Abstract
Background
The pathogenesis of obesity is reportedly related to variations in the fat mass and an obesity-associated gene (FTO); however, as the number of reports increases, particularly with respect to varying ethnicities, there is a need to determine more precisely the effect sizes in each ethnic group. In addition, some reports have claimed ethnic-specific associations with alternative SNPs, and to that end there has been a degree of confusion.
Methods
We searched PubMed, MEDLINE, Web of Science, EMBASE, and BIOSIS Preview to identify studies investigating the associations between the five polymorphisms and obesity risk. Individual study odds ratios (OR) and their 95% confidence intervals (CI) were estimated using per-allele comparison. Summary ORs were estimated using a random effects model.
Results
We identified 59 eligible case-control studies in 27 articles, investigating 41,734 obesity cases and 69,837 healthy controls. Significant associations were detected between obesity risk and the five polymorphisms: rs9939609 (OR: 1.31, 95% CI: 1.26 to 1.36), rs1421085 (OR: 1.43, 95% CI: 1.33 to 1.53), rs8050136 (OR: 1.25, 95% CI: 1.13 to 1.38), rs17817449 (OR: 1.54, 95% CI: 1.41 to 1.68), and rs1121980 (OR: 1.34, 95% CI: 1.10 to 1.62). Begg's and Egger's tests provided no evidence of publication bias for the polymorphisms except rs1121980. There is evidence of higher heterogeneity, with I2 test values ranging from 38.1% to 84.5%.
Conclusions
This meta-analysis suggests that FTO may represent a low-penetrance susceptible gene for obesity risk. Individual studies with large sample size are needed to further evaluate the associations between the polymorphisms and obesity risk in various ethnic populations.
Background
Obesity is becoming a worldwide epidemic in modern society. It is prevalent in individuals of both genders and of all ages, socio-economic strata, and ethnic groups. It is estimated that the total number of overweight adults has reached more than 1.1 billion worldwide, including 312 million obese individuals and that about 10% of children are classified as overweight or obese [1,2]. The worldwide obesity epidemic is mainly caused by environmental factors, including excessive food intake and a lack of physical activity [3]. Obesity may lead to cardiovascular disease, type 2 diabetes, and several cancers. Overweight and inactivity account for an estimated one-quarter to a one-third of cancers of the breast, colon, endometrium, kidney, and esophagus [2].
In 2007, FTO (fat mass and obesity-associated gene) was first discovered in a genome-wide association study (GWAS) for type 2 diabetes [4], and, almost simultaneously, two other teams independently reported that the FTO gene was associated with obesity (or obesity-related traits) in a GWAS and a genetic association study [5,6]. The FTO gene, which is located on chromosome 16q12.2 and has nine exons, is strongly conserved across various vertebrate species (for example, fish and chicken) and emerged 450 million years ago [7]. FTO is mainly expressed in the hypothalamus and encodes a 2-oxoglutarate-dependent nucleic acid demethylase. It may play important roles in the management of energy homeostasis [7,8], nucleic acid demethylation, and the regulation of body fat masses by lipolysis [9].
The FTO gene has recently attracted much attention in obesity research. Previous genetic association-based studies have shown that SNPs in the FTO gene are associated with increased body mass index (BMI) [5,10,11], and/or other metabolic-related traits, such as higher fasting insulin [12], glucose [12], triglycerides [12], lower HDL cholesterol [12], waist circumference [11,13], and weight [5]. For example, Scuteri et al. [5] showed that SNP rs9930506 within FTO was associated with BMI, total body weight, and hip circumference; many variants have been reported to be associated with obesity, including rs9939609, rs1421085, rs8050136, rs17817449, and rs1121980. These single nucleotide polymorphisms (SNPs) lie within a 47-kilobase linkage disequilibrium (LD) block encompassing parts of the first two introns and exon 2 of FTO. The transcriptional start site of the retinitis pigmentosa GTPase regulator-interacting protein 1-like (RPGRIP1L) gene is also near the 5' end of FTO [4]. Based on this observation, Fawcett et al. [14] argued that the association signal could be due to a correlation between FTO intronic SNPs and RPGRIP1L.
Of the SNPs that were reported to associate with obesity, rs9939609 has been of particular interest since it was discovered by Frayling et al. [4] The associations of other SNPs in the FTO gene with obesity (or overweight) have been replicated in large European populations [5,6]. In some different ethnic groups, however, such as the Chinese [15] and Oceanic populations [16], no association was observed between rs9939609 and obesity by either genetic association studies or GWAS. However, as more studies are reported, particularly with respect to various ethnicities, there is a need to determine more precisely the effect sizes in each major racial group and to investigate the minor allele frequency (MAF) and the LD patterns of the SNPs across different ethnicities. In addition, some reports have claimed ethnic-specific associations with alternative SNPs, and to that end there has been a degree of confusion. Therefore, we conducted meta-analyses of all available data.
Methods
Publication search
We searched PubMed, MEDLINE, Web of Science, EMBASE, and BIOSIS Preview up to October 2010 for studies concerning the association between FTO polymorphisms and obesity (or obesity-related traits including body weight, leptin levels, subcutaneous fat, fat mass, hip and waist circumference, lean mass, body height, and BMI). There are no limits on language. Two search themes were combined using the Boolean operator "and". The first theme was ("obesity" (Mesh) OR "overweight" (Mesh) OR "body mass index" (Mesh) OR "obesity, morbid" (Mesh) OR "morbid obesity" OR "morbid obesities" OR "BMI" OR "body weight" OR "leptin levels" OR "subcutaneous fatness" OR "fat mass" OR "hip circumference" OR "waist circumference" OR "lean mass" OR "body height"). The second theme was ("rs9939609" OR "rs8050136" OR "rs1421085" OR "rs17817449" OR "rs1121980" OR "FTO" OR "FTO protein, human" (Substance Name) OR "fat mass and obesity associated" (Substance Name)). Meta-analysis articles were not excluded because several original studies were often combined in meta-analysis articles on genetic association studies.
Selection
Genetic association studies and GWASs with case-control subjects in which the case subjects were obese and the control subjects were healthy were included. At least two studies had to be available for each SNP. Studies were excluded if the control subjects violated the Hardy-Weinberg Equilibrium (HWE).
Data extraction
Two reviewers (SHP and YMZ) independently searched the databases. The search results were then evaluated by five reviewers (SHP, YMZ, FYX, XBR, and XBL). Disagreements were resolved by discussions among the reviewers. Information on gender, author name, country, ethnicity, year of publication, mean age of examination, mean BMI (calculated as weight in kilograms divided by height in meters squared), and genotypes (for example, TT/TA/AA) were retrieved. MAF and P-values of the HWE were calculated from the above genotype data. Adult individuals with BMI ≥30 kg/m2 were defined as case subjects of obesity, and individuals with BMI <30 kg/m2 were considered control subjects. The criteria for childhood obesity described by Cole et al. [17] were adopted.
Statistical analysis
To maximize the number of studies included, analyses were conducted by calculating the ORs under per-allele comparison. We found that an additive genetic model was always employed in most of the genetic association studies concerning the association between FTO and obesity risk. Therefore, the overall effects were also calculated in an additive genetic model in this meta-analysis. For comparison, the ORs in dominant and recessive genetic models were also calculated. Recently, Zintzaras [18] reported a novel method to calculate the generalized odds ratio (ORG), which may overcome the shortcomings of multiple model testing or erroneous model specification. Thus, the ORG calculations were also performed.
All statistical tests were performed with Stata 10.0 software (Stata, College Station, TX, USA). The effect sizes were calculated by the DerSimonian-Laird method, which is a random effect model because a modest or higher heterogeneous status was found in both allelic comparison and additive genetic models.
The Cochran's χ2 test (Q test) was used to evaluate heterogeneity between studies, and a threshold P-value of 0.1 was considered statistically significant. The I2 test was also conducted to evaluate heterogeneity between studies [19]. Begg's and Egger's tests were both used to test the significance of the publication bias, with a P-value < 0.1 considered as significant. We used the software developed by Guo et al. [20] to test (using exact test [21]) the significance of the HWE in control subjects, with a threshold P-value of 0.05 and 0.001 considered statistically significant for candidate gene association studies and GWAS [22,23], respectively. Heterogeneity is considered higher when I2 >50% and much higher when I2 >75% [24], and higher heterogeneity is a common phenomenon in genetic association studies [25,26]. To estimate heterogeneity, meta-regression was performed with the mean age of control subjects (Age_Control) and the mean BMI of control subjects (BMI_Control) as the covariates [27].
Analyses of sensitivity, subgroup, and LD pattern
To identify the source of the heterogeneity between studies, we performed sensitivity analyses by including and excluding some studies. Sensitivity analyses were done sequentially for all of the SNPs and all of the studies (or by some subgroups of the studies).
We sub-grouped the studies into six groups (Caucasian, Asian, Hispanic, African, South American, and Mixed) on rs9939609, four groups (Caucasian, Asian, African, and Mixed) on rs8050136, and two groups (Caucasian, Asian) on rs1121980.
The LD patterns of the SNPs were investigated using the HapMap Database (http://hapmap.ncbi.nlm.nih.gov/) and HaploView software (Whitehead Institute for Biomedical Research, Cambridge, USA) [28].
Results
Characteristics of the included studies
According to the search strategy, 170, 142, 160, 167, and 171 articles (a total of 810 articles) were retrieved from PubMed, MEDLINE, Web of Science, EMBASE, and BIOSIS Preview, respectively. After the first screening, in which the abstracts or titles were read, 753 articles were excluded, and 59 articles underwent further review. After reviewing these articles, 30 additional articles were excluded, which left a total of 27 articles for inclusion in this meta-analysis. According to the PRISMA guidelines [29], the flow diagram is shown in Figure 1. A total of 21 articles [4,15,30-48] included 30 studies on rs9939609; five articles [6,33,41,49,50] included eleven studies on rs1421085; seven articles [15,30,31,43,46,51,52] included nine studies on; rs8050136, two articles [6,33] included six studies on rs17817449; and three articles [37,43,53] included three studies on rs1121980. Not counting the overlapping literature, 27 articles were obtained (Figure 1). In the 30 studies concerning rs9939609, subgroup analyses were performed. The 30 studies were divided into six subgroups according to ethnicity, as follows: studies with Caucasian populations [4,33,36,41-45,47], Asian populations [15,31,32,35,37,46,48], Hispanic groups [31,38,39], South American [40], African [31], and studies using mixed ethnic populations [34]. In all of the included studies, the genotype distributions in the control subjects are consistent with the HWE. Three studies by Song et al. [31] (concerning rs9939609 and rs8050136) and one study by Price et al. [33] were excluded due to the deviation from the HWE (with a P-value of less than 0.05).
Figure 1.
Flow diagram of the study selection for the meta-analyses.
Unfortunately, some articles were excluded because the associations were between one or more SNPs and obesity-related traits rather than obesity itself (for example, ref. [5,10,11,16,54,55]). There are five included articles [15,34,51-53] with OR values and other detailed information but without genotype data. We attempted to obtain the genotyped data from these studies by e-mail, but we received no response from the authors.
The clinical characteristics of the included studies are shown in Table 1. The genotype data, as well as the ORs under both per-allele comparisons and the additive genetic model, are shown in Table 2. The ORs under dominant and recessive genetic models and the ORG are shown in Table S1 (see Additional file 1). These studies were published between 2007 and 2010. A total of 59 studies relating to all five SNPs were included, involving a total of 41,734 obesity cases and 69,837 healthy controls.
Table 1.
Clinical characteristics of the included studies
| Study | Publication year | Country | Ethnicity | Case | Control | ||
|---|---|---|---|---|---|---|---|
| Mean age (SD) (years) |
Mean BMI (SD) (kg/m2) |
Mean age (SD) (years) |
Mean BMI (SD) (kg/m2) | ||||
| Frayling_a [4]a | 2007 | UK | Caucasian | NAg | NA | 28.4 (4.7) | 22.7 |
| Frayling_b | 2007 | UK | Caucasian | NA | NA | 31.0 | 24.4 |
| Frayling_c | 2007 | UK | Caucasian | NA | NA | 40.6 (6.1) | 25.6 |
| Frayling_d | 2007 | UK | Caucasian | NA | NA | 56.7 (4.5) | 26.4 |
| Frayling_e | 2007 | UK | Caucasian | 60.1 (8.8) | 32.8 | 59.7 (9.0) | 26.1 |
| Frayling_f | 2007 | UK | Caucasian | NA | NA | 68.8 (5.5) | 27.2 |
| Frayling_g | 2007 | UK | Caucasian | NA | NA | 74.3 (6.9) | 27.2 |
| Hinney [43]b | 2007 | Germany | Caucasian | 14.4 (3.7) | 33.4 (6.8) | 26.1 (5.8) | 18.3 (1.1) |
| Dina_a [6] | 2007 | France | Caucasian | 44 (12) | 47 (7.6) | 51 (10) | 22.8 (2) |
| Dina_b | 2007 | France | Caucasian | 10.6 (3.5) | NA | 49 (5.5) | 22.3 (2.3) |
| Dina_c | 2007 | France | Caucasian | 11.8 (3.1) | 29.8 (5.8) | 22.0 (3.7) | 21.1 (2.0) |
| Dina_d | 2007 | Switzerland | Caucasian | NA | 46.7 (5.5) | NA | NA |
| Dina_e | 2007 | Germany | Caucasian | 11.6 (2.7) | NA | 11.7 (1.7) | NA |
| Song_H [31]c | 2007 | USA | Hispanic | NA | NA | 61.1 (6.71) | 30.9 (7.1) |
| Song_AP | 2007 | USA | Asian | NA | NA | 60.2 (6.77) | 29.0 (5.78) |
| Song_B | 2007 | USA | African | NA | NA | 63.8 (7.58) | 24.7 (4.49) |
| Villalobos [40] | 2008 | Mexico | South American | 43.9 (13.1) | 39.4 (8.6) | NA | NA |
| Chang [35] | 2008 | China-Taiwan | Asian | 37.0 (0.56) | 38.9 (8.2) | 61.1 (0.33) | 24.0 (2.9) |
| Hotta [37] | 2008 | Japan | Asian | 49.1 (14.2) | 34.5 (5.4) | 48.2 (16.5) | 21.7 (2.1) |
| Li [15] | 2008 | China | Asian | NA | NA | 58.6 (6.0) | 24.2 |
| Peeters [41] | 2008 | Belgium | Caucasian | NA | 38.3 (0.3) | NA | NA |
| Jacobsson_2008 [42] | 2008 | Sweden | Caucasian | 12.6 (3.3) | 35.4 (6.6) | 17.1 (0.8) | 21.1 (2.6) |
| Muler [44]d | 2008 | Germany | Caucasian | 10.7 (3.1) | 28.9 (5.2) | 24.6 (2.6) | 21.8 (1.1) |
| Andreasen_a [45] | 2008 | Denmark | Caucasian | NA | NA | 46.2 (7.9) | 26.3 (4.6) |
| Andreasen_be | 2008 | Denmark | Caucasian | NA | NA | 60.0 (6.8) | 28.6 (4.9) |
| Andreasen_c | 2008 | Denmark | Caucasian | NA | NA | 58.5 (8.1) | 26.5 (4.2) |
| Grant_C [51] | 2008 | USA | Caucasian | 2-18 | BMI ≥95th percentile | 2-18 | BMI<95th percentile |
| Grant_AA | 2008 | USA | African | 2-18 | BMI ≥95th percentile | 2-18 | BMI<95th percentile |
| Price [33] | 2008 | USA | Caucasian | 41.1 (9.4) | 49.1 (8.8) | 42.6 (8.8) | 20.8 (1.8) |
| Legry [36] | 2008 | France | Caucasian | 52.9 (8.2) | 33.8 (3.5) | 48.9 (8.5) | 22.5 (1.8) |
| Gonzalez [39] | 2009 | Spain | Hispanic | 58.0 (11.0) | 33.0 (3.3) | 54.0 (11.0) | 25.8 (2.6) |
| Zabena [38] | 2009 | Spain | Hispanic | 44.8 (12.5) | 45.5 (7.4) | 45.5 (7.3) | 24.3 (2.4) |
| Tabara [32]f | 2009 | Japan | Asian | NA | NA | 61.0 (14.0) | 23.4 (3.2) |
| Attaoua [49] | 2009 | France | Caucasian | 39.1 (11.4) | 39.4 (3.8) | 39.6 (11.6) | 23.8 (2.6) |
| Meyre_FC [50] | 2009 | France | Caucasian | 11.5 (3.1) | 29.0 (9.8) | 21.0 (4.5) | 20.74 (2.2) |
| Meyre_FA | 2009 | France | Caucasian | 41.1 (10.9) | 49.4 (7.8) | 49.1 (5.5) | 22.3 (2.3) |
| Meyre_SA | 2009 | Switzerland | Caucasian | 42.4 (11.5) | 43.7 (7.2) | NA | NA |
| Meyre_GC | 2009 | Germany | Caucasian | 11.8 (2.9) | 29.7 (9.2) | 11.8 (2.9) | 18.15 (8.1) |
| Thorleifsson [52] | 2009 | Mixed | Mixed | NA | NA | NA | NA |
| Willer [34] | 2009 | Mixed | Mixed | NA | NA | NA | NA |
| Renstrom [53] | 2009 | Sweden | Caucasian | 52.6 (9.5)h | >30 | 52.6 (9.5) h | 18.5-24.9 |
| Jacobsson_2009 [47] | 2009 | Sweden | Caucasian | 50.0 (0) | 32.1 (1.9) | 50.0 (0) | 24.8 (1.8) |
| Karasawa [48]f | 2010 | Japan | Asian | NA | NA | 63.0 (1.2) | 23.5 (3.2) |
| Liu [46] | 2010 | China | Asian | NA | NA | 58.0 (9.0) | 24.5 (3.2) |
| Cheung [30] | 2010 | China | Asian | 46.3 (11.9) | 32.2 (4.9) | 45.0 (12.5) | 21.2 (1.2) |
a: The article by Frayling et al. contains seven studies, namely Frayling _a (ALSPAC), Frayling _b (NFBC1966), Frayling _c (Oxford Biobank), Frayling _d (Caerphilly), Frayling _e (EPIC-Norfolk), Frayling _f (BWHHS), and Frayling _g (InCHIANTI). Please refer to the original article [4] for details.
b: Early-onset, extremely obese cases (mean BMI Z score 4.63 ± 2.27) and healthy, underweight controls (mean BMI Z score -1.38 ± 0.35; BMI <15thage percentile).
c: The age and BMI values are reported as the mean value of all case and control subjects.
d: Obese cases are reported as the 99.5th percentile of the median age and gender-specific BMI. Lean controls are reported as the 51.0st percentile of the median age and gender-specific BMI.
e: The article by Andreasen et al. contains three studies, including Andreasen_a (Population-based, Inter99 study sample), Andreasen_b
(ADDITION study cohort), and Andreasen_c (SDC control group). For details, please refer to the original article [45].
f: In Japan, obesity is defined as a condition in which the individuals had a BMI ≥25 kg/m2 according to the guidelines of the Japanese Society for the Study of obesity [68].
g: Not available.
h: The mean age of all subjects (non-diabetic individuals).
Table 2.
Genotypic distributions and ORs for the association between five polymorphisms and obesity
| Study | N (case/control) | OR [95% CI], P-value | ||||||
|---|---|---|---|---|---|---|---|---|
| Subtotal | MAF | PHWE | 11a | 12 | 22 | Additive model | Allelic comparison | |
| rs9939609 | ||||||||
| Caucasian: | ||||||||
| Frayling_a | 5,380 (353/5,027) | 0.48/0.39 | 0.513 | 107/1,879 | 157/2,407 | 89/741 | 1.43 (1.23 to 1.67) | 1.43 (1.23 to 1.67) |
| Frayling_b | 3,072 (415/2,657) | 0.45/0.37 | 0.185 | 132/1,060 | 193/1,211 | 90/386 | 1.36 (1.18 to 1.57) | 1.37 (1.18 to 1.59) |
| Frayling_c | 487 (126/361) | 0.42/0.38 | 0.575 | 45/138 | 56/175 | 25/48 | 1.21 (0.90 to 1.62) | 1.21 (0.91 to 1.61) |
| Frayling_d | 623 (200/423) | 0.41/0.34 | 0.447 | 67/189 | 101/182 | 32/52 | 1.37 (1.07 to 1.75) | 1.37 (1.08 to 1.76) |
| Frayling_e | 2,511 (1,588/923) | 0.43/0.38 | 0.625 | 485/351 | 847/443 | 256/129 | 1.24 (1.10 to 1.40) | 1.22 (1.09 to 1.37) |
| Frayling_f | 1,875 (840/1,035) | 0.42/0.37 | 0.547 | 274/421 | 427/471 | 139/143 | 1.26 (1.10 to 1.44) | 1.25 (1.10 to 1.43) |
| Frayling_g | 461 (212/249) | 0.47/0.42 | 0.514 | 66/82 | 92/127 | 54/40 | 1.25 (0.97 to 1.61) | 1.26 (0.97 to 1.63) |
| Andreasen_a | 3,724 (1,045/2,679) | 0.45/0.39 | 0.103 | 313/993 | 517/1,309 | 215/377 | 1.33 (1.20 to 1.48) | 1.32 (1.19 to 1.47) |
| Andreasen_b | 4,671 (2,819/1,852) | 0.43/0.39 | 0.221 | 911/684 | 1,385/904 | 523/264 | 1.21 (1.11 to 1.31) | 1.20 (1.11 to 1.31) |
| Andreasen_c | 377 (104/273) | 0.45/0.39 | 0.076 | 30/93 | 55/145 | 19/35 | 1.27(0.91 to 1.80) | 1.25 (0.90 to 1.72) |
| Muler | 697 (519/178) | 0.50/0.44 | 0.763 | 140/56 | 238/86 | 141/36 | 1.24 (0.98 to 1.57) | 1.26 (0.99 to 1.60) |
| Price | 1,047 (527/520) | 0.48/0.36 | 0.084 | 152/207 | 242/257 | 133/56 | 1.69 (1.41 to 2.02) | 1.69 (1.42 to 2.02) |
| Peeters | 1,367 (1,099/268) | 0.46/0.40 | 0.703 | 316/94 | 553/133 | 230/41 | 1.28 (1.05 to 1.56) | 1.28 (1.05 to 1.55) |
| Jacobsson_2008 | 960 (450/510) | 0.48/0.42 | 0.716 | 133/174 | 206/244 | 111/92 | 1.24 (1.04 to 1.48) | 1.25 (1.05 to 1.50) |
| Legry | 3,367 (598/2,769) | 0.46/0.41 | 0.050 | 171/924 | 307/1,395 | 120/450 | 1.20 (1.05 to 1.36) | 1.19 (1.05 to 1.35) |
| Jacobsson_2009 | 1,115 (54/1,061) | 0.44/0.39 | 0.218 | 16/411 | 29/483 | 9/167 | 1.22 (0.83 to 1.80) | 1.23 (0.83 to 1.82) |
| Hinney | 926 (484/442) | 0.50/0.39 | 0.690 | 123/164 | 241/214 | 120/64 | 1.57 (1.30 to 1.90) | 1.57 (1.30 to 1.88) |
| Asian: | ||||||||
| Song_AP | 240 (77/163) | 0.20/0.17 | 0.409 | 50/114 | 23/43 | 4/6 | 1.23 (0.76 to 1.97) | 1.24 (0.76 to 2.03) |
| Chang | 2,135 (610/1,525) | 0.17/0.13 | 0.355 | 425/1,158 | 167/347 | 18/20 | 1.38 (1.15 to 1.66) | 1.37 (1.14 to 1.5) |
| Hotta | 2,423 (919/1,504) | 0.24/0.19 | 0.393 | 534/1,005 | 334/443 | 51/56 | 1.37 (1.19 to 1.58) | 1.66 (1.31 to 2.11) |
| Tabara | 1,718 (214/1,504) | 0.22/0.16 | 0.438 | 128/1,063 | 77/408 | 9/33 | 1.54 (1.20 to 1.99) | 1.53 (1.19 to 1.95) |
| Karasawa | 2,639 (794/1,845) | 0.23/0.20 | 0.373 | 477/1,203 | 271/566 | 46/76 | 1.22 (1.06 to 1.41) | 1.23 (1.06 to 1.42) |
| Liu | 1,167 (276/891) | 0.14/0.11 | 0.396 | 201/701 | 70/181 | 5/8 | 1.38 (1.04 to 1.83) | 1.36 (1.03 to 1.80) |
| Li | 3,210 (398/2,812) | NA/0.12 | NA | NA | NA | NA | 0.90 (0.70 to 1.15) | NA |
| Hispanic: | ||||||||
| Song_H | 415 (139/276) | 0.27/0.31 | 0.157 | 75/138 | 53/107 | 11/31 | 0.85 (0.62 to 1.16) | 0.84 (0.61 to 1.15 |
| Gonzalez | 725 (207/518) | 0.45/0.37 | 0.925 | 62/203 | 104/244 | 41/71 | 1.38 (1.09 to 1.74) | 1.37 (1.09 to 1.73) |
| Zabena | 255 (75/180) | 0.52/0.34 | 1.0 | 19/78 | 34/81 | 22/21 | 2.04 (1.38 to 3.02) | 2.09 (1.42 to 3.07) |
| African: | ||||||||
| Song_B | 1,114 (365/749) | 0.47/0.45 | 0.160 | 113/239 | 161/351 | 91/159 | 1.09 (0.92 to 1.30) | 1.10 (0.92 to 1.31) |
| South American: | ||||||||
| Villalobos | 788 (364/424) | 0.25/0.17 | 0.598 | 209/297 | 127/114 | 28/13 | 1.66 (1.31 to 2.11) | 1.7 (1.33 to 2.17) |
| Mixed: | ||||||||
| Willer | 19,367 (5,261/14,106) | NA | NA | NA | NA | NA | NA | 1.25 (1.19 to 1.31) |
| Total N (rs9939609) | 68,856 (21,132/47,724) | Pooled ORs | 1.31 (1.25 to 1.37) | 1.31 (1.26 to 1.36) | ||||
| rs1421085 | ||||||||
| Causian: | ||||||||
| Dina_a | 3,557 (867/2,690) | 0.52/0.41 | 0.218 | 200/944 | 425/1,273 | 242/473 | 1.55 (1.39 to 1.73) | 1.57 (1.41 to 1.75) |
| Dina_b | 1,678 (699/979) | 0.51/0.42 | 0.896 | 173/323 | 334/481 | 192/175 | 1.42 (1.24 to 1.63) | 1.43 (1.25 to 1.64) |
| Dina_c | 1,001 (482/519) | 0.51/0.41 | 0.205 | 119/187 | 233/237 | 130/95 | 1.47 (1.24 to 1.75) | 1.50 (1.26 to 1.79) |
| Dina_d | 1,018 (504/514) | 0.52/0.46 | 0.051 | 123/161 | 235/233 | 146/120 | 1.26 (1.07 to 1.49) | 1.29 (1.08 to 1.53) |
| Dina_e | 551 (283/268) | 0.53/0.40 | 0.637 | 62/246 | 142/343 | 79/110 | 1.69 (1.38 to 2.06) | 1.67 (1.37 to 2.04) |
| Peeters | 1,367 (1,099/268) | 0.48/0.41 | 0.900 | 300/93 | 544/129 | 255/46 | 1.31 (1.08 to 1.59) | 1.31 (1.09 to 1.59) |
| Attaoua | 248 (119/129) | 0.55/0.41 | 0.585 | 24/47 | 58/59 | 37/23 | 1.78 (1.24 to 2.54) | 1.81 (1.27 to 2.59) |
| Meyre_FC | 1,036 (491/545) | 0.49/0.41 | 0.077 | 137/197 | 231/244 | 123/104 | 1.31 (1.11 to 1.55) | 1.33 (1.12 to 1.59) |
| Meyre_FA | 859 (132/727) | 0.51/0.43 | 0.820 | 31/238 | 67/359 | 34/130 | 1.42 (1.09 to 1.85) | 1.41 (1.09 to 1.84) |
| Meyre_SA | 1,346 (1,028/318) | 0.50/0.46 | 0.055 | 286/100 | 460/141 | 282/77 | 1.13 (0.96 to 1.34) | 1.15 (0.96 to 1.37) |
| Meyre_GC | 1,080 (370/710) | 0.55/0.45 | 0.081 | 76/227 | 181/328 | 113/155 | 1.47 (1.23 to 1.75) | 1.50 (1.25 to 1.79) |
| Total N (rs1421085) | 13,741 (6,074/7,667) | Pooled ORs | 1.41 (1.31 to 1.51) | 1.43 (1.33 to 1.53) | ||||
| rs8050136 | ||||||||
| Caucasian: | ||||||||
| Hinney | 928 (486/442) | 0.50/0.39 | 0.842 | 123/165 | 240/212 | 123/65 | 1.58 (1.31 to 1.91) | 1.59 (1.32 to 1.91) |
| Grant_C | 2,688 (418/2,270) | 0.45/0.39 | NA | NA | NA | NA | NA | 1.27 (1.09 to 1.47) |
| Asian: | ||||||||
| Song_AP | 240 (77/163) | 0.20/0.18 | 0.425 | 52/111 | 20/45 | 5/7 | 1.09 (0.68 to 1.73) | 1.10 (0.67 to 1.78) |
| Cheung | 1,159 (468/691) | 0.16/0.12 | 0.463 | 326/535 | 131/149 | 11/7 | 1.48 (1.16 to 1.89) | 1.46 (1.15 to 1.85) |
| Liu | 1,180 (278/902) | 0.15/0.11 | 0.489 | 201/716 | 73/178 | 4/8 | 1.43 (1.08 to 1.91) | 1.42 (1.07 to 1.87) |
| Li | 3,210 (398/2,812) | NA/0.12 | NA | NA | NA | NA | 0.91 (0.71 to 1.16) | NA |
| African: | ||||||||
| Song_B | 1,114 (365/749) | 0.42/0.42 | 0.454 | 133/257 | 161/355 | 71/137 | 0.98 (0.82 to 1.17) | 0.98 (0.82 to 1.17) |
| Grant_AA | 2,002 (578/1,424) | 0.45/0.44 | NA | NA | NA | NA | NA | 1.05 (0.91 to 1.21) |
| Mixed: | ||||||||
| Thorleifsson | 22,277 (13,785/8,492) | NA | NA | NA | NA | NA | NA | 1.27 (1.21 to 1.32) |
| Total N (rs8050136) | 34,798 (16,853/17,945) | Pooled ORs | 1.22 (0.99 to 1.51) | 1.25 (1.13 to 1.38) | ||||
| rs17817449 | ||||||||
| Caucasian: | ||||||||
| Dina_a [6] | 3,548 (873/2,675) | 0.49/0.60 | 0.968 | 212/948 | 426/1,288 | 235/439 | 1.55 (1.39 to 1.72) | 1.55 (1.39 to 1.73) |
| Dina_b | 1,589 (689/900) | 0.47/0.59 | 0.600 | 164/337 | 320/489 | 205/164 | 1.59 (1.38 to 1.83) | 1.60 (1.40 to 1.84) |
| Dina_c | 1,010 (485/525) | 0.49/0.60 | 0.173 | 120/196 | 236/237 | 129/92 | 1.52 (1.28 to 1.81) | 1.55 (1.30 to 1.85) |
| Dina_d | 1,035 (516/519) | 0.50/0.55 | 0.374 | 138/164 | 243/246 | 135/109 | 1.21 (1.02 to 1.43) | 1.22 (1.03 to 1.45) |
| Dina_e | 972 (281/691) | 0.46/0.58 | 0.755 | 58/231 | 142/341 | 81/119 | 1.65 (1.35 to 2.01) | 1.63 (1.34 to 1.99) |
| Price | 1,049 (527/522) | 0.49/0.36 | 0.087 | 145/205 | 247/259 | 135/58 | 1.72 (1.44 to 2.06) | 1.72 (1.44 to 2.05) |
| Total N (rs17817449) | 9,203 (3,371/5,832) | Pooled ORs | 1.53 (1.40 to 1.67) | 1.54 (1.41 to 1.68) | ||||
| rs1121980 | ||||||||
| Caucasian: | ||||||||
| Hinney | 926 (484/442) | 0.53/0.41 | 0.694 | 104/153 | 247/218 | 133/71 | 1.66 (1.37 to 2.01) | 1.64 (1.37 to 1.97) |
| Renstrom | 1,723 (353/1,370) | NA | NA | NA | NA | NA | NA | 1.15 (1.05 to 1.25) |
| Asian: | ||||||||
| Hotta | 2,451 (927/1,524) | 0.26/0.21 | 0.593 | 499/947 | 367/504 | 61/73 | 1.32 (1.15 to 1.51) | 1.32 (1.16 to 1.51) |
| Total N (rs1121980) | 5,100 (1,764/3,336) | Pooled ORs | 1.46 (1.17 to 1.83) | 1.34 (1.10 to 1.62) | ||||
| Total Subjects (duplicated subjects were excluded) | 112,327 (42,616/69,711) | |||||||
a: 11, 12, and 22 denote TT, TA, and AA for rs9939609; TT, TC, and CC for rs1421085; CC, CA, and AA for rs8050136; TT, TG, and GG for rs17817449; and GG, GA, and AA for rs1121980.
We estimated the MAF in the five polymorphisms from the control subjects of all studies identified for inclusion in the present meta-analysis. Across all studies, the MAFs ranged between 11% and 45%, 40% and 46%, 11% and 44%, 36% and 60%, and 21% and 44% for rs9939609, rs1421085, rs8050136, rs17817449, and rs1121980, respectively.
Meta-analysis of FTO gene SNP rs9939609
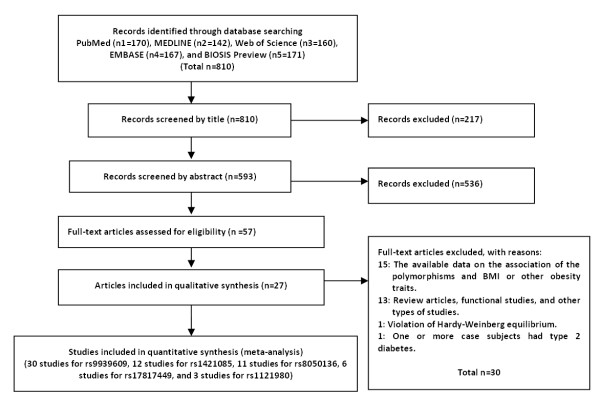
Under per-allele comparison, the OR is not available from the study by Li et al., leaving 29 studies for further consideration. A total of 21 out of 29 studies reported a significant, positive association between obesity and the rs9939609 genotype (Table 2, Figure 2). A significant association between obesity and rs9939609 was detected, with an overall OR of 1.31 (95% CI: 1.26 to 1.36) under per-allele comparison, and there is evidence of heterogeneity (I2 = 44.0%). The Begg's test (P = 0.39), and Egger's test (P = 0.17) provided no evidence of publication bias.
Figure 2.
Forest plot of the association between obesity and the rs9939609 polymorphism under per-allele comparison. On the left, the name of the first author of the study is followed by the publication year in parentheses. The size of the black box corresponding to each study is proportional to the sample size, and the horizontal line shows the corresponding 95% CI of the odds ratio (OR). The overall ORs are based on a random-effects model shown by the diamonds. The solid, vertical line represents the null result.
When stratifying the data by ethnicity, no evidence of a significant association was observed in the three studies with Hispanic ethnic groups (summary OR: 1.33, 95% CI: 0.84 to 2.10) under per-allele comparison, with the largest heterogeneity in all six ethnic groups (I2 = 85.2%, P = 0.001) (Figure 2). We observed overall ORs of 1.30 (95% CI: 1.24 to 1.37) and 1.35 (95% CI: 1.27 to 1.44) in the Caucasian and Asian ethnic groups for which more studies had been performed under per-allele comparison.
By using meta-regression, we detected a significant correlation between the mean control BMI and effect size in an allelic comparison (P = 0.03) (see Additional file 1: Figure S1).
Very similar results were obtained using an additive genetic model, with an overall OR of 1.31 (95% CI: 1.25 to 1.37) and evidence of heterogeneity (I2 = 52.8%) (Table 2, Additional file 1: Figure S2).
Meta-analysis of FTO SNPs rs1421085, rs8050136, s17817449, and rs1121980
rs1421085
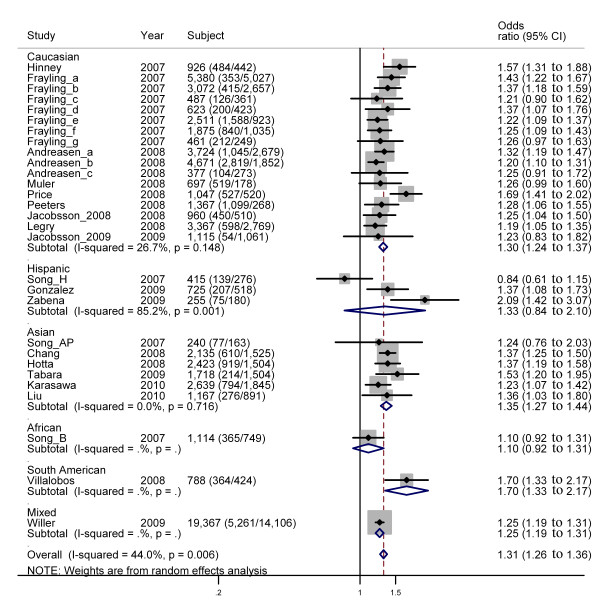
A total of 11 out of 12 studies reported a significant, positive association between obesity and the rs1421085 genotype (Table 2, Figure 3A). Also under per-allele comparison, a significant association between obesity risk and rs1421085 was found (overall OR = 1.43, 95% CI: 1.33 to 1.53), and there is evidence of heterogeneity (I2 = 38.1%). Begg's test (P = 0.76) and Egger's test (P = 0.84) provided no evidence of publication bias.
Figure 3.
Forest plot of the association between obesity and the two SNPs under per-allele comparison. On the left, the name of the first author of the study is followed by the publication year in parentheses. The size of the black box corresponding to each study is proportional to the sample size, and the horizontal line shows the corresponding 95% CI of the odds ratio (OR). The overall ORs are based on a random-effects model shown by the diamonds. The solid, vertical line represents the null result. (A) rs1421085; (B) rs8050136.
By using meta-regression, no significant correlation was found between the mean control BMI and effect size in an allelic comparison (P = 0.50).
rs8050136
Overall, 11 studies have investigated the association of rs8050136 with obesity. Of the 11 studies, the OR (under per-allele comparison) of the study by Li et al. is not available. Of the remaining 10 studies, 5 reported a significant, positive association between obesity and the rs8050136 genotype (Table 2, Figure 3B). A significant association between obesity risk and rs8050136 was found (overall OR = 1.25, 95% CI: 1.13 to 1.38) under per-allele comparison, and there is evidence of higher heterogeneity (I2 = 72.0%). Begg's test (P = 1.0) and Egger's test (P = 0.87) provided no evidence of publication bias.
By using meta-regression, no significant correlation was found between the mean control BMI and effect size in an allelic comparison (P = 0.60).
rs17817449
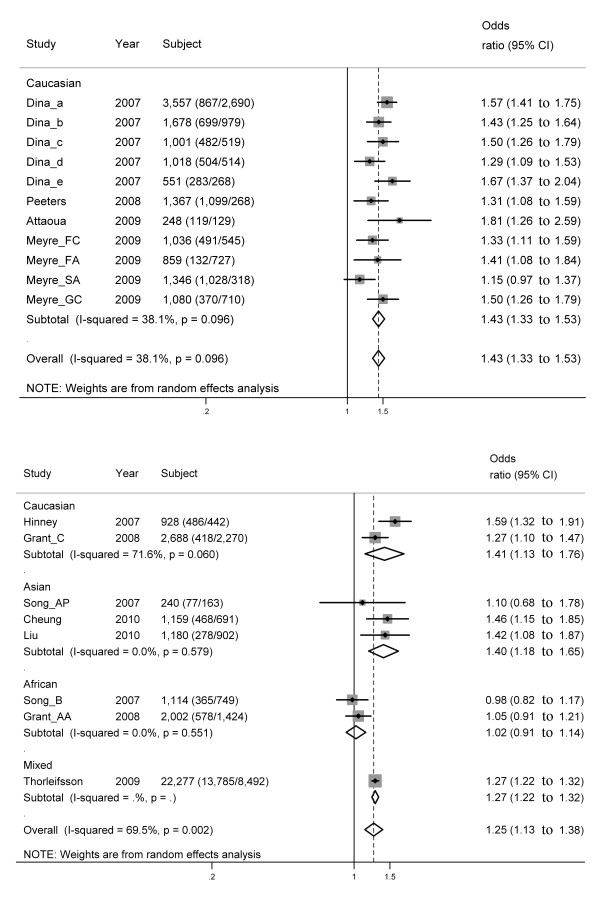
All six of the studies involving rs17817449 reported a significant, positive association between obesity and rs17817449 (Table 2, Figure 4A). A significant association between obesity risk and rs17817449 was found (overall OR = 1.54, 95% CI: 1.41 to 1.68) for obesity and rs17817449 under per-allele comparison, and there is evidence of higher heterogeneity (I2 = 45.2%). Begg's test (P = 0.85) and Egger's test (P = 0.90) provided no evidence of publication bias.
Figure 4.
Forest plot of the association between obesity and the two SNPs under per-allele comparison. On the left, the name of the first author of the study is followed by the publication year in parentheses. The size of the black box corresponding to each study is proportional to the sample size, and the horizontal line shows the corresponding 95% CI of the odds ratio (OR). The overall ORs are based on a random-effects model shown by the diamonds. The solid, vertical line represents the null result. (A) rs17917449; (B) rs1121980.
By using meta-regression, no significant correlation was found between the mean control BMI and effect size in an allelic comparison (P = 0.57).
rs1121980
All three studies reported a significant, positive association between obesity and the rs1121980 SNP (Table 2, Figure 4B). A significant association between obesity risk and rs1121980 was found (overall OR = 1.34, 95% CI: 1.10 to 1.62) under per-allele comparison, with evidence of greater heterogeneity (I2 = 84.5%). Begg's test (P = 0.12) provided no evidence of publication bias, whereas the funnel plot (not shown) and Egger's test (P = 0.09) showed evidence of publication bias.
By using meta-regression, no significant correlation was found between the mean control BMI and effect size in an allelic comparison (P = 0.28).
Discussion
To our knowledge, this meta-analysis is the first one to investigate the associations between FTO polymorphisms and obesity risk across different ethnic groups. This meta-analysis investigated the associations between five FTO polymorphisms (rs9939609, rs1421085, rs8050136, rs17817449, and rs1121980) and obesity risk in 41,734 cases and 69,837 controls from 59 studies, counting the cases and control subjects from each study only once. We found significant evidence for a modest increase in the risk of obesity associated with the five polymorphisms in various ethnic populations (Figures 2, 3, 4). However, subgroup analyses showed that in some ethnic populations, for example, rs9939609 in Hispanic and African, rs1121980 in Caucasian, and rs8050136 in Asian and African, significant associations were not found between the SNPs and obesity risk (see Additional file 1: Figure S3). These results may be due to the differences in MAF or LD patterns across different ethnic populations.
Interestingly, we found that MAF values are very similar across all five SNPs. For example, the MAFs in the study by Price et al. are 0.36 for rs9939609 and 0.36 for rs17817449. There are also ethnic variations of the MAF in the five polymorphisms. Caucasian, US Hispanics, and US Africans have higher MAFs (>31%), whereas Asian populations (for example, Chinese and Japanese) and South Americans have lower MAFs (between 11% and 20%). When stratifying the studies on rs9939609 into subgroups, the MAFs are different in various populations. For example, the MAF in control subjects is 0.34 to 0.44 in Caucasians, 0.11 to 0.20 in Asians, 0.31 to 0.37 in Hispanics, and 0.17 in South Americans, indicating that the MAF in Asian and South American populations is less than half of that in Caucasian and Hispanic populations. The population differences in MAF and LD structure probably have arisen through evolutionary divergence [56].
For rs9939609, we conducted sensitivity analyses. After the exclusion of the three children's studies [42-44], there is a small increase in overall heterogeneity (I2 = 44.6%, P = 0.008 when exclusion of the three studies vs. I2 = 44.0%, P = 0.006 when inclusion of all available studies) (see Additional file 1: Figure S4). Similarly, a slight increase in overall heterogeneity was observed after the exclusion of the two GWASs [4,43] (I2 = 51.6%, P = 0.003 with the exclusion of the two studies vs. I2 = 44.0%, P = 0.006 with the inclusion of all available studies). These results indicate that the children's studies or GWASs are not the sources of the heterogeneity. On the other hand, with the exclusion of the study by Price et al., the heterogeneity decreased rapidly (I2 = 0.0%, P = 0.626 with the exclusion of the study vs. I2 = 44.0%, P = 0.006 with the inclusion of all available studies) in the Caucasian subgroup, indicating that the study by Price et al. is the main source of heterogeneity in the Caucasian subgroup. By using an additive genetic model, coupled with either the inclusion or exclusion of the study by Li et al., the I2 value in the heterogeneity test declined sharply (61.7% vs. 15.6%). Similarly, removing the study by Chang et al. caused the I2 value to increase a little (61.7% vs. 67.0%), suggesting that the study by Li et al. is the main source of heterogeneity in the Asian ethnic group.
All of the studies on rs1421085 are from Caucasian populations, but when the five studies were excluded (two from Switzerland, two from Germany, and one from the USA) and the remaining seven studies were analyzed (six from France and one from Belgium), the I2 decreased sharply from 38.1% to 0.0%. Similarly, the same tendency was found with rs17817449 (data not shown), suggesting that geographic stratification in an ethnic subgroup is one of the sources of the between-study heterogeneity.
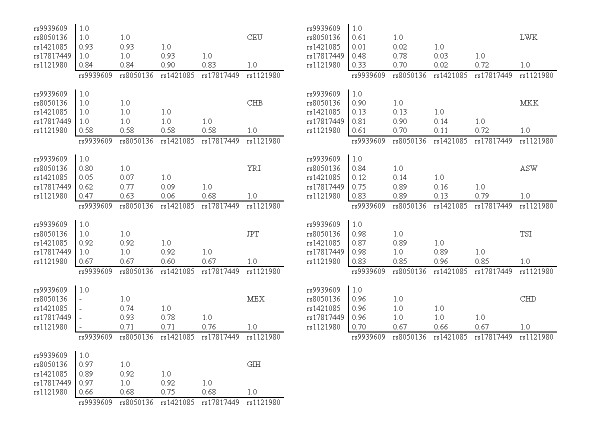
We investigated the LD structure of the five SNPs in all available ethnic populations (Figure 5). We found that there are substantial differences in the LD structures in various ethnic populations. Similar ethnic populations showed similar patterns (Figure 5), including CEU with TSI; YRI, MKK, and LWK with ASW; and CHB and CHD with JPT. All five SNPs are in strong LD with each other in Caucasians (Figure 5). For example, in one Caucasian population, all five SNPs showed strong LD (r2 ≥ 0.83), and in this case, rs9939609 may be selected as a TagSNP for the other four SNPs. Similarly, in three different populations, including Caucasian, East Asian and Gujarati Indians, four of the five SNPs (rs9939609, rs8050136, rs1421085, and rs17817449) are also in strong LD, indicating that rs9939609 can be used as a TagSNP for the other three SNPs.
Figure 5.
LD pattern of the five SNPs obtained from HapMap database ASW. African ancestry in Southwest USA; CEU, Utah residents with Northern and Western European ancestry from the CEPH collection; CHB, Han Chinese in Beijing, China; CHD, Chinese in Metropolitan Denver, Colorado; GIH, Gujarati Indians in Houston, Texas; JPT, Japanese in Tokyo, Japan; LWK, Luhya in Webuye, Kenya; MEX, Mexican ancestry in Los Angeles, California; MKK, Maasai in Kinyawa, Kenya; TSI, Tuscan in Italy; YRI, Yoruban in Ibadan, Nigeria.
Although the five SNPs are in strong LD with each other in Caucasians, there are obvious differences in the effect size across the five SNPs: 1.3 (1.24 to 1.37) for rs9939609, 1.43 (1.33 to 1.53) for rs1421085, 1.41 (1.13 to 1.76) for rs8050136, 1.54 (1.41 to 1.68) for rs17819449, and 1.36 (0.96 to 1.92) for rs1121980 (Figures 2, 3, 4). The first four SNPs reported significant associations, and the last one reported non-significant associations. On the other hand, the five SNPs are not in strong LD across all the ethnic groups. Therefore, no SNP can be treated as tagSNP across all the ethnic groups. Considering these facts, we showed all the results concerning all the five SNPs without selection of tagSNPs. In fact, some studies with very small sample size were included in this meta-analysis, for example, only 240 subjects were included in the study by Song et al. (Song_AP, Table 2). This fact may be one of the reasons for the inconsistent results across the five SNPs in Caucasians. To address this issue, more studies with larger sample sizes are needed to provide more precise evidence.
The heterogeneities of all five SNPs are higher in the meta-analysis. There are many reasons for heterogeneity in genetic association studies, and these reasons can be divided into two categories: (1) differential biases due to population stratification, misclassification of clinical outcome, differences in BMI, genotyping errors and over-estimation of genetic effects in the first study and (2) differences in the pattern of LD structure across populations [57]. Using sensitivity analyses, we showed that population stratification may be the reason for higher heterogeneity in studies focused on rs1421085, indicating that population stratification can also exist within a subgroup of the same ethnic background (Caucasians). In the meta-regression analysis, a significant association was detected between the mean control BMI and effect size on rs9939609, suggesting that the difference in BMI value between the subjects may also be a source of heterogeneity.
Analyses of statistical power constitute a crucial step in the design of genetic association studies. Whereas conventional statistical power calculations for case-control studies disregard many basic elements of analytic complexity and can seriously underestimate true sample size requirements. Many methods have been proposed to address the sample size issue in genetic association study [58-60]. However, owing to many factors, such as financial pressure, difficulties in sample collection, or neglect of the importance of sample size, too few samples were employed in many genetic association studies, for example, Andreasen_c with total subjects of 377 and Song_AP with total subjects of 240.
Limitations
Inherent limitations of this meta-analysis should be pointed out, including the following: (1) Based on the statistical tests, the heterogeneities are higher in the results of this meta-analysis. There may be many potential reasons for the higher heterogeneities, including differences in genetic susceptibility across ethnic groups and measurement error of BMI. (2) Gene-gene and gene-environment interactions could not be addressed in this meta-analysis. Actually, obesity is a complex trait, and many genes are related to obesity [61], such as MC4R [50,62,63], MAF [50], and NPC1 [50]. On the other hand, lifestyle factors, including diet and physical inactivity, are important contributors to weight gain and obesity [64-66] (3) It may be unreliable to use Begg's and Egger's test as the criteria for publication bias. Although publication bias was not found, it may exist due to the fact that some studies with non-significant associations were not published, and some articles published in other languages (not English) were not obtained and included in this meta-analysis [67]. (4) The Haplotype of the polymorphisms was not performed due to the lack of the related information.
Conclusions
This meta-analysis suggests that FTO may represent a low-penetrance susceptible gene for obesity risk. Individual studies with large sample sizes are needed to further evaluate the associations between the polymorphisms and obesity risk in various ethnic populations.
Abbreviations
BMI: body mass index; CI: confidence interval; GWAS: genome-wide association study; HWE: Hardy-Weinberg Equilibrium; LD: linkage disequilibrium; MAF: minor allele frequency; OR: odds ratio; ORG: generalized odds ratio; SNP: single nucleotide polymorphism.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SHP, YMZ and MDL contributed to the design of the study. SHP, XBR, and XBL carried out the screening procedure. SHP, YMZ and FYX performed the statistical analysis. SHP drafted the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Table S1, Figure S1, Figure S2, Figure S3, and Figure S4.
Contributor Information
Sihua Peng, Email: pengsihua@zju.edu.cn.
Yimin Zhu, Email: zhuym@zju.edu.cn.
Fangying Xu, Email: xfy@zju.edu.cn.
Xiaobin Ren, Email: xiaobinren80@126.com.
Xiaobo Li, Email: oboaixil@126.com.
Maode Lai, Email: lmp@zju.edu.cn.
Acknowledgements and Funding
We would like to thank Dr. Shulin Zhuang for helpful discussions on the manuscript. This study was supported by The "Eleventh Five-Year" Science and Technology Support Plan of the Ministry of Science and Technology of China (2009BA180B00) and a grant from the Natural Science Foundation of Zhejiang Province (Y2090081).
References
- Hossain P. Obesity and diabetes in the developing world - a growing challenge. New England Journal of Medicine. 2007;356:973–973. doi: 10.1056/NEJMp068177. [DOI] [PubMed] [Google Scholar]
- Haslam DW, James WPT. Obesity. Lancet. 2005;366:1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- Hill JO, Peters JC. Environmental contributions to the obesity epidemic. Science. 1998;280:1371–1374. doi: 10.1126/science.280.5368.1371. [DOI] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JRB, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJF, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CNA, Doney ASF, Morris AD, Smith GD, Hattersley AT, McCarthy MI, Control WTC. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orru M, Usala G, Dei M, Lai S, Maschio A, Busonero F, Mulas A, Ehret GB, Fink AA, Weder AB, Cooper RS, Galan P, Chakravarti A, Schlessinger D, Cao A, Lakatta E, Abecasis GR. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. Plos Genetics. 2007;3:1200–1210. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, Carlsson LMS, Kiess W, Vatin V, Lecoeur C, Delplanque J, Vaillant E, Pattou F, Ruiz J, Weill J, Levy-Marchal C, Horber F, Potoczna N, Hercberg S, Le Stunff C, Bougneres P, Kovacs P, Marre M, Balkau B, Cauchi S, Chevre JC, Froguel P. Variation in FTO contributes to childhood obesity and severe adult obesity. Nature Genetics. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Hagglund M, Olszewski PK, Stephansson O, Jacobsson JA, Olszewska AM, Levine AS, Lindblom J, Schioth HB. The obesity gene, FTO, is of ancient origin, up-regulated during food deprivation and expressed in neurons of feeding-related nuclei of the brain. Endocrinology. 2008;149:2062–2071. doi: 10.1210/en.2007-1457. [DOI] [PubMed] [Google Scholar]
- Stratigopoulos G, Padilla SL, Leduc CA, Watson E, Hattersley AT, McCarthy MI, Zeltser LM, Chung WK, Leibel RL. Regulation of Fto/Ftm gene expression in mice and humans. American Journal of Physiology-Regulatory Integrative and Comparative Physiology. 2008;294:R1185–R1196. doi: 10.1152/ajpregu.00839.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlen K, Sjolin E, Hoffstedt J. The common rs9939609 gene variant of the fat mass- and obesity-associated gene FTO is related to fat cell lipolysis. Journal of Lipid Research. 2008;49:607–611. doi: 10.1194/jlr.M700448-JLR200. [DOI] [PubMed] [Google Scholar]
- Cornes BK, Lind PA, Medland SE, Montgomery GW, Nyholt DR, Martin NG. Replication of the association of common rs9939609 variant of FTO with increased BMI in an Australian adult twin population but no evidence for gene by environment (G × E) interaction. International Journal of Obesity. 2009;33:75–79. doi: 10.1038/ijo.2008.223. [DOI] [PubMed] [Google Scholar]
- Tan JT, Dorajoo R, Seielstad M, Sim XL, Ong RTH, Chia KS, Wong TY, Saw SM, Chews SK, Aung T, Tai ES. FTO variants are associated with obesity in the Chinese and Malay populations in Singapore. Diabetes. 2008;57:2851–2857. doi: 10.2337/db08-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freathy RM, Timpson NJ, Lawlor DA, Pouta A, Ben-Shlomo Y, Ruokonen A, Ebrahim S, Shields B, Zeggini E, Weedon MN, Lindgren CM, Lango H, Melzer D, Ferrucci L, Paolisso G, Neville MJ, Karpe F, Palmer CN, Morris AD, Elliott P, Jarvelin MR, Smith GD, McCarthy MI, Hattersley AT, Frayling TM. Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected given its effect on BMI. Diabetes. 2008;57:1419–1426. doi: 10.2337/db07-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do R, Bailey SD, Desbiens K, Belisle A, Montpetit A, Bouchard C, Perusse L, Vohl MC, Engert JC. Genetic variants of FTO influence adiposity, insulin sensitivity, leptin levels, and resting metabolic rate in the Quebec Family Study. Diabetes. 2008;57:1147–1150. doi: 10.2337/db07-1267. [DOI] [PubMed] [Google Scholar]
- Fawcett KA, Barroso I. The genetics of obesity: FTO leads the way. Trends in Genetics. 2010;26:266–274. doi: 10.1016/j.tig.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HX, Wu Y, Loos RJF, Hu FB, Liu Y, Wang J, Yu Z, Lin X. Variants in the fat mass- and obesity-associated (FTO) gene are not associated with obesity in a Chinese Han population. Diabetes. 2008;57:264–268. doi: 10.2337/db07-1130. [DOI] [PubMed] [Google Scholar]
- Ohashi J, Naka I, Kimura R, Natsuhara K, Yamauchi T, Furusawa T, Nakazawa M, Ataka Y, Patarapotikul J, Nuchnoi P, Tokunaga K, Ishida T, Inaoka T, Matsumura Y, Ohtsuka R. FTO polymorphisms in oceanic populations. Journal of Human Genetics. 2007;52:1031–1035. doi: 10.1007/s10038-007-0198-2. [DOI] [PubMed] [Google Scholar]
- Cole TJ, Bellizzi MC, Flegal KM, Dietz WH. Establishing a standard definition for child overweight and obesity worldwide: international survey. British Medical Journal. 2000;320:1240–1243. doi: 10.1136/bmj.320.7244.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zintzaras E. The Generalized Odds Ratio as a Measure of Genetic Risk Effect in the Analysis and Meta-Analysis of Association Studies. Statistical Applications in Genetics and Molecular Biology. 2010;9 doi: 10.2202/1544-6115.1542. [DOI] [PubMed] [Google Scholar]
- Higgins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. British Medical Journal. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo SW, Thompson EA. Performing the exect test of Hardy-weinberg proportion for multiple alllels. Biometrics. 1992;48:361–372. doi: 10.2307/2532296. [DOI] [PubMed] [Google Scholar]
- Zintzaras E, Lau J. Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. Journal of Clinical Epidemiology. 2008;61:634–645. doi: 10.1016/j.jclinepi.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Wittke-Thompson JK, Pluzhnikov A, Cox NJ. Rational inferences about departures from Hardy-Weinberg equilibrium. American Journal of Human Genetics. 2005;76:967–986. doi: 10.1086/430507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DG, Kraft P. Quantification of the power of Hardy-Weinberg equilibrium testing to detect genotyping error. Human Heredity. 2006;61:10–14. doi: 10.1159/000091787. [DOI] [PubMed] [Google Scholar]
- Kavvoura FK, Ioannidis JPA. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Human Genetics. 2008;123:1–14. doi: 10.1007/s00439-007-0445-9. [DOI] [PubMed] [Google Scholar]
- Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865–872. doi: 10.1016/S0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- van Houwelingen HC, Arends LR, Stijnen T. Advanced methods in meta-analysis: multivariate approach and meta-regression. Statistics in Medicine. 2002;21:589–624. doi: 10.1002/sim.1040. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Moher D, Liberati A, Tetzlaff J, Altman DG, Grp P. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Medicine. 2009;6 [PMC free article] [PubMed] [Google Scholar]
- Cheung CYY, Tso AWK, Cheung BMY, Xu AM, Ong KL, Fong CHY, Wat NMS, Janus ED, Sham PC, Lam KSL. Obesity Susceptibility Genetic Variants Identified from Recent Genome-Wide Association Studies: Implications in a Chinese Population. Journal of Clinical Endocrinology & Metabolism. 2010;95:1395–1403. doi: 10.1210/jc.2009-1465. [DOI] [PubMed] [Google Scholar]
- Song YQ, You NC, Hsu YH, Howard BV, Langer RD, Manson JE, Nathan L, Niu TH, Tinker LF, Liu SM. FTO Polymorphisms Are Associated With Obesity but Not Diabetes Risk in Postmenopausal Women. Obesity. 2008;16:2472–2480. doi: 10.1038/oby.2008.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara Y, Osawa H, Guo H, Kawamoto R, Onuma H, Shimizu I, Takara Y, Nishida W, Yamamoto M, Makino H, Kohara K, Miki T. Prognostic significance of FTO genotype in the development of obesity in Japanese: the J-SHIPP study. Int J Obes (Lond) 2009;33:1243–1248. doi: 10.1038/ijo.2009.161. [DOI] [PubMed] [Google Scholar]
- Price RA, Li WD, Zhao HY. FTO gene SNPs associated with extreme obesity in cases, controls and extremely discordant sister pairs. Bmc Medical Genetics. 2008;9:4. doi: 10.1186/1471-2350-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM. et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Liu PH, Lee WJ, Chang TJ, Jiang YD, Li HY, Kuo SS, Lee KC, Chuang LM. Common variation in the fat mass and obesity-associated (FTO) gene confers risk of obesity and modulates BMI in the Chinese population. Diabetes. 2008;57:2245–2252. doi: 10.2337/db08-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legry V, Cottel D, Ferrieres J, Arveiler D, Andrieux N, Bingham A, Wagner A, Ruidavets JB, Ducimetiere P, Amouyel P, Meirhaeghe A. Effect of an FTO polymorphism on fat mass, obesity, and type 2 diabetes mellitus in the French MONICA Study. Metabolism-Clinical and Experimental. 2009;58:971–975. doi: 10.1016/j.metabol.2009.02.019. [DOI] [PubMed] [Google Scholar]
- Hotta K, Nakata Y, Matsuo T, Kamohara S, Kotani K, Komatsu R, Itoh N, Mineo I, Wada J, Masuzaki H, Yoneda M, Nakajima A, Miyazaki S, Tokunaga K, Kawamoto M, Funahashi T, Hamaguchi K, Yamada K, Hanafusa T, Oikawa S, Yoshimatsu H, Nakao K, Sakata T, Matsuzawa Y, Tanaka K, Kamatani N, Nakamura Y. Variations in the FTO gene are associated with severe obesity in the Japanese. Journal of Human Genetics. 2008;53:546–553. doi: 10.1007/s10038-008-0283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabena C, Gonzalez-Sanchez JL, Martinez-Larrad MT, Torres-Garcia A, Alvarez-Fernandez-Represa J, Corbaton-Anchuelo A, Perez-Barba M, Serrano-Rios M. The FTO Obesity Gene. Genotyping and Gene Expression Analysis in Morbidly Obese Patients. Obesity Surgery. 2009;19:87–95. doi: 10.1007/s11695-008-9727-0. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Sanchez JL, Zabena C, Martinez-Larrad MT, Martinez-Calatrava MJ, Perez-Barba M, Serrano-Rios M. Variant rs9939609 in the FTO gene is associated with obesity in an adult population from Spain. Clinical Endocrinology. 2009;70:390–393. doi: 10.1111/j.1365-2265.2008.03335.x. [DOI] [PubMed] [Google Scholar]
- Villalobos-Comparan M, Dorantes MTF, Villarreal-Molina MT, Rodriguez-Cruz M, Sanchez-Munoz F, Dominguez-Lopez A, Froguel P, Tusie-Luna MT, Aguilar-Salinas C, Canizales-Quinteros S. The FTO Gene is Associated with Adulthood Obesity in the Mexican Population. Obesity. 2008;16:S326–S326. doi: 10.1038/oby.2008.367. [DOI] [PubMed] [Google Scholar]
- Peeters A, Beckers S, Verrijken A, Roevens P, Peeters P, Van Gaal L, Van Hul W. Variants in the FTO gene are associated with common obesity in the Belgian population. Molecular genetics and metabolism. 2008;93:481–484. doi: 10.1016/j.ymgme.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Jacobsson JA, Danielsson P, Svensson V, Klovins J, Gyllensten U, Marcus C, Schioth HB, Fredriksson R. Major gender difference in association of FTO gene variant among severely obese children with obesity and obesity related phenotypes. Biochemical and Biophysical Research Communications. 2008;368:476–482. doi: 10.1016/j.bbrc.2008.01.087. [DOI] [PubMed] [Google Scholar]
- Hinney A, Nguyen TT, Scherag A, Friedel S, Bronner G, Muller TD, Grallert H, Illig T, Wichmann HE, Rief W, Schafer H, Hebebrand J. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS ONE. 2007;2:e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller TD, Hinney A, Scherag A, Nguyen TT, Schreiner F, Schafer H, Hebebrand J, Roth CL, Reinehr T. Fat mass and obesity associated gene (FTO): no significant association of variant rs9939609 with weight loss in a lifestyle intervention and lipid metabolism markers in German obese children and adolescents. Bmc Medical Genetics. 2008;9:85. doi: 10.1186/1471-2350-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen CH, Stender-Petersen KL, Mogensen MS, Torekov SS, Wegner L, Andersen G, Nielsen AL, Albrechtsen A, Borch-Johnsen K, Rasmussen SS, Clausen JO, Sandbaek A, Lauritzen T, Hansen L, Jorgensen T, Pedersen O, Hansen T. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57:95–101. doi: 10.2337/db07-0910. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu Z, Song Y, Zhou D, Zhang D, Zhao T, Chen Z, Yu L, Yang Y, Feng G, Li J, Zhang J, Liu S, Zhang Z, He L, Xu H. Meta-analysis Added Power to Identify Variants in FTO Associated With Type 2 Diabetes and Obesity in the Asian Population. Obesity. 2010;18:1619–1624. doi: 10.1038/oby.2009.469. [DOI] [PubMed] [Google Scholar]
- Jacobsson JA, Riserus U, Axelsson T, Lannfelt L, Schioth HB, Fredriksson R. The common FTO variant rs9939609 is not associated with BMI in a longitudinal study on a cohort of Swedish men born 1920-1924. Bmc Medical Genetics. 2009;10 doi: 10.1186/1471-2350-10-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasawa S, Daimon M, Sasaki S, Toriyama S, Oizumi T, Susa S, Kameda W, Wadad K, Muramatsu M, Fukao A, Kubota I, Kawata S, Kayama T, Kato T. Association of the Common Fat Mass and Obesity Associated (FTO) Gene Polymorphism with Obesity in a Japanese Population. Endocrine Journal. 2010;57:293–301. doi: 10.1507/endocrj.K09E-305. [DOI] [PubMed] [Google Scholar]
- Attaoua R, Ait El Mkadem S, Lautier C, Kaouache S, Renard E, Brun JF, Fedou C, Gris JC, Bringer J, Grigorescu F. Association of the FTO gene with obesity and the metabolic syndrome is independent of the IRS-2 gene in the female population of Southern France. Diabetes Metab. 2009;35:476–483. doi: 10.1016/j.diabet.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Meyre D, Delplanque J, Chevre JC, Lecoeur C, Lobbens S, Gallina S, Durand E, Vatin V, Degraeve F, Proenca C, Gaget S, Korner A, Kovacs P, Kiess W, Tichet J, Marre M, Hartikainen AL, Horber F, Potoczna N, Hercberg S, Levy-Marchal C, Pattou F, Heude B, Tauber M, McCarthy MI, Blakemore AI, Montpetit A, Polychronakos C, Weill J, Coin LJ, Asher J, Elliott P, Jarvelin MR, Visvikis-Siest S, Balkau B, Sladek R, Balding D, Walley A, Dina C, Froguel P. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- Grant SFA, Li M, Bradfield JP, Kim CE, Annaiah K, Santa E, Glessner JT, Casalunovo T, Frackelton EC, Otieno FG, Shaner JL, Smith RM, Imielinski M, Eckert AW, Chiavacci RM, Berkowitz RI, Hakonarson H. Association analysis of the FTO gene with obesity in children of Caucasian and African ancestry reveals a common tagging SNP. PLoS ONE. 2008;3:e1746. doi: 10.1371/journal.pone.0001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- Renstrom F, Payne F, Nordstrom A, Brito EC, Rolandsson O, Hallmans G, Barroso I, Nordstrom P, Franks PW, Consortium G. Replication and extension of genome-wide association study results for obesity in 4923 adults from northern Sweden. Human Molecular Genetics. 2009;18:1489–1496. doi: 10.1093/hmg/ddp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Kang K, Zhang CL, van Dam RM, Kraft P, Hunter D, Lee CH, Hu FB. Fat Mass- and Obesity-Associated (FTO) Gene Variant Is Associated With Obesity Longitudinal Analyses in Two Cohort Studies and Functional Test. Diabetes. 2008;57:3145–3151. doi: 10.2337/db08-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig BJ, Fulford AJ, Sirugo G, Rayco-Solon P, Hattersley AT, Frayling TM, Prentice AM. FTO gene variation and measures of body mass in an African population. Bmc Medical Genetics. 2009;10 doi: 10.1186/1471-2350-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ, Bouchard C. FTO: the first gene contributing to common forms of human obesity. Obes Rev. 2008;9:246–250. doi: 10.1111/j.1467-789X.2008.00481.x. [DOI] [PubMed] [Google Scholar]
- Nakaoka H, Inoue I. Meta-analysis of genetic association studies: methodologies, between-study heterogeneity and winner's curse. Journal of Human Genetics. 2009;54:615–623. doi: 10.1038/jhg.2009.95. [DOI] [PubMed] [Google Scholar]
- Gordon D, Finch SJ. Factors affecting statistical power in the detection of genetic association. Journal of Clinical Investigation. 2005;115:1408–1418. doi: 10.1172/JCI24756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Vega FM, Gordon D, Su XP, Scafe C, Isaac H, Gilbert DA, Spier EG. Power and sample size calculations for genetic case/control studies using gene-centric SNP maps: Application to human chromosomes 6, 21, and 22 in three populations. Human Heredity. 2005;60:43–60. doi: 10.1159/000087918. [DOI] [PubMed] [Google Scholar]
- Menashe I, Rosenberg PS, Chen BE. PGA: power calculator for case-control genetic association analyses. Bmc Genetics. 2008;9 doi: 10.1186/1471-2156-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CG, Walley AJ, Froguel P. The genetics of human obesity. Nature Reviews Genetics. 2005;6:221–234. doi: 10.1038/nrg1556. [DOI] [PubMed] [Google Scholar]
- Larsen LH, Echwald SM, Sorensen TIA, Andersen T, Wulff BS, Pedersen O. Prevalence of mutations and functional analyses of melanocortin 4 receptor variants identified among 750 men with juvenile-onset obesity. Journal of Clinical Endocrinology & Metabolism. 2005;90:219–224. doi: 10.1210/jc.2004-0497. [DOI] [PubMed] [Google Scholar]
- Larsen LH, Rose CS, Sparso T, Overgaard J, Torekov SS, Grarup N, Jensen DP, Albrechtsen A, Andersen G, Ek J, Glumer C, Borch-Johnsen K, Jorgensen T, Hansen T, Pedersen O. Genetic analysis of the estrogen-related receptor alpha and studies of association with obesity and type 2 diabetes. International Journal of Obesity. 2007;31:365–370. doi: 10.1038/sj.ijo.0803408. [DOI] [PubMed] [Google Scholar]
- Gillis LJ, Kennedy LC, Gillis AM, Bar-Or O. Relationship between juvenile obesity, dietary energy and fat intake and physical activity. International Journal of Obesity. 2002;26:458–463. doi: 10.1038/sj.ijo.0801967. [DOI] [PubMed] [Google Scholar]
- Barbeau P, Stallmann-Jorgensen IS, Gutin B, Hatfield-Laube JL, Humphries MC, Johnson MH. General and visceral adiposity in black and white adolescents and their relation with reported physical activity and diet. International Journal of Obesity. 2007;31:622–629. doi: 10.1038/sj.ijo.0803587. [DOI] [PubMed] [Google Scholar]
- Besson H, Ekelund U, Luan J, May AM, Sharp S, Travier N, Agudo A, Slimani N, Rinaldi S, Jenab M, Norat T, Mouw T, Rohrmann S, Kaaks R, Bergmann M, Boeing H, Clavel-Chapelon F, Boutron-Ruault MC, Overvad K, Andreasen EL, Johnsen NF, Halkjaer J, Gonzalez C, Rodriguez L, Sanchez MJ, Arriola L, Barricarte A, Navarro C, Key TJ, Spencer EA, Orfanos P, Naska A, Trichopoulou A, Manjer J, Wirfalt E, Lund E, Palli D, Agnoli C, Vineis P, Panico S, Tumino R, Bueno-De-Mesquita HB, van den Berg SW, Odysseos AD, Riboli E, Wareham NJ, Peeters PH. A cross-sectional analysis of physical activity and obesity indicators in European participants of the EPIC-PANACEA study. International Journal of Obesity. 2009;33:497–506. doi: 10.1038/ijo.2009.25. [DOI] [PubMed] [Google Scholar]
- Lau J, Ioannidis JPA, Terrin N, Schmid CH, Olkin I. Evidence based medicine - The case of the misleading funnel plot. British Medical Journal. 2006;333:597–600. doi: 10.1136/bmj.333.7568.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa Y, Nakamura T, Takahashi M, Ryo M, Inoue S, Ikeda Y, Ohno M, Sakata T, Fukagawa K, Saitoh Y, Sato Y, Shirai K, Miyazaki S, Tokunaga K, Yamanouchi K, Shibasaki T, Nagai M. O Japan Soc Study, New criteria for 'obesity disease' in Japan. Circulation Journal. 2002;66:987–992. doi: 10.1253/circj.66.987. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, Figure S1, Figure S2, Figure S3, and Figure S4.