a whole prostate histological section from four separate slide images of histology quadrants. HistoStitcher© is demonstrated to be fast, memory efficient, and capable of producing better quality stitches of very high resolution images.
| Stitching Method | Image Size | Operation Time | Result | Output |
|---|---|---|---|---|
| Manipulation in Photoshop | Medium resolution (4x mag.) | 56 minutes | Partial failure due to memory requirements | Partially stitched image only |
| Manipulation in Photoshop | Low resolution (1x mag.) | 4 minutes | Success with some gaps, inconsistencies | Low-resolution stitched image only |
| HistoStitcher© | High resolution (8x mag.) | 6 minutes | Success with negligible gaps, overlap | All stitched images and fully-parameterized transformations |
5.3. Experiment 3: Evaluation of Stitching Quality via Multiple Experts
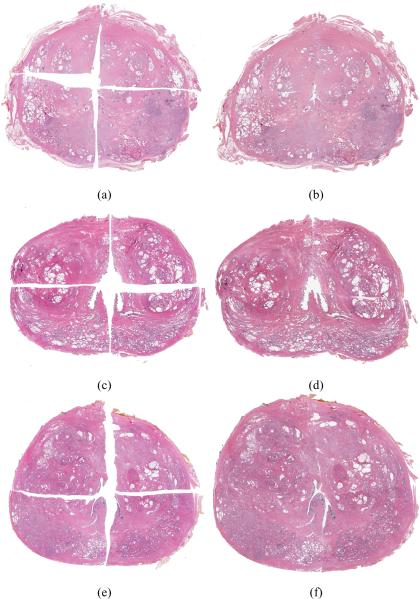
On 6 additional high resolution studies, both HistoStitcher© and manual stitching were used to reassemble the quadrants at 4× resolution (approximately 160MB per quadrant). A different computer workstation with 8GB of RAM and an Intel Core 2 Quad CPU was used for manual stitching in this experiment, while the same desktop computer with 6GB of RAM was used to run HistoStitcher. The results of the manual Photoshop stitching of 3 studies are shown in Figures 6(a), 6(c), and 6(e). The results of HistoStitcher© for the same 3 studies are shown in Figures 6(b), 6(d), and 6(f). Three independent expert pathologists scored each section using the 6 point scoring system described in Section 4. The average scores for the six sections are presented in Table 3. Section numbers 6, 1, and 4 correspond to the three rows in Figure 6. In addition to the 6 point scoring system, all three experts consistently identified the reconstructions generated by HistoStitcher as the higher quality stitch for each of the 6 sections in this experiment.
Figure 6.
Three prostate histology sections reconstructed using Photoshop (left column) and HistoStitcher (right column). Stitching quality scores for these sections are given Table 3 under section numbers 6 ((a) and (b)), 1 ((c) and (d)), and 4 ((e) and (f)).
Table 3.
Average and standard deviation of scores for 6 sections reconstructed using both Photoshop and HistoStitcher.
| Section Number | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Photoshop | 4.0 ± 1.0 | 3.33 ± 1.15 | 3.33 ± 0.58 | 4.33 ± 1.15 | 3.67 ± 0.58 | 4.0 ± 1.0 |
| HistoStitcher© | 5.0 ± 1.0 | 4.33 ± 1.15 | 5.33 ± 0.58 | 6.0 ± 0.0 | 4.33 ± 0.58 | 6.0 ± 0.0 |
6. Concluding Remarks
Reassembly of whole histological sections from smaller tissue fragments is necessary to facilitate improved pathologist annotation, especially when pathologies span multiple fragments, and multimodal image fusion such as registration of WMH with MRI. To achieve accurate reconstruction of sections with variable tissue loss and uneven deformations along the incisions between fragments, an expert-guided interactive process is necessary. In this paper, we address this need for an efficient histology reassembly tool and present HistoStitcher©, a graphical software package for combining images, that offers a powerful image alignment engine with flexible spatial transformation options. The program was demonstrated for the successful reconstruction of a whole histological section of the prostate from four “quadrants”, requiring only identification of pairs of anatomical landmarks along the boundaries via mouse clicks. Note that the reconstructed whole mount sections obtained by HistoStitcher© were generated by a naïve user, yet were obtained with greater accuracy and efficiency than the sections that were reconstructed via Photoshop by an expert pathologist. Using formalized criteria for stitching quality and a 6 point scoring scheme, which assesses the alignment and continuity of anatomical structures important for disease annotation, three independent expert pathologists evaluated the reconstructions of 6 prostate studies. Each of the reconstructed sections generated via HistoStitcher scored higher than the corresponding reconstructions generated by an expert pathologist using Photoshop. Further, the ability of HistoStitcher© to operate more efficiently on higher resolution images compared to Photoshop can be valuable in many applications. Although we demonstrated HistoStitcher© on prostate histology sections, the program can be applied to other applications such as reassembly of liver biopsy or breast lumpectomy histology fragments. Not only is HistoStitcher© efficient and flexible, with multiple options for constrained transformation, but the interface is intuitive enough to be used by relatively inexperienced users with minimal domain expertise.
Acknowledgment
The authors wish to thank Dr. Jason Hipp, MD, PhD, Clinical Pathologist at the Department of Pathology, University of Michigan Medical School for his valuable contributions. This work was made possible via grants from the Department of Defense Prostate Cancer Research Program (W81XWH-08-1-0072), Wallace H. Coulter Foundation, National Cancer Institute (Grant Nos. R01CA136535-01, R01CA140772-01, and R03CA143991-01), and The Cancer Institute of New Jersey.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Monaco JP, Tomaszewski JE, Feldman MD, Hagemann I, Moradi M, Mousavi P, Boag A, Davidson C, Abolmaesumi P, Madabhushi A. High-throughput detection of prostate cancer in histological sections using probabilistic pairwise markov models. Med Image Anal. 2010 doi: 10.1016/j.media.2010.04.007. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Doyle S, Tomaszewski J, Feldman M, Madabhushi A. Hierarchical boosted bayesian ensemble for prostate cancer detection from digitized histopathology. IEEE Transactions on Biomedical Engineering. 2010;57(7) doi: 10.1109/TBME.2010.2053540. accepted. [DOI] [PubMed] [Google Scholar]
- [3].Madabhushi A. Digital pathology image analysis: opportunities and challenges. Imaging in Medicine. 2009;1(1):7–10. doi: 10.2217/IIM.09.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chappelow J, Viswanath S, Monaco J, Rosen M, Tomaszewski J, Feldman M, Madabhushi A. Improving supervised classification accuracy using non-rigid multimodal image registration: detecting prostate cancer. SPIE: Medical Imaging. 2008;6915:69150V. [Google Scholar]
- [5].Vos PC, Hambrock T, van de Kaa CAH, Ftterer JJ, Barentsz JO, Huisman HJ. Computerized analysis of prostate lesions in the peripheral zone using dynamic contrast enhanced mri. Med Phys. 2008;35(3):888–899. doi: 10.1118/1.2836419. [DOI] [PubMed] [Google Scholar]
- [6].Madabhushi A, Feldman MD, et al. Automated detection of prostatic adenocarcinoma from high-resolution ex vivo MRI. IEEE Trans. Med. Imag. 2005;24:1611–1625. doi: 10.1109/TMI.2005.859208. [DOI] [PubMed] [Google Scholar]
- [7].Chappelow J, Madabhushi A, Rosen M, Tomaszeweski J, Feldman M. Multimodal image registration of ex vivo 4 Tesla MRI with whole mount histology for prostate cancer detection. SPIE. 2007;6512:65121S1–8. [Google Scholar]
- [8].Jacobs M, Windham J, et al. Registration and warping of magnetic resonance images to histological sections. Med. Phys. 1999;26(8):1568–1578. doi: 10.1118/1.598671. [DOI] [PubMed] [Google Scholar]
- [9].Taylor LS, Porter BC, Nadasdy G, di Sant'Agnese PA, Pasternack D, Wu Z, Baggs RB, Rubens DJ, Parker KJ. Three-dimensional registration of prostate images from histology and ultrasound. Ultrasound Med Biol. 2004;30(2):161–168. doi: 10.1016/j.ultrasmedbio.2003.10.005. [DOI] [PubMed] [Google Scholar]
- [10].Ou Y, Shen D, Feldman M, Tomaszewski J, Davatzikos C. Non-rigid registration between histological and mr images of the prostate: A joint segmentation and registration framework. Computer Vision and Pattern Recognition Workshop. 2009:125–132. [Google Scholar]
- [11].Jia J, Tang C-K. Image stitching using structure deformation. IEEE Trans Pattern Anal Mach Intell. 2008;30(4):617–631. doi: 10.1109/TPAMI.2007.70729. [DOI] [PubMed] [Google Scholar]
- [12].Suen ST, Lam EY, Wong KK. Photographic stitching with optimized object and color matching based on image derivatives. Opt Express. 2007;15(12):7689–7696. doi: 10.1364/oe.15.007689. [DOI] [PubMed] [Google Scholar]
- [13].Zomet A, Levin A, Peleg S, Weiss Y. Seamless image stitching by minimizing false edges. IEEE Trans Image Process. 2006;15(4):969–977. doi: 10.1109/tip.2005.863958. [DOI] [PubMed] [Google Scholar]
- [14].Rankov V, Locke RJ, Edens RJ, Barber PR, Vojnovic B. Conchello J-A, Cogswell CJ, Wilson T, editors. An algorithm for image stitching and blending. SPIE: Three-Dimensional and Multidimensional Microscopy: Image Acquisition and Processing XII, Vol. 5701, SPIE. 2005:190–199. [Google Scholar]
- [15].Chow SK, Hakozaki H, Price DL, MacLean NAB, Deerinck TJ, Bouwer JC, Martone ME, Peltier ST, Ellisman MH. Automated microscopy system for mosaic acquisition and processing. J Microsc. 2006;222(Pt 2):76–84. doi: 10.1111/j.1365-2818.2006.01577.x. [DOI] [PubMed] [Google Scholar]
- [16].Ma B, Zimmermann T, Rohde M, Winkelbach S, He F, Lindenmaier W, Dittmar KEJ. Use of autostitch for automatic stitching of microscope images. Micron. 2007;38(5):492–499. doi: 10.1016/j.micron.2006.07.027. [DOI] [PubMed] [Google Scholar]
- [17].Steckhan D, Bergen T, Wittenberg T, Rupp S. efficient large scale image stitching for virtual microscopy. Conf Proc IEEE Eng Med Biol Soc. 2008;2008:4019–4023. doi: 10.1109/IEMBS.2008.4650091. [DOI] [PubMed] [Google Scholar]
- [18].da Gama Leitao HC, Stolfi J. A multiscale method for the reassembly of two-dimensional fragmented objects. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2002;24(9):1239–1251. [Google Scholar]
- [19].Tsamoura E, Pitas I. Automatic color based reassembly of fragmented images and paintings. IEEE Transactions on Image Processing. 2010;19(3):680–690. doi: 10.1109/TIP.2009.2035840. [DOI] [PubMed] [Google Scholar]
- [20].Amigoni F, Gazzani S, Podico S. A method for reassembling fragments in image reconstruction. Proc. Int. Conf. Image Processing ICIP 2003. 2003;3:581–4. [Google Scholar]
- [21].Sagiroglu MS, Ercil A. A texture based matching approach for automated assembly of puzzles. Proc. 18th Int. Conf. Pattern Recognition ICPR 2006. 2006;3:1036–1041. doi:10.1109/ICPR.2006.184. [Google Scholar]
- [22].Gower JC, Dijksterhuis GB. Procrustes Problems. Oxford University Press; 2004. [Google Scholar]
- [23].Trefethen LN, Bau D. Numerical linear algebra, Society for Industrial and Applied Mathematics. 1997 [Google Scholar]