Abstract
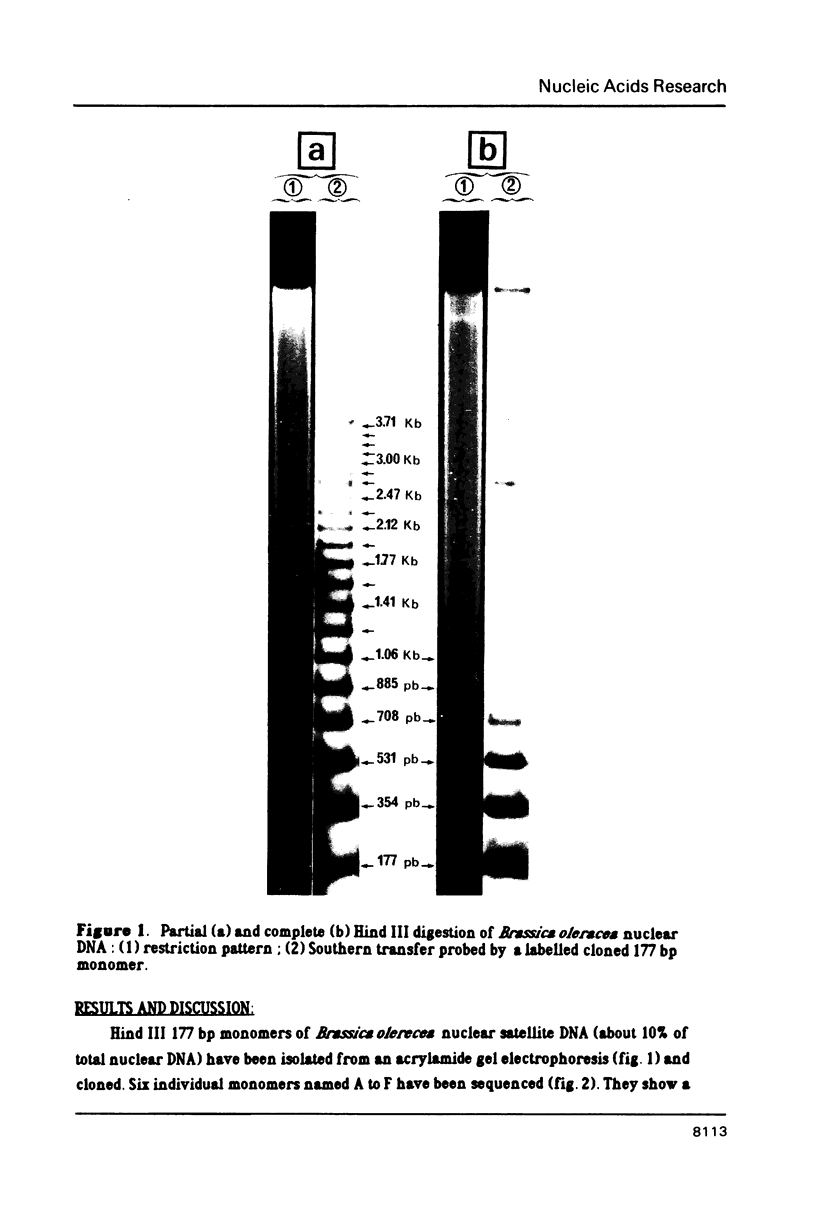
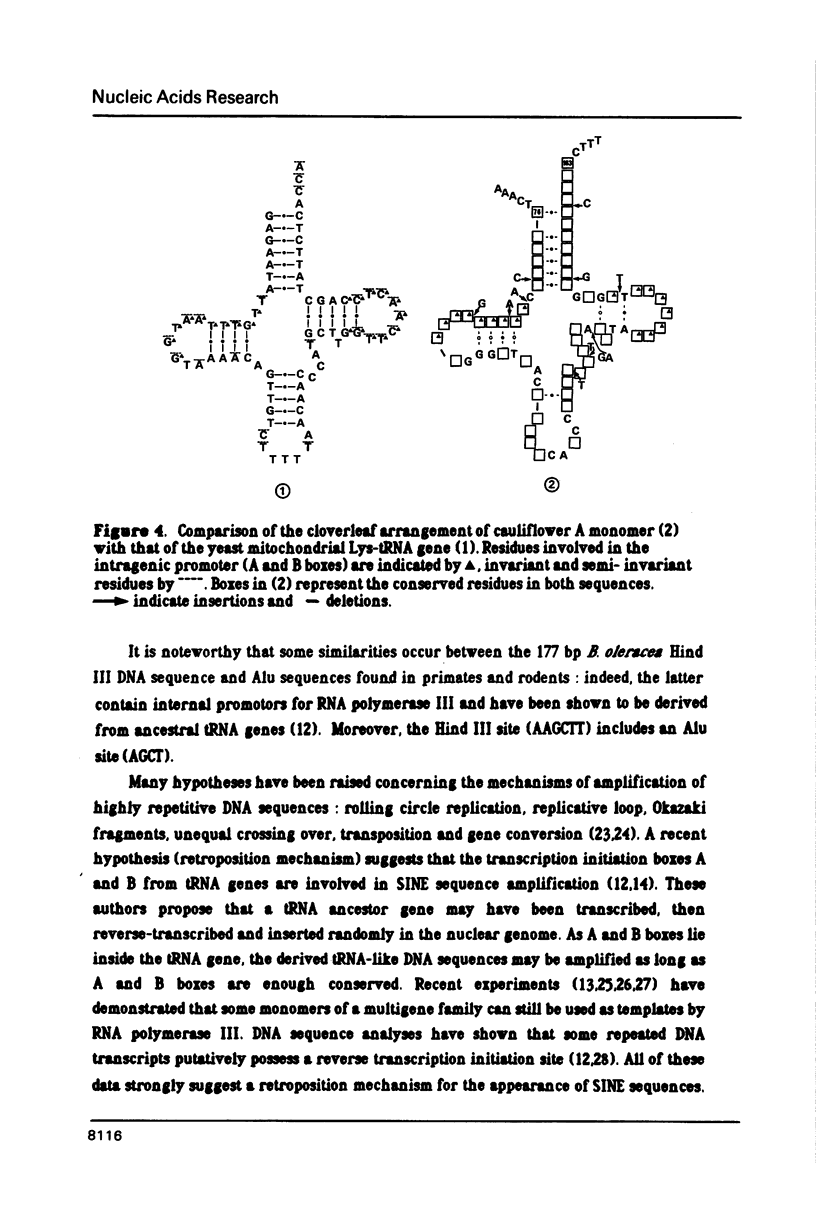
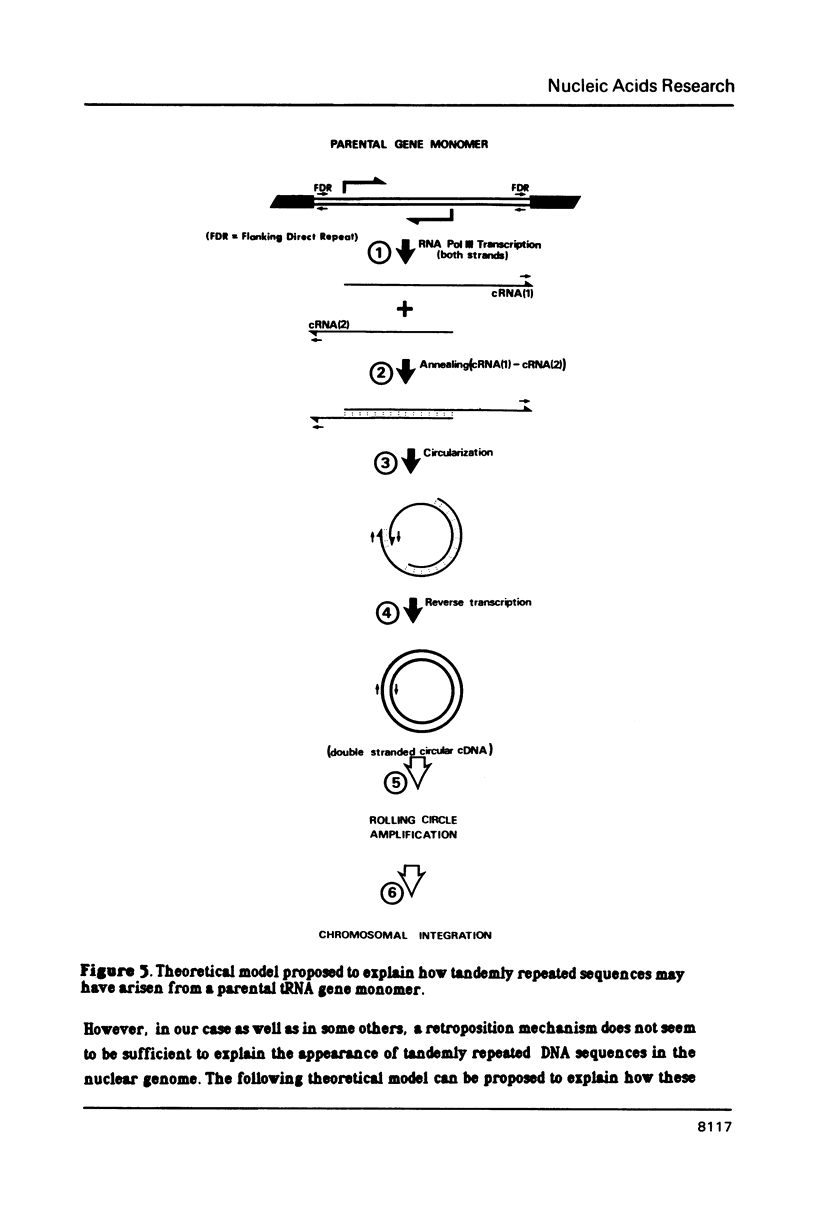
Several monomers (177 bp) of a tandemly arranged repetitive nuclear DNA sequence of Brassica oleracea have been cloned and sequenced. They share up to 95% homology between one another and up to 80% with other satellite DNA sequences of Cruciferae, suggesting a common ancestor. Both strands of these monomers show more than 50% homology with many tRNA genes; the best homologies have been obtained with Lys and His yeast mitochondrial tRNA genes (respectively 64% and 60%). These results suggest that small tandemly repeated DNA sequences of plants may have evolved from a tRNA gene ancestor. These tandem repeats have probably arisen via a process involving reverse transcription of polymerase III RNA intermediates, as is the case for interspersed DNA sequences of mammalians. A model is proposed to explain the formation of such small tandemly repeated DNA sequences.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackerman E. J. Molecular cloning and sequencing of OAX DNA: an abundant gene family transcribed and activated in Xenopus oocytes. EMBO J. 1983;2(8):1417–1422. doi: 10.1002/j.1460-2075.1983.tb01600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews D. L., Millstein L., Hamkalo B. A., Gottesfeld J. M. Competition between Xenopus satellite I sequences and Pol III genes for stable transcription complex formation. Nucleic Acids Res. 1984 Oct 25;12(20):7753–7769. doi: 10.1093/nar/12.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedbrook J. R., Jones J., O'Dell M., Thompson R. D., Flavell R. B. A molecular description of telometic heterochromatin in secale species. Cell. 1980 Feb;19(2):545–560. doi: 10.1016/0092-8674(80)90529-2. [DOI] [PubMed] [Google Scholar]
- Bendich A. J., Anderson R. S. Novel properties of satellite DNA from muskmelon. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1511–1515. doi: 10.1073/pnas.71.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonitz S. G., Tzagoloff A. Assembly of the mitochondrial membrane system. Sequences of yeast mitochondrial tRNA genes. J Biol Chem. 1980 Oct 10;255(19):9075–9081. [PubMed] [Google Scholar]
- Bos J. L., Osinga K. A., Van der Horst G., Borst P. Nucleotide sequence of the mitochondrial structural genes for cysteine-tRNA and histidine-tRNA of yeast. Nucleic Acids Res. 1979 Jul 25;6(10):3255–3266. doi: 10.1093/nar/6.10.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels G. R., Deininger P. L. Repeat sequence families derived from mammalian tRNA genes. 1985 Oct 31-Nov 6Nature. 317(6040):819–822. doi: 10.1038/317819a0. [DOI] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Endoh H., Okada N. Total DNA transcription in vitro: a procedure to detect highly repetitive and transcribable sequences with tRNA-like structures. Proc Natl Acad Sci U S A. 1986 Jan;83(2):251–255. doi: 10.1073/pnas.83.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemleben V., Leweke B., Roth A., Stadler J. Organization of highly repetitive satellite DNA of two Cucurbitaceae species (Cucumis melo and Cucumis sativus). Nucleic Acids Res. 1982 Jan 22;10(2):631–644. doi: 10.1093/nar/10.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Saigo K., Yamada K., Kuchino Y. Identification and nucleotide sequence determination of a potential primer tRNA for reverse transcription of a Drosophila retrotransposon, 297. Nucleic Acids Res. 1986 Apr 11;14(7):3031–3043. doi: 10.1093/nar/14.7.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Saigo K., Yamada K., Kuchino Y. Identification and nucleotide sequence determination of a potential primer tRNA for reverse transcription of a Drosophila retrotransposon, 297. Nucleic Acids Res. 1986 Apr 11;14(7):3031–3043. doi: 10.1093/nar/14.7.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A., Yakura K., Tanifuji S. Sequence analysis of Vicia faba repeated DNA, the FokI repeat element. Nucleic Acids Res. 1984 Aug 24;12(16):6415–6426. doi: 10.1093/nar/12.16.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maio J. J., Brown F. L., Musich P. R. Subunit structure of chromatin and the organization of eukaryotic highly repetitive DNA: recurrent periodicities and models for the evolutionary origins of repetitive DNA. J Mol Biol. 1977 Dec 15;117(3):637–655. doi: 10.1016/0022-2836(77)90062-6. [DOI] [PubMed] [Google Scholar]
- Peacock W. J., Dennis E. S., Rhoades M. M., Pryor A. J. Highly repeated DNA sequence limited to knob heterochromatin in maize. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4490–4494. doi: 10.1073/pnas.78.7.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair J., Wells R., Deumling B., Ingle J. The complexity of satellite deoxyribonucleic acid in a higher plant. Biochem J. 1975 Jul;149(1):31–38. doi: 10.1042/bj1490031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A computer program to search for tRNA genes. Nucleic Acids Res. 1980 Feb 25;8(4):817–825. [PMC free article] [PubMed] [Google Scholar]
- Wakefield L., Ackerman E., Gurdon J. B. The activation of RNA synthesis by somatic nuclei injected into amphibian oocytes. Dev Biol. 1983 Feb;95(2):468–475. doi: 10.1016/0012-1606(83)90048-9. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]