Abstract
Receptor tyrosine kinases (RTKs) regulate critical cell signaling pathways, yet the properties of their cognate ligands that influence receptor activation are not fully understood. There is great interest in parsing these complex ligand-receptor relationships using engineered proteins with altered binding properties. Here we focus on the interaction between two engineered epidermal growth factor (EGF) mutants and the EGF receptor (EGFR), a model member of the RTK superfamily. We found that EGF mutants with faster kinetic on-rates stimulate increased EGFR activation compared to wild-type EGF. These findings support previous predictions that faster association rates correlate with enhanced receptor activity.
Keywords: epidermal growth factor, protein engineering, receptor activation, receptor tyrosine kinase, kinetic binding rate
1. Introduction
Epidermal growth factor (EGF) is a 53 amino-acid polypeptide that stimulates a variety of cellular processes, including proliferation, survival, and differentiation [1]. Binding of EGF to its receptor (EGFR, ErbB1) induces receptor dimerization, leading to activation of the receptor intracellular tyrosine kinase domain and initiation of pleiotropic downstream signaling pathways [2]. After activation, EGF ligand-receptor complexes are rapidly internalized and trafficked through the cell, where they are ultimately targeted for recycling or degradation [3]. Therefore, EGFR signaling potency is regulated by both ligand binding properties and receptor trafficking patterns [4]; however, understanding the interplay between these processes remains a significant challenge. Although some studies have found a correlation between EGF binding affinity and biological activity, recent findings have demonstrated that this trend is not always present [5–9]. Other subtleties of EGF-EGFR interactions, such as binding kinetics and pH sensitivity, can also influence the magnitude and duration of the signaling response. Computational modeling studies suggested the importance of binding on-rates to receptor activation [10], as the rapid internalization of EGFR can impose a limit on activation that is independent of binding off-rates [11]. Furthermore, the pH sensitivity of the EGF-EGFR interaction influences the fraction of internalized receptor that is recycled back to the cell surface for continued signaling [3,12].
To develop a molecular toolkit for studying EGF-EGFR interactions, we previously used combinatorial methods to engineer EGF mutants with four- to 30-fold increased receptor binding affinity [13]. Here, we selected two of these clones, mutant 28 (m28) and mutant 123 (m123) (Table 1), for further characterization due to their high soluble expression levels in yeast. We showed that these mutants retain their binding specificities for EGFR compared to other ErbB family members, have stronger EGFR binding affinities resulting from increased kinetic on-rates, and interact with EGFR with different pH sensitivities. These unique binding properties translated into enhanced EGFR activation responses. To our knowledge this work is the first experimental validation of ligands with faster binding on-rates exhibiting increased receptor activation.
Table 1.
Amino acid sequences of wild-type EGF, mutant 28, and mutant 123. Mutations are underlined in bold type.
| Clone | Sequence |
|---|---|
| EGFwt | NSDSECPLSHDGYCLHDGVCMYIEALDKYACNCVVGYIGERCQYRDLKWWELR |
| m28 | NSDSECPLSHDGYCLHGGVCMYIKAVDRYACNCVVGYIGERCQYRDLTWWGPR |
| m123 | NSYSECPPSYDGYCLHDGVCRYIEALDSYACNCVVGYAGERCQYRDLRWWGRR |
2. Materials and methods
For additional materials and methods, see Supplementary Methods.
2.1 Cell binding assays
Equilibrium receptor binding affinities were measured on NR6WT and BJ-5ta fibroblast cells after incubation with EGF (three-fold dilutions from 200 nM to 10 pM) for 6 hrs at 4 °C. Cells were labeled with a FITC-conjugated antibody directed against an N-terminal FLAG epitope tag on EGF and analyzed using a FACSCalibur flow cytometer (BD Biosciences). Receptor binding off-rates were measured using NR6WT cells pretreated for 20 min with 100 µM phenylarsine oxide to inhibit EGFR internalization. Cells were incubated with 25 nM EGF for 10 min at 37 °C, washed, and incubated in serum-free medium at 37 °C for times ranging from 30 min to 7 h. The level of EGF persisting on the cell surface was measured by flow cytometry as above.
2.2 Surface plasmon resonance assays
EGF binding interactions with immobilized human (hEGFR) and murine EGFR (mEGFR) were analyzed by surface plasmon resonance (SPR) using a Biacore 3000 instrument (Biacore Life Sciences). Kinetic experiments were performed at 25 °C in degassed running buffer. EGF at various concentrations (two-fold dilutions from 400 nM to 780 pM) were flowed over EGFR-immobilized surfaces at 30 µL/min for 2 min. Final sensorgrams were analyzed with BIAevaluation software (Biacore Life Sciences) and simultaneously fit for affinity and kinetic parameters using a 1:1 Langmuir binding model.
2.3 EGFR activation and immunoblotting
BJ-5ta fibroblasts were pretreated with Na3VO4 phosphatase inhibitor and incubated with EGF (five-fold dilutions from 20 nM to 6.4 pM) for 15 min at 37 °C. Cells were lysed with 100 µL lysis buffer supplemented with 1 mM Na3VO4 and protease inhibitors. Cell lysates were resolved by SDS-PAGE under reducing conditions and analyzed by western blot with primary antibodies directed against actin or phosphorylated or total EGFR and a horseradish peroxidase-conjugated secondary antibody. Western blots were developed using chemiluminescence and imaged using a Chemidoc System (BioRad).
2.4 EGFR downregulation assays
BJ-5ta fibroblasts were treated with 0.1 nM EGF for times ranging from 15 min to 6 h. Post-stimulation, cells were fixed with 1.5% paraformaldehyde, and cell-surface EGFR was analyzed by flow cytometry using a primary antibody directed against EGFR and a secondary R-phycoerythrin-conjugated antibody.
3. Results
3.1 EGF mutants bind specifically to EGFR and not to other ErbB receptors
EGFR is one of four receptors in the ErbB family, which also includes ErbB2, ErbB3, and ErbB4. We measured the ErbB binding specificity of m28 and m123 compared to wild-type EGF (EGFwt), using stably-transfected CHO cells individually expressing each of the four ErbB receptors. We found that EGFwt and the engineered mutants bound specifically to EGFR but not to other ErbB receptor family members (Fig. S1), demonstrating that the amino acid mutations conferring high-affinity binding to EGFR do not alter binding specificity.
3.2 EGF mutants bind cell surface EGFR with higher affinity than wild-type EGF
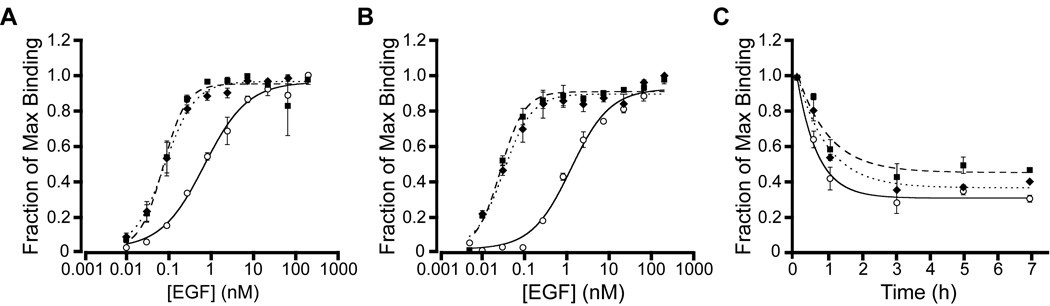
We next determined equilibrium binding affinities (KD) of EGFwt, m28, and m123 to EGFR expressed on fibroblasts and confirmed that the mutants bound with stronger affinity (Fig. 1A and B, and Table 2). Compared to EGFwt, m28 and m123 bound eight-fold more tightly to EGFR on NR6WT cells. On BJ-5ta cells, m28 and m123 bound 37- and 33-fold more tightly, respectively, than EGFwt. We also found that the kinetic off-rates (koff) of binding of EGFwt and mutants to NR6WT cells was comparable (Fig. 1C and Table 2). Based on the empirically observed KD and koff values, expected on-rates (kon) of receptor binding were determined (KD=koff /kon) to be approximately four- and six-fold faster for m28 and m123, respectively, compared to EGFwt.
Fig. 1.
Binding of wild-type EGF, mutant 28, and mutant 123 to EGFR expressed on the cell surface. Binding titrations of EGF to EGFR on (A) NR6WT and (B) BJ-5ta cells. (C) Off-rates of EGF binding to EGFR on NR6WT cells. EGFwt (⭘, solid line), m28 (■, dashed line), and m123 (♦, dotted line). Binding experiments were performed in triplicate and error bars denote standard error of the mean.
Table 2.
Equilibrium binding affinities and kinetic rates of wild-type EGF, mutant 28, and mutant 123 binding to cell-surface EGFR. Numbers in parenthesis denote fold-change over EGFwt.
| NR6WT Cells | EGFwt | m28 | m123 |
|---|---|---|---|
| KD (pM) | 600 ± 200 (--) | 80* ± 20 (8) | 80* ± 50 (8) |
| koff (s−1) × 10−4 | 4.3 ± 0.8 (--) | 2.60 ± 0.08 (1.7) | 2.8 ± 0.4 (1.5) |
| kon (M−1s−1) ×105 | 7 (--) | 30 (4) | 40 (6) |
| BJ-5ta Cells | EGFwt | m28 | m123 |
| KD (pM) | 1100 ± 200 (--) | 30* ± 6 (37) | 34* ± 7 (33) |
Statistical significance (p < 0.05) compared to EGFwt.
3.3 EGF mutants bind EGFR extracellular domain with faster kinetic on-rates than wild-type EGF
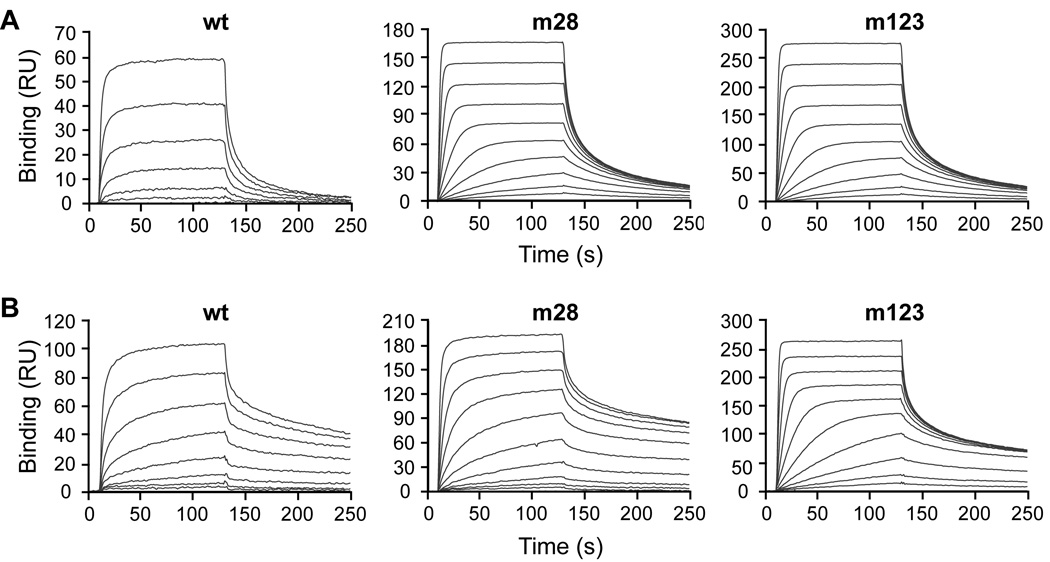
Real-time interactions of wild-type and mutant EGF with hEGFR and mEGFR were analyzed by SPR (Fig. 2 and Table 3). In these experiments, m28 and m123 bound 15- and 18-fold more tightly to hEGFR, respectively, than EGFwt. In addition, EGFwt bound to mEGFR with higher affinity than hEGFR. Compared to EGFwt, m28 and m123 bound four- and eight-fold more tightly to mEGFR, respectively. Since the EGF mutants were affinity-matured against human EGFR, it was not surprising that the difference between wild-type and mutant EGF binding to mEGFR was not as great as for hEGFR. For both mutants, improvements in KD over EGFwt resulted primarily from increased kon rather than decreased koff (Table 3), in agreement with cell surface studies above. Differences observed in absolute values of binding parameters from cell surface and SPR studies occur due to the removal of membrane constraints in SPR experiments, which use only EGFR extracellular domain [14].
Fig. 2.
Binding of wild-type EGF, mutant 28, and mutant 123 to the extracellular domain of human and murine EGFR. EGF binding to (A) hEGFR or (B) mEGFR was measured by surface plasmon resonance. Binding experiments were performed in triplicate and representative titration series sensorgrams are shown.
Table 3.
Equilibrium binding affinities and kinetic rates of wild-type EGF, mutant 28, and mutant 123 binding to human and murine EGFR extracellular domain by SPR. Numbers in parenthesis denote fold-change over EGFwt.
| Human EGFR | EGFwt | m28 | m123 |
|---|---|---|---|
| KD (nM) | 90 ± 10 (--) | 6* ± 2 (15) | 4.9* ± 0.3 (18) |
| koff (s−1) × 10−3 | 18 ± 1 (--) | 11* ± 2 (1.6) | 12.4* ± 0.6 (1.5) |
| kon (M−1s−1) × 105 | 2.0 ± 0.2 (--) | 20* ± 8 (10) | 25* ± 1 (13) |
| Murine EGFR | EGFwt | m28 | m123 |
| KD (nM) | 18 ± 1 (--) | 4.5* ± 0.2 (4.0) | 2.27* ± 0.05 (7.9) |
| koff (s−1) × 10−3 | 2.7 ± 0.2 (--) | 2.14* ± 0.08 (1.3) | 4.0* ± 0.1 (0.7) |
| kon (M−1s−1) × 105 | 1.47 ± 0.03 (--) | 4.76* ± 0.07 (3.2) | 17.6* ± 0.8 (12.0) |
Statistical significance (p < 0.05) compared to EGFwt.
The pH sensitivity of the binding interaction of EGF ligands (200 nM) with hEGFR and mEGFR was measured by SPR over pH values ranging from 5.0 to 8.5. Measurement of the steady-state binding responses for each ligand-receptor pair across various pH values revealed that the interactions of EGFwt and m28 with EGFR were sensitive to changes in pH, while the binding of m123 with EGFR was much less so (Fig. S2).
3.4 EGF mutants more strongly activate EGFR compared to wild-type EGF
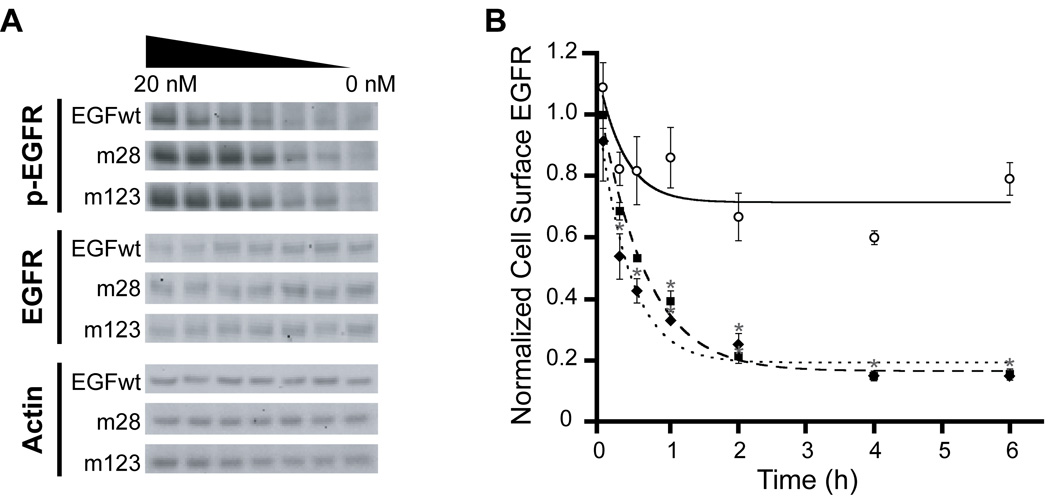
We next measured the ability of wild-type and mutant EGF to activate EGFR on fibroblasts. We found that m28 and m123 more strongly stimulated EGFR phosphorylation in these cells at lower concentrations than EGFwt (Fig. 3A). Since EGFR is rapidly internalized into the cell upon activation of the intracellular tyrosine kinase domain [15], downregulation of the receptor can serve as a surrogate measurement for receptor activation. Treatment of fibroblasts with m28 and m123 induced significantly increased EGFR downregulation compared to EGFwt, further indicating that the EGF mutants more strongly activate cell-surface EGFR (Fig. 3B).
Fig. 3.
Activation of EGFR by wild-type EGF, mutant 28, and mutant 123. (A) Western blot analysis of phosphorylated EGFR (p-EGFR, top panel), total EGFR (EGFR, middle panel) and actin loading control (Actin, lower panel) in BJ-5ta cells after treatment with EGF. (B) Downregulation of cell-surface EGFR in BJ-5ta cells in response to EGF stimulation. EGFwt (⭘, solid line), m28 (■, dashed line), or m123 (♦, dotted line). Experiments were performed in triplicate and error bars denote standard error of the mean. (*) Statistical significance (p < 0.05) compared to EGFwt.
4. Discussion
We measured the EGFR binding affinities and kinetic rate constants of two previously identified EGF mutants, m28 and m123 [13]. We showed through cell surface measurements and SPR that m28 and m123 have increased binding affinity for EGFR predominantly due to increased kinetic on-rates. The increased association rates of m28 and m123 were surprising since both mutants were discovered by screening combinatorial libraries under equilibrium binding conditions, which typically isolates mutants with decreased dissociation rates. We showed that m28 and m123 elicited increased EGFR activation compared to EGFwt, as measured by phosphorylation of the receptor tyrosine kinase domain and receptor downregulation. Interestingly, m123 had stronger binding at low pH compared to m28 and EGFwt. This difference implied that m123 might induce more intracellular receptor degradation compared to m28, yet both mutants exhibited greater levels of EGFR downregulation compared to EGFwt, suggesting that cell surface binding events drive this biological response.
Because of its important biological role, there has been much interest in EGF mutants with enhanced cell signaling for applications in wound healing and regenerative medicine [16,17]. Previous attempts at engineering EGF [5,7–9] had mixed success, but highlighted the complex relationship between ligand binding and EGFR activation. While some studies concluded that receptor activation is directly proportional to equilibrium binding affinity [5,9], others found that equivalent or enhanced potency can be attained by EGF mutants with weaker receptor binding interactions than EGFwt [7,8]. Computational studies of cellular signaling and trafficking processes coupled to the activation of transmembrane receptors have attempted to explain these inconsistencies by highlighting the importance of receptor binding on-rates [10,18].
To our knowledge, our work represents the first experimental corroboration of the effects of increased ligand binding on-rates with enhanced receptor activation. We demonstrated that EGF mutants with faster association rates, but nearly equivalent dissociation rates, more strongly activated EGFR compared to EGFwt. Collectively, these studies suggest that while receptor activity is linked to ligand binding, the magnitude of the response can be altered solely by differences in the association rate of the interaction. Furthermore, these results suggest a general strategy for engineering ligands that stimulate enhanced receptor activity [19].
While m28 and m123 stimulate increased EGFR activation, this attribute may not correlate with enhanced agonistic potential, since increased EGFR activation and downregulation could result in decreased or unaffected biological outcomes due to signal attenuation [3]. This consideration is also coupled with unaltered (m28) or decreased (m123) pH binding sensitivities, which are expected to influence receptor recycling. In future studies, it will therefore be interesting to explore the effects of increased EGFR phosphorylation and downregulation on downstream signaling and biological processes such as cellular migration and proliferation.
Supplementary Material
Acknowledgements
We thank Luo Luo Zheng, Eugene Hur, and Bradley French for technical assistance and Michael Eckart and Agustin Sanchez (Stanford PAN Facility) for assistance with Biacore. We are grateful to Alan Wells (U Pittsburgh) for providing the NR6WT cell line, Ross Mikkelsen (Virginia Commonwealth U) for providing the CHO+ErbB3 cells, and Yosef Yarden (Weizmann Institute) for providing the plasmids for ErbB transfection. This work was supported by a Wallace H. Coulter Foundation Translational Research Partnership Award. The following fellowship support is acknowledged: NIH Interdisciplinary Regenerative Medicine Training Grant T90 DK070103 (SEB and JLL), California Breast Cancer Research Program 13GB-0161 (JLL), NSF and Stanford Bio-X (SSL), Bio-X Lubert Stryer and ARCS Fellowship (BHL), and Gerald J. Lieberman Fellowship (JLL).
Abbreviations
- EGF
epidermal growth factor
- EGFR
EGF receptor
- RTK
receptor tyrosine kinase
- SPR
surface plasmon resonance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wells A. EGF receptor. Int J Biochem Cell Biol. 1999;31:637–643. doi: 10.1016/s1357-2725(99)00015-1. [DOI] [PubMed] [Google Scholar]
- 2.Lemmon MA. Ligand-induced ErbB receptor dimerization. Exp Cell Res. 2009;315:638–648. doi: 10.1016/j.yexcr.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiley HS. Trafficking of the ErbB receptors and its influence on signaling. Exp Cell Res. 2003;284:78–88. doi: 10.1016/s0014-4827(03)00002-8. [DOI] [PubMed] [Google Scholar]
- 4.Lazzara MJ, Lauffenburger DA. Quantitative modeling perspectives on the ErbB system of cell regulatory processes. Exp Cell Res. 2009;315:717–725. doi: 10.1016/j.yexcr.2008.10.033. [DOI] [PubMed] [Google Scholar]
- 5.Mullenbach GT, et al. Modification of a receptor-binding surface of epidermal growth factor (EGF): analogs with enhanced receptor affinity at low pH or at neutrality. Protein Eng. 1998;11:473–480. doi: 10.1093/protein/11.6.473. [DOI] [PubMed] [Google Scholar]
- 6.Souriau C, Gracy J, Chiche L, Weill M. Direct selection of EGF mutants displayed on filamentous phage using cells overexpressing EGF receptor. Biol Chem. 1999;380:451–458. doi: 10.1515/BC.1999.059. [DOI] [PubMed] [Google Scholar]
- 7.Coco WM, et al. Growth factor engineering by degenerate homoduplex gene family recombination. Nat Biotechnol. 2002;20:1246–1250. doi: 10.1038/nbt757. [DOI] [PubMed] [Google Scholar]
- 8.Reddy CC, Niyogi SK, Wells A, Wiley HS, Lauffenburger DA. Engineering epidermal growth factor for enhanced mitogenic potency. Nat Biotechnol. 1996;14:1696–1699. doi: 10.1038/nbt1296-1696. [DOI] [PubMed] [Google Scholar]
- 9.Souriau C, Fort P, Roux P, Hartley O, Lefranc MP, Weill M. A simple luciferase assay for signal transduction activity detection of epidermal growth factor displayed on phage. Nucleic Acids Res. 1997;25:1585–1590. doi: 10.1093/nar/25.8.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoeberl B, Eichler-Jonsson C, Gilles ED, Muller G. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat Biotechnol. 2002;20:370–375. doi: 10.1038/nbt0402-370. [DOI] [PubMed] [Google Scholar]
- 11.Wiley HS, Shvartsman SY, Lauffenburger DA. Computational modeling of the EGF-receptor system: a paradigm for systems biology. Trends Cell Biol. 2003;13:43–50. doi: 10.1016/s0962-8924(02)00009-0. [DOI] [PubMed] [Google Scholar]
- 12.Roepstorff K, Grandal MV, Henriksen L, Knudsen SL, Lerdrup M, Grovdal L, Willumsen BM, van Deurs B. Differential effects of EGFR ligands on endocytic sorting of the receptor. Traffic. 2009;10:1115–1127. doi: 10.1111/j.1600-0854.2009.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cochran JR, Kim YS, Lippow SM, Rao B, Wittrup KD. Improved mutants from directed evolution are biased to orthologous substitutions. Protein Eng Des Sel. 2006;19:245–253. doi: 10.1093/protein/gzl006. [DOI] [PubMed] [Google Scholar]
- 14.Brown PM, Debanne MT, Grothe S, Bergsma D, Caron M, Kay C, O'Connor-McCourt MD. The extracellular domain of the epidermal growth factor receptor. Studies on the affinity and stoichiometry of binding, receptor dimerization and a binding-domain mutant. Eur J Biochem. 1994;225:223–233. doi: 10.1111/j.1432-1033.1994.00223.x. [DOI] [PubMed] [Google Scholar]
- 15.Wiley HS, Herbst JJ, Walsh BJ, Lauffenburger DA, Rosenfeld MG, Gill GN. The role of tyrosine kinase activity in endocytosis, compartmentation, and down-regulation of the epidermal growth factor receptor. J Biol Chem. 1991;266:11083–11094. [PubMed] [Google Scholar]
- 16.Moss AJ, Sharma S, Brindle NP. Rational design and protein engineering of growth factors for regenerative medicine and tissue engineering. Biochem Soc Trans. 2009;37:717–721. doi: 10.1042/BST0370717. [DOI] [PubMed] [Google Scholar]
- 17.Berlanga-Acosta J, Gavilondo-Cowley J, López-Saura P, González-López T, Castro-Santana MD, López-Mola E, Guillén-Nieto G, Herrera-Martinez L. Epidermal growth factor in clinical practice – a review of its biological actions, clinical indications and safety implications. International Wound Journal. 2009;6:331–346. doi: 10.1111/j.1742-481X.2009.00622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haugh JM. Mathematical model of human growth hormone (hGH)-stimulated cell proliferation explains the efficacy of hGH variants as receptor agonists or antagonists. Biotechnol Prog. 2004;20:1337–1344. doi: 10.1021/bp0499101. [DOI] [PubMed] [Google Scholar]
- 19.Jones DS, Silverman AP, Cochran JR. Developing therapeutic proteins by engineering ligand-receptor interactions. Trends Biotechnol. 2008;26:498–505. doi: 10.1016/j.tibtech.2008.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.